

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0103.9
(705 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
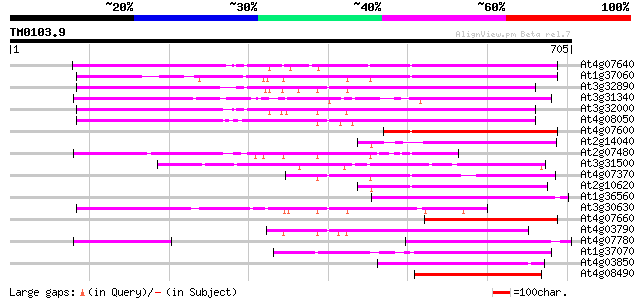
Score E
Sequences producing significant alignments: (bits) Value
At4g07640 putative athila transposon protein 271 1e-72
At1g37060 Athila retroelment ORF 1, putative 266 4e-71
At3g32890 Athila ORF 1, putative 229 3e-60
At3g31340 Athila ORF 1, putative 220 2e-57
At3g32000 unknown protein 219 5e-57
At4g08050 212 5e-55
At4g07600 156 3e-38
At2g14040 putative retroelement pol polyprotein 154 1e-37
At2g07480 F9A16.15 154 2e-37
At3g31500 hypothetical protein 152 6e-37
At4g07370 149 5e-36
At2g10620 putative Athila retroelement ORF1 protein 149 5e-36
At1g36560 hypothetical protein 149 7e-36
At3g30630 hypothetical protein 147 3e-35
At4g07660 putative athila transposon protein 139 7e-33
At4g03790 putative athila-like protein 135 1e-31
At4g07780 putative athila transposon protein 129 6e-30
At1g37070 hypothetical protein 129 6e-30
At4g03850 putative transposon protein 121 2e-27
At4g08490 putative athila-like protein 120 2e-27
>At4g07640 putative athila transposon protein
Length = 866
Score = 271 bits (692), Expect = 1e-72
Identities = 204/628 (32%), Positives = 310/628 (48%), Gaps = 35/628 (5%)
Query: 79 NAELVRLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVF 138
+A+ +L+LFPFSL D A W + P SI TW+D K F K F +L+N+I F
Sbjct: 82 SADGFKLRLFPFSLGDKAHIWEKNLPHDSIITWDDCKKAFLSKFFSNARTARLRNEISGF 141
Query: 139 KQEDTENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALS 198
Q+ E+ EA ERFK +CP H T + Y G+ R L+ A++G F+
Sbjct: 142 SQKTGESFCEAWERFKGYTNQCPHHGFTKASMLSTLYRGVLPRIRMLLDTASNGNFQNKD 201
Query: 199 AQAGWNLINKMAESAVNSTNDRQNRRGVLEIEAYDRMVASNKQLSQQMTTIQRQFQAAKI 258
+ GW L++ +A+S N N+ +R ++ D+ K L+ ++ I Q
Sbjct: 202 VEEGWELVDNLAQSDGN-YNEDCDRTVRDTADSDDKHRKEIKALNDKLDRILLNQQKHVH 260
Query: 259 SNVDSIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNN 318
VD G G EEV + N+Q+ YN N+PN S+R N
Sbjct: 261 FLVDDEQFQVQDGE------GNQL-EEVSYINNNQSG--YKGYNNFKTNNPNLSYRSTNI 311
Query: 319 FNQGGN----NQRQYSNQRFQSQNSG--PQQNQDQGSGN-----GKKSLEELMENFINKA 367
N Q+Q N+ F N G P+Q Q QG+ G K+L E
Sbjct: 312 ANPQDQVYPPQQQQVQNKPFVPYNQGFVPKQ-QFQGNYQPPPPPGGKALPTREEPKTVTE 370
Query: 368 DTSFKNHEAAIKSLETQVG-----QMAKQMSERPPGMFPSDTVINPKENCSAITLRNIEP 422
D+ ++ E SLE Q Q E+P + + P +N + T R I P
Sbjct: 371 DSEDQDGEDL--SLEKDQADKPHEQPLDQSLEQPLDLSLEQPLDLPLDNVTRPTTRPIFP 428
Query: 423 LGENKKKQKEKEEEKRKVELENKFTKVLFPPFPTNIAKRRLEKQFSKFISMFKKLRVELP 482
+ + K KV + + L PFP K +K + F K++ + +P
Sbjct: 429 AASATAPKPITVKNKEKVFVPPPYKPEL--PFPGRHKKALADKYRAMFAKNIKEVELRIP 486
Query: 483 FSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKR-KDPGSFTLP 541
+ L +P KF+K+++ + R+ E ++ L+ CSAI+Q+K+ PK+ DPGSFTLP
Sbjct: 487 LVDALALIPDSHKFLKDLIVE--RIQEVQGMVVLSHGCSAIIQKKIIPKKLSDPGSFTLP 544
Query: 542 VNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVL 601
+ G R LCDLG+SV+LM LS+ +RL + K + + L LADRS+ P G++++
Sbjct: 545 CSLGPLAFNRCLCDLGASVSLMALSVAKRLGFTQYKSSNISLILADRSVRIPHGLLKNFG 604
Query: 602 VRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVA-DEKITFTI 660
+ +G E P DFV+++MDE+ K PLILGRPFLAT+ A I+V G I L + D ++TF +
Sbjct: 605 ITIGAVEIPTDFVVLEMDEEPKDPLILGRPFLATAGAMIDVKKGKIDLSLGKDFRMTFDV 664
Query: 661 FDLKPKPVEKNDVFLVEMMDEWSDEKLK 688
D KP + +F ++ MD+ +DE L+
Sbjct: 665 KDAMKKPTIEGQLFWIKEMDQLADELLE 692
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 266 bits (679), Expect = 4e-71
Identities = 207/702 (29%), Positives = 317/702 (44%), Gaps = 132/702 (18%)
Query: 84 RLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQEDT 143
+L+LFPFSL D A +W S PQGSIT+W D K F K F + +L+NDI F Q +
Sbjct: 87 KLRLFPFSLGDKAHQWEKSLPQGSITSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNN 146
Query: 144 ENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQAGW 203
E +EA ERFK +CP H + L + A++G F + GW
Sbjct: 147 ETFYEAWERFKGYQTQCPHHEMLL-------------------DTASNGNFLNKDVEDGW 187
Query: 204 NLINKMAESAVNSTNDRQNRRGVLEIEAYDRMVA----SNKQLSQQMTTIQRQFQAAKIS 259
++ +A+S N D YDR + S+++ ++M + + +
Sbjct: 188 EVVENLAQSDGNYNED------------YDRSIRTSSDSDEKHRREMKAMNDKLDKLLLM 235
Query: 260 NVDSIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQN-- 317
IH ++ T EEV + N Y+ +N +NHPN S+R N
Sbjct: 236 QQKHIHFLGDDETLQVQDGETLQLEEVSYVQNQGG--YNKGFNNFKQNHPNLSYRSTNVA 293
Query: 318 -------------------NFNQG----------GNNQRQYSNQRFQSQNSGPQ------ 342
+NQG GN Q+Q F Q P
Sbjct: 294 NRQDQVYPSQQQNQPKPFVPYNQGQGYVPKQQYQGNYQQQLPPPGFTQQQQQPALTTPDS 353
Query: 343 ------QNQDQGSGNGKKSLEELMENFINKADTSFKNHEAAIKSLETQVGQMAKQMSERP 396
Q QG G L + M NK D S+ + +++L +++ + Q
Sbjct: 354 DLKNMLQQILQGQATGAMDLSKRMAEIHNKVDCSYNDINIKVEALTSKIRYIEGQTGSTA 413
Query: 397 PGMF--PSD-TVINPKENCSAITLRNIEPL-----------------GENKKKQKEKEEE 436
F PS ++ N KE AITLR+ + L GE+ + E+
Sbjct: 414 APKFTGPSGKSMSNSKEYAHAITLRSGKELPTKESPNQNTEDSLDQDGEDFCQNGNSAEK 473
Query: 437 KRKVELENKFTKVLFP----------------------------PFPTNIAKRRLEKQFS 468
+ + ++ T+ L P PFP K ++K +
Sbjct: 474 AIEEPILHQPTRPLAPAASPLVEKPAAAKTKENVFIPPPYKPPLPFPGRFKKVMIQKYKA 533
Query: 469 KFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKL 528
K L V +P + L +P K++K+++++ R+ E ++ L+ ECSAI+Q+K+
Sbjct: 534 LLEKQLKNLEVTMPLVDCLALIPDSNKYVKDMITE--RIKEVQGMVVLSHECSAIIQQKI 591
Query: 529 PPKRK-DPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLAD 587
PK+ DPGSFTLP G + LCDLG+SV+LMPL + ++L + KP + L LAD
Sbjct: 592 IPKKLGDPGSFTLPCALGPLAFSKCLCDLGASVSLMPLPVAKKLGFNKYKPCNISLILAD 651
Query: 588 RSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTI 647
RS+ G++ED+ V +G E P DFV+++MDE+ K PLILGRPFLA ++A I+V G I
Sbjct: 652 RSVRISHGLLEDLPVMIGVVEVPTDFVVLEMDEEPKDPLILGRPFLARARAIIDVKKGKI 711
Query: 648 SLRVA-DEKITFTIFDLKPKPVEKNDVFLVEMMDEWSDEKLK 688
L + D K+TF I + KP + ++F +E MD +D+ L+
Sbjct: 712 DLNLGRDLKMTFDITNTMKKPTIEGNIFWIEEMDMLADKMLE 753
>At3g32890 Athila ORF 1, putative
Length = 755
Score = 229 bits (585), Expect = 3e-60
Identities = 187/637 (29%), Positives = 289/637 (45%), Gaps = 84/637 (13%)
Query: 84 RLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQEDT 143
+L+LFPFSL D A W + P SITTW+D K F K F +L+N+I F Q+
Sbjct: 40 KLRLFPFSLGDKAHIWEKNLPHDSITTWDDCKKAFLSKFFSNARTARLRNEISGFSQKTR 99
Query: 144 ENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQAGW 203
E+ EA ERFK +CP H T + Y G+ R L+ +++G F+ + GW
Sbjct: 100 ESFCEAWERFKGYTNQCPHHGFTKASLLSTLYRGVLLRIRMLLDTSSNGNFQNKDVEEGW 159
Query: 204 NLINKMAESAVNSTNDRQNRRGVLEIEAYDRMVASNKQLSQQMTTIQRQFQAAKISNVDS 263
L+ +A+S N N+ +R + + M+ ++Q +QFQ + +
Sbjct: 160 ELVENLAQSDGN-YNEDCDRNEIKALNDKLDMILLSQQKHVHFLVDDKQFQ-VQYGEGNQ 217
Query: 264 IHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNNFN--- 320
+ EEV + N+Q YN N+PN S+R N N
Sbjct: 218 L-------------------EEVSYIINNQGG--YKGYNNFKTNNPNLSYRSTNVANPPQ 256
Query: 321 --QGGN------NQRQYSNQRFQSQNSGP-------QQNQDQGSGNG--KKSLEELME-- 361
QG N NQ Q+FQ P QQNQ + + K+ +++L+E
Sbjct: 257 QQQGQNKPFVPYNQGFVPKQQFQGNYQPPPPPGFTTQQNQGPAAPDAEMKQMVQQLLEVQ 316
Query: 362 ------------NFINKADTSFKNHEAAIKSLETQV----GQMAKQMSERPPGMFPSDTV 405
+K D S+ + A +++L T+V GQ A + + G+ P ++
Sbjct: 317 ASSSMEIAKKLSELHHKLDCSYNDMNAKVEALNTKVRYLEGQSASTSTPKVTGL-PGKSI 375
Query: 406 INPKE--NCSAITLRNIEPL-----------------GENKKKQKEKEEEKRKVELENKF 446
NPKE AIT+ + L GE + +E+ + +F
Sbjct: 376 QNPKEYATAHAITICHDRELPTRHVPDFITGDSDVQDGEASTQSTSEEKAAIIERMVKRF 435
Query: 447 TKVLFP--PFPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKK 504
P P K +EK S ++ +P EVL +P K ++ ++ ++
Sbjct: 436 KPTPLPSSALPWTFRKAWMEKYKSVASKQLDEIEAVMPLIEVLNLIPDPHKDVRNLILER 495
Query: 505 RRLSEENEIIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMP 564
++ +++ A +R + K +DPGSFTLP + G LCDLG+SV+LMP
Sbjct: 496 IKMYHDSDDESDATPSRASDKRIVQEKLEDPGSFTLPCSIGELAFSDCLCDLGASVSLMP 555
Query: 565 LSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKI 624
LS+ RL + KP + L LADRS P+G+++D+ V + E P DFV++DM+ + K
Sbjct: 556 LSVARRLEFIQYKPCDLTLILADRSFRKPFGMLKDLPVMINGVEVPTDFVVLDMEVEHKD 615
Query: 625 PLILGRPFLATSQAKINVGNGTISLRVADE-KITFTI 660
PLILGRP LA+ A I+V G ISL + K+ F I
Sbjct: 616 PLILGRPLLASVGAVIDVREGKISLNLGKHIKLQFGI 652
>At3g31340 Athila ORF 1, putative
Length = 781
Score = 220 bits (561), Expect = 2e-57
Identities = 174/611 (28%), Positives = 280/611 (45%), Gaps = 71/611 (11%)
Query: 81 ELVRLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQ 140
++++ +LF FSL D A WL S ++ +WED F + F ++ L+N I F+Q
Sbjct: 132 DIIKCRLFIFSLADNAHRWLKSLDPINLRSWEDYKAAFLGQYFTQSRTAILRNKISSFQQ 191
Query: 141 EDTENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQ 200
TE+ EA ERFK R+CP H + + FY G+ + + L+ A++G+F +
Sbjct: 192 GGTESFPEAWERFKDYYRECPHHGFSRATLISTFYQGVDKAYKMALDTASNGDFMTKTET 251
Query: 201 AGWNLINKMAESAVNST--NDRQNRRGVLEIEAYDRMVASNKQLSQQMTTIQRQFQAAKI 258
LI +A S +N DR NR G E++ ++ + ++ Q + Q+ +
Sbjct: 252 EATKLIKNLAASNINHNVDYDRSNRGGGGELK---QLAELSVKVEQLLRRDQKVINFCED 308
Query: 259 SNVDSIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNN 318
S+ +H S +C E+ + + + + NFS +
Sbjct: 309 SSKGMVH------QEYSGDCSEDLQAEMNFVNGIRYIQLRQAHKV-----QNFS---PTD 354
Query: 319 FNQGGNNQRQYSNQRFQSQNSGPQQNQDQGSGNGKKSLEELMENFINKADTSFKNHEAAI 378
F G + NS QQ + G + +++ N + F + +
Sbjct: 355 FRIKGTIR----------DNSNLQQILE-GLKKNAADINVKVDSMYNDLNVKFATLSSHV 403
Query: 379 KSLETQVGQMAKQMSERPPGMF-----PSDTVINPKENCSAITLRNIEPLGENKKKQKEK 433
K+LE QV Q+ S RP G P + + + +A+ +E L E+K +
Sbjct: 404 KTLENQVSQVVSA-SMRPAGAHSGKEEPREIDVAKQVETNAVV---VETLVEDKIVE--- 456
Query: 434 EEEKRKVELENKFTKVLFPPFPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQY 493
++E E K+ FP I R+ E+ ++ ++E Q
Sbjct: 457 DDEPLSEEPPPYVPKLPFPGRERQIQSRKREE-----------------YALLVESQRQ- 498
Query: 494 AKFMKEILSKKRRLSEENEII---ELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQV 550
K+ +L++ E++ E+ CSAI +P K DPGSF LP G S
Sbjct: 499 --------QKEAQLTDVVEVLAGKEMVSTCSAIPPATIPEKLGDPGSFVLPCRIGKSAFE 550
Query: 551 RALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFP 610
R LCDLG+ VNLMPLSM +RL + KP+ + L LADRS+ P G+ E+V VRVG+F P
Sbjct: 551 RCLCDLGAGVNLMPLSMSKRLGITNFKPSRISLILADRSVRFPVGLAENVHVRVGDFYIP 610
Query: 611 VDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVADEKITFTIFDLKPKPVEK 670
DFV++++D++ PL LGRPFL T A I+V TI+L++ D + F + + P +
Sbjct: 611 TDFVVLELDKEPHDPLTLGRPFLNTVGAIIDVRRSTINLQIGDFALEFDMKGTRKNPTIE 670
Query: 671 NDVFLVEMMDE 681
F V+ DE
Sbjct: 671 GHAFSVDTNDE 681
>At3g32000 unknown protein
Length = 839
Score = 219 bits (557), Expect = 5e-57
Identities = 179/631 (28%), Positives = 280/631 (44%), Gaps = 67/631 (10%)
Query: 84 RLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQEDT 143
+L+LFPFSL D A W + P SITT +D K F K F +L+N+I F Q+
Sbjct: 87 KLRLFPFSLGDKALIWEMNLPHDSITTRDDCKKAFLSKFFSNDRTARLRNEISGFSQKIG 146
Query: 144 ENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQAGW 203
E+ EA ERFK +CP H T + Y G+ R L+ A++G F+ + GW
Sbjct: 147 ESFCEAWERFKDYTNQCPHHGFTKASLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGW 206
Query: 204 NLINKMAESAVNSTNDRQNR-RGVLEIEAYDR--MVASNKQLSQQMTTIQRQFQAAKISN 260
L++ +A+S N D RG + + R + A N +L + + + Q+
Sbjct: 207 ELVDNLAQSDGNYNEDCDRTVRGTADSDDKHRKEIKALNDKLDRILLSQQKHVDFLVDDE 266
Query: 261 VDSIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNNFN 320
+ G G EEV + N+Q YN N+PN S+R N N
Sbjct: 267 QYQVQDGE----------GNQL-EEVSYINNNQGG--YKGYNNFKTNNPNLSYRSTNVAN 313
Query: 321 QGGN----NQRQYSNQRFQSQNSG--------------------PQQNQD---------- 346
Q+Q N+ F N G PQQNQ
Sbjct: 314 PQDQVYPPQQQQGQNKPFVPYNQGFVPKQQFQGNYQQPPPPGFAPQQNQGPATPDAEMKQ 373
Query: 347 ------QGSGNGKKSLEELMENFINKADTSFKNHEAAIKSLETQV----GQMAKQMSERP 396
QG + + + + +K D S+ + A +++L T+V GQ A + +
Sbjct: 374 MVQQLLQGQASSSMEIAKKLSELHHKLDCSYNDLNAKVEALNTKVRYLEGQSASTSAPKV 433
Query: 397 PGMFPSDTVINPKENCSAITLRNIE---PLGENKKKQKEKEEEKRKVE-LENKFTKVLFP 452
G+ D+ + E + + + +E G Q EE+ +E + +F P
Sbjct: 434 TGLPGKDSDVQEGEAFTQVEVSVVEFNHSAGSRHLIQSTSEEKAAIIERMVKRFKPTPLP 493
Query: 453 P--FPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEE 510
P K +E+ S ++ +P EVL +P K ++ ++ ++ ++ +
Sbjct: 494 SRALPWTFRKSWMERYKSVAAKQPDEIEAVMPLMEVLNLIPDPHKDVRNLILERIKMYHD 553
Query: 511 NEIIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFER 570
++ A +R + K +DPGSF+LP + G LCDLG+ V+LMP S+ R
Sbjct: 554 SDDESDATPSRAADKRIVQEKLEDPGSFSLPCSIGEFAFSDCLCDLGAFVSLMPCSVARR 613
Query: 571 LNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGR 630
L + KP + L LADRS P+G+++D+ V + E P DFV++DM+ + K PLILGR
Sbjct: 614 LEFIQYKPCDLTLILADRSSRKPFGMLKDLPVMINGVEVPTDFVVLDMEVEHKDPLILGR 673
Query: 631 PFLATSQAKINVGNGTISLRVADE-KITFTI 660
PFLA+ A I+V G I L + K+ F I
Sbjct: 674 PFLASVGAVIDVREGKIGLNLGKHIKLQFGI 704
>At4g08050
Length = 1428
Score = 212 bits (540), Expect = 5e-55
Identities = 176/625 (28%), Positives = 275/625 (43%), Gaps = 59/625 (9%)
Query: 84 RLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQEDT 143
+L+LFPFSL D A W + SITTW+D K F K F +L+N+I F Q+
Sbjct: 87 KLRLFPFSLGDKAHIWEKNLSHDSITTWDDYKKAFLSKFFSNARTARLRNEIYGFSQKTG 146
Query: 144 ENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQAGW 203
E+ EA ERFK +CP H+ T + Y G+ R L+ A++G F+ + GW
Sbjct: 147 ESFCEAWERFKGYTNQCPHHSFTKASLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGW 206
Query: 204 NLINKMAESAVNSTNDRQNR-RGVLEIEAYDR--MVASNKQLSQQMTTIQRQFQAAKISN 260
L+ +A+S N D RG + + R + A N +L + + + Q+
Sbjct: 207 ELVENLAQSDGNYNEDCDRTVRGTADSDDKHRKEIKALNDKLDRILLSQQKHVHFLVDDE 266
Query: 261 VDSIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNNFN 320
+ G G E Y ++ + QN P+ YN G+ F Q
Sbjct: 267 QYQVQDGE--GNQLEE---VYLPQQKQ----GQNKPFV-LYNQGFVPKQQFQGNYQPPPP 316
Query: 321 QGGNNQRQYSNQRFQSQNSGPQQNQDQGSGNGKKSLEELMENFINKADTSFKNHEAAIKS 380
G Q+ + ++ + QG + + + + +K D S+ + A +++
Sbjct: 317 PGFVTQQNQCHAAPDAKMKQMVKQLLQGQASSSMEIAKKLSELHHKLDCSYNDLNAKVEA 376
Query: 381 LETQV----GQMAKQMSERPPGMFPSDTVINPKENCSA-------------------ITL 417
L T+V GQ A S + + P ++ NPKE +A IT
Sbjct: 377 LNTKVRYLEGQSASTFSPKVTRL-PGKSIQNPKEYATAHAITICHDRELPTRHVLDLITR 435
Query: 418 RNIEPLGENKKK------------------QKEKEEEKRKVE-LENKFTKVLFP--PFPT 456
N GE + Q EE+ +E + +F P P
Sbjct: 436 DNDVQEGEASTQVEASVVEFNHSAGSRHLTQSTSEEKAAIIERMVKRFKPTPLPLRALPW 495
Query: 457 NIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIEL 516
K +E+ S ++ +P EVL + K ++ ++ ++ ++ +++
Sbjct: 496 TFRKAWMERYKSVAAKQLDEIEAVMPLMEVLNLILDPHKVVRNLILERIKMYHDSDDESD 555
Query: 517 TEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGEL 576
A +R + K +DPGSFTLP + G LCDLG+SVNLMPLSM RL +
Sbjct: 556 ATPSRAADKRIVQEKLEDPGSFTLPCSIGEFAFSDCLCDLGASVNLMPLSMARRLEFIQY 615
Query: 577 KPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATS 636
KP + L LADRS +G+++D+ V + E P DFV++DM+ + K PLILGRPFLA+
Sbjct: 616 KPCDLTLILADRSSRKHFGMLKDLPVMINGVEVPTDFVVLDMEVEHKDPLILGRPFLASV 675
Query: 637 QAKINVGNGTISLRVADE-KITFTI 660
A I+V G I L + K+ F I
Sbjct: 676 GAVIDVKEGKIGLNLGKHIKLQFNI 700
>At4g07600
Length = 630
Score = 156 bits (395), Expect = 3e-38
Identities = 90/221 (40%), Positives = 137/221 (61%), Gaps = 4/221 (1%)
Query: 470 FISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLP 529
F K++ + +P + L + KF+K+++ + R+ E ++ L+ ECSAI+Q+K+
Sbjct: 2 FAKNIKEVELRIPLVDALALILDTHKFLKDLIVE--RIQEVQGMVVLSHECSAIIQKKIV 59
Query: 530 PKR-KDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADR 588
PK+ DPGSFTLP G R LCDLG+ V+ MPLS+ +RL + K + L LADR
Sbjct: 60 PKKLSDPGSFTLPCFLGTVAFNRCLCDLGALVSPMPLSIAKRLGFTQYKSCNISLILADR 119
Query: 589 SIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTIS 648
S+ G+++++ +R+G E DFVI++MDE+ K PLIL RPFLAT+ A I+V G I
Sbjct: 120 SVRISHGLLKNLPIRIGAAEISTDFVILEMDEEPKDPLILRRPFLATAGAMIDVKKGKID 179
Query: 649 LRVA-DEKITFTIFDLKPKPVEKNDVFLVEMMDEWSDEKLK 688
L + D ++TF I D KP + +F +E MD+ +DE L+
Sbjct: 180 LNLGKDFRMTFDIKDAMKKPNIEGQLFWIEEMDQLADELLE 220
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 154 bits (390), Expect = 1e-37
Identities = 101/256 (39%), Positives = 130/256 (50%), Gaps = 42/256 (16%)
Query: 438 RKVELENKFTKVLFPP------FPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMP 491
RK LE K K + PP FP K + EK++S F E++ ++
Sbjct: 4 RKQTLEEKAKKTVLPPYVPKLPFPGRQRKIQREKEYSLF-------------DEIMRQLQ 50
Query: 492 QYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVR 551
+Y+ ECSAILQ +P KR+DPGSF LP G R
Sbjct: 51 RYSP-----------------------ECSAILQNVIPVKREDPGSFVLPSRIGEYTFDR 87
Query: 552 ALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPV 611
LCDLG+ V+LMP S+ +RL PT M L L DRSI P GV EDV VRVG F P
Sbjct: 88 CLCDLGAGVSLMPFSVAKRLGDTNFTPTKMSLVLGDRSISFPVGVAEDVQVRVGNFYIPT 147
Query: 612 DFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVADEKITFTIFDLKPKPVEKN 671
DFVII++DE+ + LILGRPFL A I+V I+LR+ D F + + KP +
Sbjct: 148 DFVIIELDEEPRHRLILGRPFLNIVAALIDVRKSKINLRIGDIVQEFNMERIMSKPTTEC 207
Query: 672 DVFLVEMMDEWSDEKL 687
F V++MDE +E L
Sbjct: 208 QTFWVDIMDELVNELL 223
>At2g07480 F9A16.15
Length = 1012
Score = 154 bits (389), Expect = 2e-37
Identities = 149/548 (27%), Positives = 228/548 (41%), Gaps = 94/548 (17%)
Query: 81 ELVRLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQ 140
E +L+LFPFSL D A W + P SI TW+D K F K F + +L+++I F Q
Sbjct: 79 ESFKLRLFPFSLGDKAHLWEKTLPVKSIDTWDDCKKAFLAKFFSNSRKARLRSEISGFNQ 138
Query: 141 EDTENLHEALERFKKLLRKCPQH-NLTLGVQVERFYDGLADSARSNLEAAASGEFEALSA 199
+++E+ EA ERFK +CP H +L + R R L+ A++G
Sbjct: 139 KNSESFSEAWERFKGYTTQCPHHESLPPQYSILR----CLPKIRMLLDTASNGNILNKDV 194
Query: 200 QAGWNLINKMAES--AVNSTNDRQNRRGVLEIEAYDRMVAS-NKQLSQQMTTIQRQFQAA 256
GW L+ +A+S N DR NR + + + + + N ++ + + QR
Sbjct: 195 AEGWELVENLAQSHRNYNEDYDRTNRGSSDSEDKHKKEIKTLNDRIDKLVLAQQRNVYYI 254
Query: 257 KISNVDSIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRN----HPNFS 312
+ + G EEV L N YN G+ N HPN S
Sbjct: 255 TEEELTQLQDGE-----------NLTIEEVSYLQN------QGGYNKGFNNYKPPHPNLS 297
Query: 313 WREQN----------------------NFNQGGNNQRQYSNQRFQSQNSGPQ-------- 342
+R N +NQG N ++ + F Q P
Sbjct: 298 YRSNNVANPQDQVYPPQNQPTQAKPFVPYNQGYNQKQNFGPPGFTQQPQQPSAQDSEMKT 357
Query: 343 --QNQDQGSGNGKKSLEELMENFINKADTSFKNHEAAIKSLETQV----GQMAKQMSERP 396
Q QG + ++++ + + D S+ + I +L T+V G +A + +
Sbjct: 358 LPQQLVQGHASCSMTMDKKLAELTTRIDCSYNDLNIKIDALNTRVKSMEGHIASTSAPKH 417
Query: 397 PGMFPSDTVINPKENCSAITLRNIEPLGENKKKQKEKEEEKRKVELENKFTKVLF----- 451
G P V NPKE AI+ N ++ ++ E + + E+E F L
Sbjct: 418 LGQLPGKAVQNPKEYAHAISTVNTSATEDSGIQEGEVLRPRSRQEIELDFFARLVERAHD 477
Query: 452 --------------PPFPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFM 497
P FP IA+ E F K MF K E+ EK+P
Sbjct: 478 PSNPIHIPSPYVPKPAFPERIAQID-EMIFQKHKMMFIKC-----IKELEEKVP-LVDTP 530
Query: 498 KEILSKKRRLSEENEIIELTEECSAILQRKLPPKR-KDPGSFTLPVNFGASKQVRALCDL 556
KE++ + R E +I+EL+ ECSAI+Q+K+ PK+ DPGSFTLP + +LCDL
Sbjct: 531 KEMIME--RPQEAQQIVELSFECSAIIQKKVIPKKLGDPGSFTLPCSLAPLVFNNSLCDL 588
Query: 557 GSSVNLMP 564
G+S++ P
Sbjct: 589 GASMDEEP 596
>At3g31500 hypothetical protein
Length = 591
Score = 152 bits (384), Expect = 6e-37
Identities = 141/527 (26%), Positives = 236/527 (44%), Gaps = 62/527 (11%)
Query: 186 LEAAASGEFEALSAQAGWNLINKMAESAVNST--NDRQNRRGVLEIEAYDRMVASNKQLS 243
L+ A++GEF + LI +A S N DR NR G E + + + A +QL
Sbjct: 3 LDTASNGEFMTKTETEATKLIENLAGSNSNHNVDYDRSNRGGGGESKQFAELSAKVEQL- 61
Query: 244 QQMTTIQRQFQAAKISNVDSIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNL 303
M Q+ + S+ +H G + F V GN QN +N N
Sbjct: 62 --MRRDQKSVNFCEDSSKGMVHQEFSGDGSEDLQAEINF---VNGYGNYQNGGSTNVENP 116
Query: 304 GWRNHPNFSWREQNNFNQGG-NNQRQYSNQRFQSQNSGPQQNQDQGSGNGKKSLEELMEN 362
+ +P + + F G N+ Y Q FQ +G G K +++++E+
Sbjct: 117 QDQVYPTQAGSQGQKFQPYGFQNKGNYQGQ-FQPP-AGTGHASSFGDNEMKLMMQQVLED 174
Query: 363 --------------FINKADTSFKNHEAAIKSLETQVGQMAKQMSERPPGMFPSDTVINP 408
N + F + +K+LE QV Q+ S RP G
Sbjct: 175 QKKNAADINVKVDSMYNDLNGKFATLSSHVKTLENQVSQIVSA-SMRPDGTHSGKVKPKG 233
Query: 409 KENCSAITLRN-----------------IEPLGENKKKQKEKEEEKRKVELENKFTKVLF 451
KE C AI ++ +E L E+K + ++E VE K+
Sbjct: 234 KEQCYAIMIQEELREIVVAKQVETNVVVVETLVEDKIVE---DDEPLSVEPPPYVPKL-- 288
Query: 452 PPFPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEEN 511
PFP + + +K++++F + K+L V LPF +++ +P Y ++K ILS KR + E
Sbjct: 289 -PFPGRERQIQRQKEYARFDEIMKQLYVRLPFLQLVLHVPSYRSYLKYILSNKRSIEEGV 347
Query: 512 EIIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERL 571
++I EE + +++ + +++ T V A K++ C + P ++ ++L
Sbjct: 348 KLISKGEEHAQLVESQ---RQQKEAQLTNVVEMLAGKEMVNTCSA-----IPPATIPKKL 399
Query: 572 NVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRP 631
+ KP+ + L LADRS+ P G+ E+V RVG F P +FV++++D++ PL LGRP
Sbjct: 400 GITNFKPSRISLILADRSVQFPMGLAENVHARVGNFYIPTNFVVLELDKEPHDPLNLGRP 459
Query: 632 FLATSQAKINVGNGTISLRVADEKITFTIFDLKPKP-----VEKNDV 673
FL T +A I+V TI+L++ D + F I + P +E +DV
Sbjct: 460 FLNTVEAIIDVRRSTINLQIGDYALEFDIKGTRKNPTIEDCIELSDV 506
>At4g07370
Length = 531
Score = 149 bits (376), Expect = 5e-36
Identities = 111/376 (29%), Positives = 182/376 (47%), Gaps = 58/376 (15%)
Query: 347 QGSGNGKKSLEELMENFINKADTSFKNHEAAIKSLETQV----GQMAKQMSERPPGMFPS 402
QG + + + + K D S+ + A +++L ++V G+ A S + G+ P
Sbjct: 11 QGQVSSSIEIAKKISELHQKLDCSYNDLNAKVEALNSKVRYLEGESASTSSPKVTGL-PG 69
Query: 403 DTVINPKENC-SAITLRNIEPLGENKKKQKEKEEEKRKV--ELENKFTKVLFP------- 452
++ NPKE ++T + GE+ +++ ++ +V + ++ T+ P
Sbjct: 70 KSIQNPKEERPKSVTEDSAYQDGEDFSLNEDQVDKPTEVLEPILDRDTRPTNPLTSSAAL 129
Query: 453 ---------------------PFPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMP 491
PFP K +K + F KK+ + +P ++ L +P
Sbjct: 130 KLVAAKNNEIAFIPPPYKPPLPFPGRHKKEVEDKYRAMFAKNIKKVELRIPLADALTHIP 189
Query: 492 QYAKFMKEILSKKRRLSEENEIIELTEECSAILQRK-LPPKRKDPGSFTLPVNFGASKQV 550
KF+K+++ + R+ E + L+ ECSAI+Q + K DPGSFTLP + G+
Sbjct: 190 DSQKFLKDLIME--RIQEVQKTTVLSHECSAIIQENDVSEKLGDPGSFTLPCSLGSLTFN 247
Query: 551 RALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFP 610
+ LCDLG SVNLMPLS+ A RS+ P G++ED+ +++ E P
Sbjct: 248 KCLCDLGPSVNLMPLSV------------------AKRSVRLPHGLLEDLPIKIRNVEVP 289
Query: 611 VDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVADE-KITFTIFDLKPKPVE 669
DFV+++MDE+ K PLILGRPFLAT I+V G I L + ++ F I D KP
Sbjct: 290 TDFVVLNMDEEPKDPLILGRPFLATVGVIIDVKQGKIDLNLGKNFEMKFDINDAMKKPTI 349
Query: 670 KNDVFLVEMMDEWSDE 685
+ FLV+ +D + E
Sbjct: 350 EEQTFLVKEVDRLAGE 365
>At2g10620 putative Athila retroelement ORF1 protein
Length = 451
Score = 149 bits (376), Expect = 5e-36
Identities = 91/248 (36%), Positives = 142/248 (56%), Gaps = 12/248 (4%)
Query: 438 RKVELENKFTKVLFPP-------FPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKM 490
+ V ++NK KV PP FP K K F K++ +++P + L +
Sbjct: 192 KPVVVKNK-EKVFVPPPYKPHPSFPGRHKKALAHKYRVMFAKNIKEVELQIPLVDALALI 250
Query: 491 PQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKR-KDPGSFTLPVNFGASKQ 549
KF+K+++ + R+ E ++ L++ECSAI+Q+K+ K+ DPGSFTLP + G
Sbjct: 251 MDSHKFLKDLIVE--RIQELQGMVVLSDECSAIIQKKIIHKKLSDPGSFTLPCSLGPLAF 308
Query: 550 VRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEF 609
+ LCDLG+S +LMPLS+ +RL + K + L LADRS+ P +E++ +R+ E
Sbjct: 309 NKCLCDLGASASLMPLSVTKRLGFTQYKSCNISLILADRSVRIPHDFLENIPIRIRAVEI 368
Query: 610 PVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVA-DEKITFTIFDLKPKPV 668
P DFV+++MDE+ K LILGRPFL T A I + G I+L + D ++TF + D KP
Sbjct: 369 PTDFVVLEMDEEPKDHLILGRPFLTTVGAMIYIKKGKINLNLGKDFRMTFDVKDTMKKPT 428
Query: 669 EKNDVFLV 676
+ + L+
Sbjct: 429 IEGQLLLM 436
Score = 38.5 bits (88), Expect = 0.013
Identities = 21/76 (27%), Positives = 36/76 (46%), Gaps = 4/76 (5%)
Query: 348 GSGNGKKSLEELMENFINKADTSFKNHEAAIKSLETQV----GQMAKQMSERPPGMFPSD 403
G +G + + + NK D S+ + +++L T+V G A +++ P
Sbjct: 11 GQASGSMEIAKKIFELHNKLDYSYNDLNVKVETLSTKVQYLEGNSASTSAQKQTSQLPGK 70
Query: 404 TVINPKENCSAITLRN 419
+ NPKE AITLR+
Sbjct: 71 AIQNPKEYAHAITLRS 86
>At1g36560 hypothetical protein
Length = 524
Score = 149 bits (375), Expect = 7e-36
Identities = 92/249 (36%), Positives = 138/249 (54%), Gaps = 6/249 (2%)
Query: 455 PTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEII 514
P K + + S ++ +P EVL +P K ++ ++ + ++ +++
Sbjct: 42 PWTFRKAWMARYKSVAAKQLNEIEAVMPLMEVLNLIPNPHKDVRNLILEWIKMYHDSDDE 101
Query: 515 ELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVG 574
A +R + K +DPGSFTLP + G+ + LCDLG+SV+LMPLS+ +RL
Sbjct: 102 SDATPSRAADKRIVQEKLEDPGSFTLPCSLGSLTFNKCLCDLGASVSLMPLSVAKRLGFN 161
Query: 575 ELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLA 634
+ K + L LAD S+ P GV+ED+ +++G+ E P DF+I++MDE+ K PLILGRPFLA
Sbjct: 162 KYKYCNISLILADGSVRQPHGVLEDLPIKIGKVEVPTDFIILNMDEEPKDPLILGRPFLA 221
Query: 635 TSQAKINVGNGTISLRVA-DEKITFTIFDLKPKPVEKNDVFLVEMMDEWSDEKLK*FFLK 693
T+ A I+V G I L + D K+ F I D KP + FLVE D DE L
Sbjct: 222 TAGAIIDVKQGKIDLNMGKDFKMKFDINDAMKKPTIEGQTFLVEETDRLVDE-----LLV 276
Query: 694 EKADASNKK 702
E +A+N K
Sbjct: 277 ELKEANNSK 285
>At3g30630 hypothetical protein
Length = 785
Score = 147 bits (370), Expect = 3e-35
Identities = 146/566 (25%), Positives = 236/566 (40%), Gaps = 70/566 (12%)
Query: 84 RLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQEDT 143
+L+LFPF L D A W + P SI TW D K F K F +L N+I F Q+
Sbjct: 87 KLRLFPFFLGDKAHIWEKNMPHDSIITWVDCKKAFLAKFFSNAKTARLINEISSFSQKTG 146
Query: 144 ENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQAGW 203
E+ EA ERFK +CP H + Y G+ R L+ A++ F+ + GW
Sbjct: 147 ESFCEAWERFKGYTNQCPHHGFKKASLLSTLYRGVLPRIRMLLDTASNENFQNKDVEEGW 206
Query: 204 NLINKMAESAVNSTND-RQNRRGVLEIEAYDRMVASNKQLSQQMTTIQRQFQAAKISNVD 262
LI +A+S N D + RG + S+ + +++ + + +S
Sbjct: 207 ELIENLAQSNDNYNKDCERTIRGTTD---------SDDKHRKEIKALNDKQDKILLSQQK 257
Query: 263 SIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNNFNQG 322
+H + ++ EEV + N N + N+PN S+R N N
Sbjct: 258 HVHFLVDDEQYQVQDGEGNQLEEVSYINNQSGYKGYNNFK---TNNPNLSYRSTNIANP- 313
Query: 323 GNNQRQYSNQRFQSQNSGPQQ--------NQDQG----SGNGKKSLEELM-ENFINKADT 369
+ Y QR Q+ + QQ Q QG N K+ ++L+ E + D
Sbjct: 314 --QDQVYPPQRQQAISGNYQQQPPPRFAPQQHQGPPAPDANMKQMAQQLLQEQASSSMDC 371
Query: 370 SFKNHEAAIKSLETQV----GQMAKQMSERPPGMFPSDTVINPKE--NCSAITLRNIEPL 423
+ + A +++L T+V GQ A + + G+ P + N KE +AIT+ + L
Sbjct: 372 RYNDLNAKVEALNTKVRYLEGQSASTSAPKVTGL-PGKPIQNQKEYATVNAITICHDREL 430
Query: 424 ----------------GENKKKQKEKEEEKRKVE-LENKFTKVLFP--PFPTNIAKRRLE 464
G + Q EE+ +E + +F P P K +E
Sbjct: 431 PTRHVKVSVVEFNHSAGSHHLIQSTSEEKVAIIERMAKRFKPTPLPSRALPWKFRKAWME 490
Query: 465 KQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECS--- 521
+ S ++ V +P EVL + K ++ ++ ++ ++ +++ +EC
Sbjct: 491 RYKSVAEKQLNEIEVVMPLMEVLNVIHDPHKDVRNLILERIKMYHDSD-----DECDANP 545
Query: 522 --AILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFE-----RLNVG 574
A +R + K +DPGS TLP + G LCDLG+SV+LMPLS+ +L
Sbjct: 546 SRAADKRIVQEKLEDPGSCTLPCSIGELAFSNCLCDLGASVSLMPLSVARSGSAYKLRCA 605
Query: 575 ELKPTMMMLQLADRSIVAPWGVVEDV 600
LK + +R +A G V DV
Sbjct: 606 RLKEEHKDHLILERPFLASVGAVIDV 631
>At4g07660 putative athila transposon protein
Length = 724
Score = 139 bits (349), Expect = 7e-33
Identities = 76/169 (44%), Positives = 109/169 (63%), Gaps = 2/169 (1%)
Query: 522 AILQRKLPPKRK-DPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTM 580
AI Q+K+ PK+ DPGSFTLP + G R LCDLG+ V+LMPLS+ +RL + K
Sbjct: 3 AITQKKIVPKKLIDPGSFTLPCSLGPLAFKRCLCDLGALVSLMPLSVAKRLGFTQYKSCN 62
Query: 581 MMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKI 640
+ L LADRS+ P + E++ +R+G + P DFV+++MDE+ K PLILGRPFLAT+ A
Sbjct: 63 ISLILADRSVRIPHSLFENLPIRIGAVDIPTDFVVLEMDEEPKDPLILGRPFLATAGAMN 122
Query: 641 NVGNGTISLRVADE-KITFTIFDLKPKPVEKNDVFLVEMMDEWSDEKLK 688
+V G I L + ++TF + D KP K +F +E +D+ +DE L+
Sbjct: 123 DVKKGKIDLNLGKYCRMTFDVKDAMKKPTIKGQLFWIEEIDQLADELLE 171
>At4g03790 putative athila-like protein
Length = 1064
Score = 135 bits (339), Expect = 1e-31
Identities = 108/383 (28%), Positives = 172/383 (44%), Gaps = 55/383 (14%)
Query: 323 GNNQRQYSNQRFQSQNSGPQ----------QNQDQGSGNGKKSLEELMENFINKADTSFK 372
GN Q Q + +N GP Q QG + + + + +K D ++
Sbjct: 146 GNYQPQPPHGFAPQENQGPTAPDAEMKQMVQQLLQGQASSSMEIPKKLSELHHKLDCNYN 205
Query: 373 NHEAAIKSLETQV----GQMAKQMSERPPGMFPSDTVINPKEN----------------- 411
+ A +++L T+V GQ A + G+ P ++ NPKE
Sbjct: 206 DLNAKVEALNTKVRYLEGQSASTSVPKVTGL-PGKSIQNPKEYATAHAITICHDRELPTR 264
Query: 412 -----------------CSAITLRNIE---PLGENKKKQKEKEEEKRKVE-LENKFTKVL 450
C+ I + IE G + Q EE+ +E + +F
Sbjct: 265 PVLDLIIGESDVQEGEACTQIEVSVIEFNHSAGSHHLIQSTSEEKAAIIERMVKRFKPTP 324
Query: 451 FPP--FPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLS 508
P P K +E+ S ++ +P EVL +P + K ++ ++ +K ++
Sbjct: 325 LPSRALPWTFRKAWIERYKSVAAKQLDEIEAVMPLMEVLNLIPDHHKDVRNLILEKIKMY 384
Query: 509 EENEIIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMF 568
+++ + +R + K +DPGSFTLP + G LCDLG+SV+LMPLSM
Sbjct: 385 HDSDDESDATPSRVVDKRIVQEKLEDPGSFTLPCSIGELAFSDFLCDLGASVSLMPLSMG 444
Query: 569 ERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLIL 628
RL KP + L LADRS P+G+++D+ V + E P DFV+++M+ K PLIL
Sbjct: 445 RRLEFIHYKPCDLTLILADRSSRKPFGMLKDLPVMINGVEVPTDFVVLNMEVKHKDPLIL 504
Query: 629 GRPFLATSQAKINVGNGTISLRV 651
GRPFLA+ A I++ G ISL +
Sbjct: 505 GRPFLASVGAVIDIREGKISLNL 527
Score = 35.4 bits (80), Expect = 0.11
Identities = 20/55 (36%), Positives = 27/55 (48%)
Query: 84 RLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVF 138
+L+LFPFSL D A W + SITTW + F + P L+ + L F
Sbjct: 87 KLRLFPFSLGDKAHIWEKNLSHDSITTWMIARRLFYQSSSPMPELQDSEMRCLAF 141
>At4g07780 putative athila transposon protein
Length = 446
Score = 129 bits (324), Expect = 6e-30
Identities = 78/209 (37%), Positives = 115/209 (54%), Gaps = 12/209 (5%)
Query: 498 KEILSKKRRLSEENEIIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLG 557
K+ +K + + E+I ++ + K DPGSFTLP + G R LCDLG
Sbjct: 247 KKFKIEKLKKDQNQEMISRVSTQETYKKKFIQEKLDDPGSFTLPCSLGPLTFNRCLCDLG 306
Query: 558 SSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIID 617
+SV+LMPLS +RL + E K + L L D S+ P G++ ++ V++G E P DFV++D
Sbjct: 307 ASVSLMPLSTAKRLGIVEYKFCNLALLLPDGSVAHPHGLIGNLPVKIGNDEIPTDFVVLD 366
Query: 618 MDEDSKIPLILGRPFLATSQAKINVGNGTISLRVAD-EKITFTIFDLKPKPVEKNDVFLV 676
DE+ K PLILGRPFLA++ A I+VGNG I L + ++ F I K + F +
Sbjct: 367 TDEEGKDPLILGRPFLASAGAVIDVGNGKIDLNLEKCIEMRFNISKASGKSTTRGQSFGI 426
Query: 677 EMMDEWSDEKLK*FFLKEKADASNKKKKE 705
++MD + E+ +A K KKE
Sbjct: 427 QVMD-----------VDEETEAVKKIKKE 444
Score = 86.3 bits (212), Expect = 5e-17
Identities = 43/123 (34%), Positives = 59/123 (47%)
Query: 81 ELVRLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQ 140
++ +L+LFPFSL D A W + P SIT+W+D K F K F +L+N+I F Q
Sbjct: 92 DMFKLRLFPFSLGDKAHHWKKTLPPDSITSWDDCKKDFLAKFFSNARTARLRNEISGFTQ 151
Query: 141 EDTENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQ 200
++ E EA ERFK CP H + Y G R L+ ++G F
Sbjct: 152 KNNETFFEASERFKSYTTYCPHHGFKKASLLRTLYRGALPKIRMLLDTTSNGNFLNNDVA 211
Query: 201 AGW 203
GW
Sbjct: 212 EGW 214
>At1g37070 hypothetical protein
Length = 590
Score = 129 bits (324), Expect = 6e-30
Identities = 108/350 (30%), Positives = 155/350 (43%), Gaps = 28/350 (8%)
Query: 332 QRFQSQNSGPQQNQDQGSGNGKKS-LEELMENFINKADTSFKNHEAAIKSLETQVGQMAK 390
+ FQ Q G Q + K + L + N + F+ + IK +E+Q A
Sbjct: 80 EHFQMQEGGNDQTVELCYIQNKGAELHNKIGCSYNDLNVKFEALNSKIKYVESQ---FAS 136
Query: 391 QMSERPPGMFPSDTVINPKENCSAITLRNIEPLGENKKKQKEKEEEKRKVELENKFTKVL 450
+ P P V NPK+ + + + +Q EE E T++
Sbjct: 137 TSALEHPQQLPGKAVQNPKDYATGNAITIHQEDESPPSRQTPHTEENMIQEGGGDSTQIA 196
Query: 451 FPPFPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEE 510
P +QF I L P KF E L K +
Sbjct: 197 APA----------TQQFIWTIKHTPLLNTHHPGK----------KFKIEKLKK----DQN 232
Query: 511 NEIIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFER 570
E+I ++ + K DPGSFTLP + G R LCDLG+SV+LMPLS +R
Sbjct: 233 QEMISSASTQETYKKKIIQEKLDDPGSFTLPCSLGPLTFNRCLCDLGASVSLMPLSTAKR 292
Query: 571 LNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGR 630
L + E K + L LAD S+ P G++E++ V++ E P DFV++D+DE K PLILGR
Sbjct: 293 LGIMEYKFCNLALLLADGSVAHPHGLIENLPVKIENVEIPTDFVVLDVDEKGKDPLILGR 352
Query: 631 PFLATSQAKINVGNGTISLRVADEKITFTIFDLKPKPVEKNDVFLVEMMD 680
PFLA++ A I+V NG I+L + K+ F I + K F V+ MD
Sbjct: 353 PFLASAGAVIDVRNGKINLNLEGIKMKFDIRESSWKSTTGVQNFGVQNMD 402
>At4g03850 putative transposon protein
Length = 334
Score = 121 bits (303), Expect = 2e-27
Identities = 74/210 (35%), Positives = 115/210 (54%), Gaps = 2/210 (0%)
Query: 463 LEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSA 522
+E+ S ++ +P EVL + K ++ ++ ++ ++ ++ A
Sbjct: 1 MERYKSVAAKQLDEIEAVMPLIEVLNLIHDPHKNVRNLILERIKMYHDSNDDSDATPSRA 60
Query: 523 ILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMM 582
+R + K DPGSFTLP + G LCDLG+SV+LMPLSM RL + +P +
Sbjct: 61 ADKRIVQEKFGDPGSFTLPCSIGELAFSDCLCDLGASVSLMPLSMARRLEFIQYRPCDLT 120
Query: 583 LQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINV 642
L LADR+ P+G+++D+ V + E P+DFV++DM+ + K PLILGRPFLA+ A I+V
Sbjct: 121 LILADRASRKPFGMLKDLPVMINGVEVPIDFVVLDMEVEHKDPLILGRPFLASVGAVIDV 180
Query: 643 GNGTISLRVADEKITFTIFDLKPKPVEKND 672
G ISL + K FD+ P E +
Sbjct: 181 REGKISLNLG--KHIMLQFDINKTPQESKE 208
>At4g08490 putative athila-like protein
Length = 587
Score = 120 bits (302), Expect = 2e-27
Identities = 67/161 (41%), Positives = 98/161 (60%), Gaps = 1/161 (0%)
Query: 509 EENEIIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMF 568
E+ + +++ E I K DPGSF L + S+ R+LCDLGSS+NLM S+
Sbjct: 233 EKAQRVDIAEYIRTITPGLTTEKLPDPGSFVLDCSISTSRFSRSLCDLGSSINLMSKSVV 292
Query: 569 ERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLIL 628
ERL + + +PT + L ADRS G++EDV V+VG P DFV++D +++ K PLIL
Sbjct: 293 ERLGMTQYRPTRITLLFADRSKRISEGILEDVPVKVGNSIIPADFVVLDYEKEPKNPLIL 352
Query: 629 GRPFLATSQAKINVGNGTISLRVADEKITFTIFDLK-PKPV 668
GR FLAT+ A+ +V G I L+V D ++ F + L+ KP+
Sbjct: 353 GRAFLATAAARFDVKRGRIFLKVCDSEMEFGMDSLELTKPI 393
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,307,670
Number of Sequences: 26719
Number of extensions: 679701
Number of successful extensions: 3581
Number of sequences better than 10.0: 139
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 82
Number of HSP's that attempted gapping in prelim test: 3283
Number of HSP's gapped (non-prelim): 262
length of query: 705
length of database: 11,318,596
effective HSP length: 106
effective length of query: 599
effective length of database: 8,486,382
effective search space: 5083342818
effective search space used: 5083342818
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0103.9


