

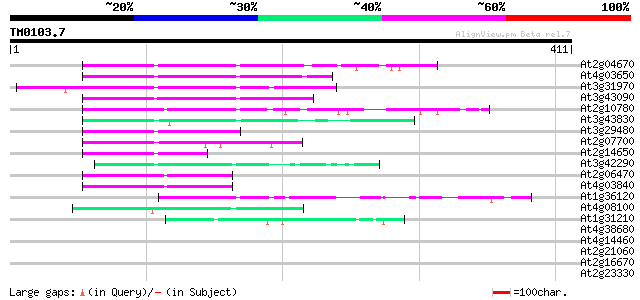
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0103.7
(411 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g04670 putative retroelement pol polyprotein 90 3e-18
At4g03650 putative reverse transcriptase 84 1e-16
At3g31970 hypothetical protein 82 4e-16
At3g43090 putative protein 79 5e-15
At2g10780 pseudogene 79 6e-15
At3g43830 putative protein 71 1e-12
At3g29480 hypothetical protein 68 1e-11
At2g07700 putative retroelement pol polyprotein 63 3e-10
At2g14650 putative retroelement pol polyprotein 60 2e-09
At3g42290 putative protein 59 4e-09
At2g06470 putative retroelement pol polyprotein 57 2e-08
At4g03840 putative transposon protein 56 3e-08
At1g36120 putative reverse transcriptase gb|AAD22339.1 49 4e-06
At4g08100 putative polyprotein 48 9e-06
At1g31210 putative reverse transcriptase 42 5e-04
At4g38680 glycine-rich protein 2 (GRP2) 37 0.026
At4g14460 retrovirus-related like polyprotein 37 0.026
At2g21060 glycine-rich protein (AtGRP2) 36 0.034
At2g16670 putative retroelement pol polyprotein 36 0.034
At2g23330 putative retroelement pol polyprotein 35 0.058
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 89.7 bits (221), Expect = 3e-18
Identities = 78/274 (28%), Positives = 114/274 (41%), Gaps = 29/274 (10%)
Query: 54 FEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPEEGE 113
F G +P+ A W ++R F + C +V A + L +A WW SV R + +
Sbjct: 138 FSSGTSPEEADSWRSRVQRNFGSSRCPAEYRVDLAVHFLEGDAHLWWRSVTAR--RRQAD 195
Query: 114 LIWEIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQFHPRYSTANDE 173
+ W F F KYFP +A R E F EL Q SV EY +F L + R D+
Sbjct: 196 MSWADFVAEFNAKYFPQEALDRMEARFLELTQGEWSVREYDREFNRLLAYAGR--GMEDD 253
Query: 174 TSKCVKFEYGLRPDIRTVVGHDQIRNFATLVKKCRIFEENEKT*KEYLKGLGVNKAPKKK 233
++ +F GLRPD+R Q A LV+ EE+ + + +GV+ A + K
Sbjct: 254 QAQMRRFLRGLRPDLRVQCRVSQYATKAALVETAAEVEEDLQR-----QVVGVSPAVQTK 308
Query: 234 EEKRKPYF*PQDQGRNQFG--RTWNPTGGFGFQGRPGGFNNTPFCGR-------CRRNGH 284
+++ P G+ G R W+ G GR G F+ CG C GH
Sbjct: 309 NTQQQVT--PSKGGKPAQGQKRKWDHPSRAGQGGRAGYFS----CGSLDHTVADCTERGH 362
Query: 285 -----RAQECIVVLGGQSGVQTGGSGNPNPTTTV 313
+ + VLG GV +G P +
Sbjct: 363 LHVKVAGGQFLAVLGRAKGVDIQIAGESMPADLI 396
>At4g03650 putative reverse transcriptase
Length = 839
Score = 84.3 bits (207), Expect = 1e-16
Identities = 57/183 (31%), Positives = 84/183 (45%), Gaps = 7/183 (3%)
Query: 54 FEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPEEGE 113
F GG +P+ A W ++ER F + C +V + L +A WW SV R + +
Sbjct: 156 FSGGTSPEEADSWRSQVERNFGSSRCPAEYRVDLTVHFLEGDAHLWWRSVTAR--RRQAD 213
Query: 114 LIWEIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQFHPRYSTANDE 173
+ W F F KYFP +A R E F EL Q SV EY KF L + R D+
Sbjct: 214 MSWADFMAEFNAKYFPREALDRMEARFLELTQGVRSVREYDRKFNRLLVYAGR--GMEDD 271
Query: 174 TSKCVKFEYGLRPDIRTVVGHDQIRNFATLVKKCRIFEENEKT*KEYLKGLGVNKAPKKK 233
++ +F GLRP++R Q A LV+ EE+ + + G+ PKK
Sbjct: 272 QAQMRRFLRGLRPNLRVRCRVSQYATKAALVETAAEVEEDL---QRQVVGVSPAVQPKKT 328
Query: 234 EEK 236
+++
Sbjct: 329 QQQ 331
>At3g31970 hypothetical protein
Length = 1329
Score = 82.4 bits (202), Expect = 4e-16
Identities = 70/238 (29%), Positives = 106/238 (44%), Gaps = 11/238 (4%)
Query: 6 MARALEQLTAFLTEQAERA-RQGAGQGAANNQEEI--YHGLDKFLKRSPPK-FEGGYNPD 61
+ + LE+L + QA A R Q A EE+ Y + + L+R + F GG +P+
Sbjct: 106 LRQLLERLPGVVPVQAPVAPRVAEVQQRAAVAEEVPSYLRMMEQLQRIGTRYFFGGTSPE 165
Query: 62 GAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPEEGELIWEIFKN 121
A W +E F + C +V A + L +A WW SV R + + W F
Sbjct: 166 EADSWKSRVEHNFGSSRCPAEYRVDLAVHFLEGDAHLWWRSVTAR--RRQAHMSWADFVA 223
Query: 122 SFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQFHPRYSTANDETSKCVKFE 181
F KYFP +A R E F EL Q SV EY +F L + R D+ ++ +F
Sbjct: 224 EFNAKYFPQEALDRMEARFLELTQGERSVREYEREFNRLLVYAGR--GMQDDQAQMRRFL 281
Query: 182 YGLRPDIRTVVGHDQIRNFATLVKKCRIFEENEKT*KEYLKGLGVNKAPKKKEEKRKP 239
GLRPD+R ++ +AT E E+ + + G+ PKK +++ P
Sbjct: 282 RGLRPDLRV---RCRVLQYATKAALVETAAEVEEDLQRQVVGVSPAVKPKKTQQQVAP 336
>At3g43090 putative protein
Length = 357
Score = 79.0 bits (193), Expect = 5e-15
Identities = 54/169 (31%), Positives = 74/169 (42%), Gaps = 3/169 (1%)
Query: 54 FEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPEEGE 113
F GG +P A +W + LE FE C + + L EA WW V G TP +
Sbjct: 131 FTGGIDPVKADDWRKLLENNFENARCPVEFQKDIIVHFLREEASHWWDGVLGN-TPVQHV 189
Query: 114 LIWEIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQFHPRYSTANDE 173
+ WE F+ F K+FP +A E +F EL+Q+ V E + L +F R E
Sbjct: 190 IFWEDFREEFNRKFFPQEAMDSLEDDFEELRQDTKKVRENERELSHLSRFSVR--AGRGE 247
Query: 174 TSKCVKFEYGLRPDIRTVVGHDQIRNFATLVKKCRIFEENEKT*KEYLK 222
S + GLRPDI T + + LV+K E +YLK
Sbjct: 248 QSMIRRLMRGLRPDILTRCASREFFSMIDLVEKAARIESGLVLEAKYLK 296
>At2g10780 pseudogene
Length = 1611
Score = 78.6 bits (192), Expect = 6e-15
Identities = 91/336 (27%), Positives = 135/336 (40%), Gaps = 69/336 (20%)
Query: 54 FEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPEEGE 113
F G +P A EW L+R F++ C E + A + L +A WW +V R E
Sbjct: 182 FMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVEKRRGDEVRS 241
Query: 114 LIWEIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQFHPRYSTANDE 173
+ F++ F +KYFP +A R E + +L Q +V EY +F L ++ R +E
Sbjct: 242 --FADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRYVGR--ELEEE 297
Query: 174 TSKCVKFEYGLRPDIRTVVGHDQIRNF---ATLVKKCRIFEENEKT*KEYLKGLGVNKAP 230
++ +F GLR +IR H +R F + LV++ + EE E + L KAP
Sbjct: 298 QAQLRRFIRGLRIEIR---NHCLVRTFNSVSELVERAAMIEEG----IEEERYLNREKAP 350
Query: 231 KKKEEKRKP-----YF*PQDQ-------------GRNQFGRTWNPTGGFGFQGRPGGFNN 272
+ + KP F D G+N G W G
Sbjct: 351 IRNNQSTKPADKKRKFDKVDNTKSDAKTGECVTCGKNHSGTCWKAIGA------------ 398
Query: 273 TPFCGRCRRNGHRAQECIVVLGGQSGV------------QTGGSGNPNPTTT----VGGG 316
CGRC H Q C + GQS V +TG P T G
Sbjct: 399 ---CGRCGSKDHAIQSCPKMEPGQSKVLGEETRTCFYCGKTGHLKRECPKLTAEKQAGQR 455
Query: 317 SNRNAN-VPPRDNRRVGHPANKGKVFAMTMEEASES 351
NR N +PP R+ A +V+ ++ EEA+++
Sbjct: 456 DNRGGNGLPPPPKRQ----AVASRVYELS-EEANDA 486
>At3g43830 putative protein
Length = 329
Score = 70.9 bits (172), Expect = 1e-12
Identities = 65/245 (26%), Positives = 93/245 (37%), Gaps = 36/245 (14%)
Query: 54 FEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPEEGE 113
F GG +P AY W LE F+++ C AS L EA+ WW V+ R E+
Sbjct: 116 FNGGTDPVKAYNWRNMLECKFKSMRCPVEFWRDLASCYLRGEAQEWWERVKQR---EQVG 172
Query: 114 LI--WEIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQFHPRYSTAN 171
+ W FK F +Y P + EM+F L+Q +V +Y +F L +F R
Sbjct: 173 CVDQWSFFKEEFTRRYLPEETIDDLEMKFLRLQQGTKTVRKYEKEFHSLERFERR---KR 229
Query: 172 DETSKCVKFEYGLRPDIRTVVGHDQIRNFATLVKKCRIFEENEKT*KEYLKGLGVNKAPK 231
E KF GLR DIR N LV+K E +G+ + +
Sbjct: 230 GEHELIHKFISGLRVDIRLCCHVRDFDNMIDLVEKAASLE------------IGLEEEAR 277
Query: 232 KKEEKRKPYF*PQDQGRNQFGRTWNPTGGFGFQGRPGGFNNTPFCGRCRRNGHRAQECIV 291
K + + G +G G C RC GH A++C +
Sbjct: 278 YK----------------RIAQEKEAMGSYGQTGHSKRRCQEVTCYRCGVAGHIARDCRM 321
Query: 292 VLGGQ 296
G+
Sbjct: 322 PFDGK 326
>At3g29480 hypothetical protein
Length = 718
Score = 67.8 bits (164), Expect = 1e-11
Identities = 41/116 (35%), Positives = 53/116 (45%), Gaps = 2/116 (1%)
Query: 54 FEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPEEGE 113
F GG +P+ A W +ER F + C +V A + L +A WW SV R +
Sbjct: 51 FSGGTSPEEADSWRSRVERNFGSSRCPVEYRVDLAVHFLEGDAHLWWKSVTTR--RRQAN 108
Query: 114 LIWEIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQFHPRYST 169
+ W F F KYFP +A R E F EL Q SV EY +F L + ST
Sbjct: 109 MSWADFVAEFNAKYFPQEALDRMEAHFLELTQGERSVREYDREFNRLLVYAECRST 164
>At2g07700 putative retroelement pol polyprotein
Length = 543
Score = 62.8 bits (151), Expect = 3e-10
Identities = 51/181 (28%), Positives = 85/181 (46%), Gaps = 23/181 (12%)
Query: 54 FEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPEEGE 113
F+G +N A +W+R+LE FE C + K A L +AR W SV R +G+
Sbjct: 53 FKGEHNTVVADKWLRDLEMNFEISRCPDNFKRQIAVNFLDKDARIWRESVTAR---NQGQ 109
Query: 114 LI-WEIFKNSFLEKYFPADAKCRKEMEFFE------LKQEAMSVGEY--AAKFEELCQFH 164
+I WE F+ F +KYFP +A+ R E +F + EA+ V + + E +
Sbjct: 110 MISWEAFRREFEKKYFPPEARDRLEQQFMKRYLYNGQDDEALMVRRFLRGLRLEIWGRLQ 169
Query: 165 P-RYSTANDETSKCVKFEYGLRPDIRT---------VVGHDQIRNF-ATLVKKCRIFEEN 213
+Y + N+ T + E P+++T +G +IR F L+ + ++ EN
Sbjct: 170 AVKYDSVNELTERADNVEEESIPELKTNLTYPEGPLEIGERRIRKFKKRLIPQVQVIWEN 229
Query: 214 E 214
+
Sbjct: 230 K 230
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 60.1 bits (144), Expect = 2e-09
Identities = 32/92 (34%), Positives = 44/92 (47%), Gaps = 2/92 (2%)
Query: 54 FEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPEEGE 113
F GG +P+ A W +ER F + C ++ A + L +A WW SV R + +
Sbjct: 156 FSGGTSPEVADSWRSRVERNFGSSRCPAEYRIDLAVHFLEGDAHLWWRSVTAR--RRQAD 213
Query: 114 LIWEIFKNSFLEKYFPADAKCRKEMEFFELKQ 145
+ W F F KYFP +A R E F EL Q
Sbjct: 214 ISWADFVAEFNAKYFPQEALDRMEAHFLELTQ 245
Score = 29.6 bits (65), Expect = 3.2
Identities = 14/30 (46%), Positives = 18/30 (59%)
Query: 353 ELIKGMCYVNGFPLKVLYDSGATHSFISHE 382
E+ G V G VL+DSGA+H FI+ E
Sbjct: 300 EVCVGTLLVGGVEAHVLFDSGASHCFITPE 329
>At3g42290 putative protein
Length = 462
Score = 59.3 bits (142), Expect = 4e-09
Identities = 58/209 (27%), Positives = 83/209 (38%), Gaps = 33/209 (15%)
Query: 63 AYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPEEGELIWEIFKNS 122
A W ++R F + C +V A + L +A WW SV R + ++ W F
Sbjct: 163 ADSWRSRVKRNFSSSRCLMEYRVDLAVHFLEEDAHLWWKSVTAR--RRQADMSWADFVVE 220
Query: 123 FLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQFHPRYSTANDETSKCVKFEY 182
F KYF D R E+ F EL SV EY +F + + D ++ +F
Sbjct: 221 FNAKYFLQDELDRMEVRFLELTLVERSVWEYDREFNRVLVY--AGWGMKDGQAELRRFMR 278
Query: 183 GLRPDIRTVVGHDQIRNFATLVKKCRIFEENEKT*KEYLKGLGVNKAPKKKEEKRKPYF* 242
GLRPD+R +CR+ ++ K L AP+K KP
Sbjct: 279 GLRPDLRV---------------RCRM---SQYALKAALVETAAEVAPRK---GGKPM-- 315
Query: 243 PQDQGRNQFGRTWNPTGGFGFQGRPGGFN 271
QG+ R+WN G GR G F+
Sbjct: 316 ---QGKK---RSWNHLSRAGQGGRAGCFS 338
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 56.6 bits (135), Expect = 2e-08
Identities = 35/110 (31%), Positives = 54/110 (48%), Gaps = 2/110 (1%)
Query: 54 FEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPEEGE 113
F G +P A EW L+R F++ C E + A + L +A WW +V R E
Sbjct: 147 FMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVEKRRGDEVRS 206
Query: 114 LIWEIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQF 163
+ F++ F +KYFP +A R E + +L Q +V EY +F L ++
Sbjct: 207 --FADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRY 254
>At4g03840 putative transposon protein
Length = 973
Score = 56.2 bits (134), Expect = 3e-08
Identities = 35/110 (31%), Positives = 54/110 (48%), Gaps = 2/110 (1%)
Query: 54 FEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSVRGRITPEEGE 113
F G +P A EW L+R F++ C E + A + L +A WW +V R E
Sbjct: 147 FMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLERDAHNWWLTVEKRRGDEVRS 206
Query: 114 LIWEIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQF 163
+ F++ F +KYFP +A R E + +L Q +V EY +F L ++
Sbjct: 207 --FADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRY 254
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 49.3 bits (116), Expect = 4e-06
Identities = 67/277 (24%), Positives = 113/277 (40%), Gaps = 63/277 (22%)
Query: 110 EEGELIWEIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQFHPRYST 169
E+ + I F F KYFP +A R E F EL + +SV EY +F+ L + R
Sbjct: 148 EQLQRIGTDFVAEFNAKYFPQEALDRMEARFLELTKGELSVREYGREFKRLLVYAGR--G 205
Query: 170 ANDETSKCVKFEYGLRPDIRTVVGHDQIRNFATLVKKCRIFEENEKT*KEYLKGLGVNKA 229
D+ ++ +F GLRPD+R ++ +AT K E E+ + + + +
Sbjct: 206 MEDDQAQMRRFLRGLRPDLRV---RCRVSQYAT--KAALTAAEVEEDLQRQVVAVSPSVQ 260
Query: 230 PKKKEEKRKPYF*PQDQGRNQFGRTWNPTGGFGFQGRPGGFNNTPFCGRCRRNGHRAQEC 289
PKK +++ P GG Q + +++ PF
Sbjct: 261 PKKIQQQVAP-----------------SKGGKPAQVQKRKWDH-PF-------------- 288
Query: 290 IVVLGGQSGVQTGGSGNPNPTTTVGGGSNRNANVPPRDNRRVGHPANKGKVFAMTMEEAS 349
GQ G + G + + + T+T GG + + G + + A++ +
Sbjct: 289 ---RAGQGG-RAGSAADDSSTSTAGGAA-----------QSAGAARSAAECAAVSTYCSG 333
Query: 350 ES----PELIKGMCYVNGFPLKVLYDSGATHSFISHE 382
+ +LI G C + VL+DSGA+H FI+ E
Sbjct: 334 TTWLYYADLISGRCRGH-----VLFDSGASHCFITPE 365
>At4g08100 putative polyprotein
Length = 1054
Score = 48.1 bits (113), Expect = 9e-06
Identities = 41/171 (23%), Positives = 67/171 (38%), Gaps = 5/171 (2%)
Query: 47 LKRSPPKFEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSV--R 104
LK P F G NPD EW +++E +F C + KV A+ + A +WW +
Sbjct: 92 LKLKIPAFHGTDNPDTYLEWEQKIELVFLCQECLQSNKVKIAATKFYNYALSWWDQLVTS 151
Query: 105 GRITPEEGELIWEIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQFH 164
R T + W K +++ P+ L Q + +V EY + E L
Sbjct: 152 RRRTRDYPIKTWNQLKFVMRKRFVPSYYHRELHQRLRNLVQGSKTVEEYFLEMETLML-- 209
Query: 165 PRYSTANDETSKCVKFEYGLRPDIRTVVGHDQIRNFATLVKKCRIFEENEK 215
R D + +F GL +I+ + ++ K +FE+ K
Sbjct: 210 -RADLQEDGEAVMSRFMGGLNREIQDRLETQHYVELEEMLHKAVMFEQQIK 259
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 42.4 bits (98), Expect = 5e-04
Identities = 44/192 (22%), Positives = 75/192 (38%), Gaps = 22/192 (11%)
Query: 115 IWEIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQFHPRYSTANDET 174
IW +F + + R+ ++ K++ SV Y +F+ +C DE+
Sbjct: 108 IWVSLAENFNKSSVAREFSLRQNLQLLSKKEKPFSV--YCREFKTICDALSSIGKPVDES 165
Query: 175 SKCVKFEYGLRPD---IRTVVGHDQIR----NFATLVKKCRIFEENEKT*KEYLKGLGVN 227
K F GL D I TV+ + F +V + + F+ ++ +E
Sbjct: 166 MKIFGFLNGLGRDYDPITTVIQSSLSKLPTPTFNDVVSEVQGFDSKLQSYEEAASVTPHL 225
Query: 228 KAPKKKEEKRKPYF*PQDQGRNQFGRTWNPTGGFGFQGRPGGFNN----------TPFCG 277
++ E P + P +GR + G+ GG+ +GR GF+ P C
Sbjct: 226 AFNIERSESGSPQYNPNQKGRGRSGQN-KGRGGYSTRGR--GFSQHQSSPQVSGPRPVCQ 282
Query: 278 RCRRNGHRAQEC 289
C R GH A +C
Sbjct: 283 ICGRTGHTALKC 294
>At4g38680 glycine-rich protein 2 (GRP2)
Length = 203
Score = 36.6 bits (83), Expect = 0.026
Identities = 22/58 (37%), Positives = 26/58 (43%), Gaps = 3/58 (5%)
Query: 259 GGFGFQGRPGGFNNTPFCGRCRRNGHRAQECIVVLGGQSGVQTGGSGNPNPTTTVGGG 316
GG G+ GR GG C +C GH A++C GG G GG G GGG
Sbjct: 114 GGGGYGGRGGGGRGGSDCYKCGEPGHMARDCSEGGGGYGG---GGGGYGGGGGYGGGG 168
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 36.6 bits (83), Expect = 0.026
Identities = 17/42 (40%), Positives = 22/42 (51%), Gaps = 3/42 (7%)
Query: 271 NNTPFCGRCRRNGHRAQECIVVLGGQSGVQTGGSG---NPNP 309
N P C C + GH Q+C V G G++TG +G PNP
Sbjct: 281 NQKPICTHCGKVGHTIQKCYKVHGYPPGMKTGNTGYTYKPNP 322
>At2g21060 glycine-rich protein (AtGRP2)
Length = 201
Score = 36.2 bits (82), Expect = 0.034
Identities = 24/58 (41%), Positives = 28/58 (47%), Gaps = 4/58 (6%)
Query: 259 GGFGFQGRPGGFNNTPFCGRCRRNGHRAQECIVVLGGQSGVQTGGSGNPNPTTTVGGG 316
GG G GR GG + C +C GH A+EC GG SG GG G + GGG
Sbjct: 122 GGRGSGGRGGGGGDNS-CFKCGEPGHMARECSQGGGGYSG---GGGGGRYGSGGGGGG 175
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 36.2 bits (82), Expect = 0.034
Identities = 26/94 (27%), Positives = 39/94 (40%), Gaps = 13/94 (13%)
Query: 263 FQGRPGGFNNTPFCGRCRRNGHRAQECIVVLG---------GQSGVQTGGSGNPNPTTTV 313
+QGR + + C C R+GH ++ C V+G +QT G G N +
Sbjct: 164 YQGRGDEKDKSMVCKHCNRSGHASESCYAVIGYPEWWGDRPRSRSLQTRGRGGTNSSGGR 223
Query: 314 GGGSNRNANVPPRDNRRVGHPANKGKVFAMTMEE 347
G G+ AN N AN +A+T E+
Sbjct: 224 GRGAAAYANRVTVPNHDTYEQAN----YALTDED 253
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 35.4 bits (80), Expect = 0.058
Identities = 22/78 (28%), Positives = 32/78 (40%), Gaps = 1/78 (1%)
Query: 272 NTPFCGRCRRNGHRAQECIVVLGGQSGVQTGGSGNPNPTTTVGGGSNRNANVPPRDNRRV 331
+T FC C R GH +C +V G + N +T G GSN + R R
Sbjct: 256 STLFCTHCNRKGHEVTQCFLV-HGYPDWWLEQNPQENQPSTRGRGSNGRGSSSGRGGNRS 314
Query: 332 GHPANKGKVFAMTMEEAS 349
P +G+ A + A+
Sbjct: 315 SAPTTRGRGRANNAQAAA 332
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,199,144
Number of Sequences: 26719
Number of extensions: 496437
Number of successful extensions: 1335
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 1268
Number of HSP's gapped (non-prelim): 82
length of query: 411
length of database: 11,318,596
effective HSP length: 102
effective length of query: 309
effective length of database: 8,593,258
effective search space: 2655316722
effective search space used: 2655316722
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0103.7


