

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0103.12
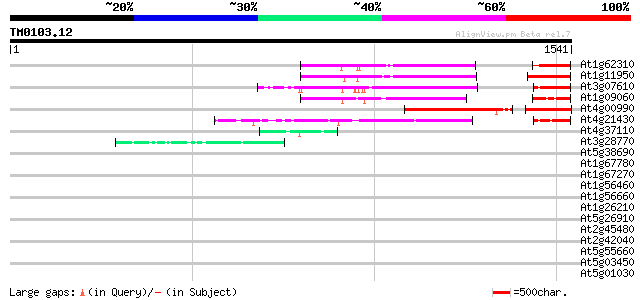
(1541 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g62310 hypothetical protein 418 e-116
At1g11950 putative DNA-binding protein 414 e-115
At3g07610 hypothetical protein 347 3e-95
At1g09060 hypothetical protein 297 4e-80
At4g00990 unknown protein 279 1e-74
At4g21430 unknown protein 223 9e-58
At4g37110 unknown protein 57 8e-08
At3g28770 hypothetical protein 47 9e-05
At5g38690 unknown protein 43 0.001
At1g67780 putative protein 43 0.001
At1g67270 unknown protein 40 0.008
At1g56460 unknown protein 39 0.030
At1g56660 hypothetical protein 38 0.052
At1g26210 hypothetical protein 37 0.089
At5g26910 putative protein 37 0.12
At2g45480 unknown protein 37 0.12
At2g42040 unknown protein 37 0.12
At5g55660 putative protein 36 0.15
At5g03450 unknown protein 36 0.15
At5g01030 unknown protein (At5g01030) 35 0.26
>At1g62310 hypothetical protein
Length = 883
Score = 418 bits (1075), Expect = e-116
Identities = 223/517 (43%), Positives = 294/517 (56%), Gaps = 45/517 (8%)
Query: 800 CHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKD 859
CHQC + ++IC++C++ +C +CI KWYP + +++ CP C +NCNC CL +
Sbjct: 209 CHQCLKGERITLLICSECEKTMFCLQCIRKWYPNLSEDDVVEKCPLCRQNCNCSKCLHLN 268
Query: 860 ISVMTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEVEASMHGS---------- 909
+ T E L YL+ LP L + Q E+E EA++ G
Sbjct: 269 GLIETSKRELAKSERRHHLQYLITLMLPFLNKLSIFQKLEIEFEATVQGKLPSEVEITAA 328
Query: 910 -PLMEEDIQFDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNG---------LHYEDI- 958
+E + D+C TSIV+ HRSCP C Y+LCL CC E+R G HY D
Sbjct: 329 ISYTDERVYCDHCATSIVDLHRSCPK--CSYELCLKCCQEIREGSLSERPEMKFHYVDRG 386
Query: 959 ------------PASGNEETIDEPPITSAWRAEINGRIPCPPKARGGCGTSILSLRRLFE 1006
S E + P + W NG I C P+ GGCG +L LRR+
Sbjct: 387 HRYMHGLDAAEPSLSSTFEDEEANPSDAKWSLGENGSITCAPEKLGGCGERMLELRRILP 446
Query: 1007 ANWVNKLVRNAEELTIQYHPPSVDLLVGCLQCHRFVVDLAQNSVRKAASRETNHDNFLYC 1066
W++ L AE Y+ ++ C +C +L RK+ASR T+ DN+L+C
Sbjct: 447 LTWMSDLEHKAETFLSSYNISP--RMLNC-RCSSLETELT----RKSASRTTSSDNYLFC 499
Query: 1067 PDAVD-MGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAF-RGANKILKEEP 1124
P+++ + + E HFQ HW +GEPVIVRN + GLSW PMVMWRA N E
Sbjct: 500 PESLGVLKEEELLHFQEHWAKGEPVIVRNALDNTPGLSWEPMVMWRALCENVNSTSSSEM 559
Query: 1125 TTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAE 1184
+ KAIDCL CEV+IN QFF+GY +GR Y N WPEMLKLKDWPPS+ FE+ PRH E
Sbjct: 560 SQVKAIDCLANCEVEINTRQFFEGYSKGRTYENFWPEMLKLKDWPPSDKFEDLLPRHCDE 619
Query: 1185 FIAMLPFSDYTHPKFGILNLATKLP-AVLKPDLGPKTYIAYGSLEELSRGDSVTKLHCDI 1243
FI+ LPF +Y+ P+ GILN+ATKLP +KPDLGPKTYIAYG +EL RGDSVTKLHCD+
Sbjct: 620 FISALPFQEYSDPRTGILNIATKLPEGFIKPDLGPKTYIAYGIPDELGRGDSVTKLHCDM 679
Query: 1244 SDAVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRD 1280
SDAVNILTHT EV Q +K L++K++ ++ D
Sbjct: 680 SDAVNILTHTAEVTLSQEQISSVKALKQKHKLQNKVD 716
Score = 145 bits (365), Expect = 2e-34
Identities = 73/106 (68%), Positives = 81/106 (75%), Gaps = 8/106 (7%)
Query: 1435 DISSNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPI 1494
+ISSN ++ ET GSA+WDIFRR+DVPKL EYL+KH KEFRH PV V HPI
Sbjct: 738 EISSN------ENEET--GSALWDIFRREDVPKLEEYLRKHCKEFRHTYCSPVTKVYHPI 789
Query: 1495 HDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRN 1540
HDQ YL +HKR+LK EYGIEPWTF Q LGEAVFIPAGCPHQVRN
Sbjct: 790 HDQSCYLTLEHKRKLKAEYGIEPWTFVQKLGEAVFIPAGCPHQVRN 835
>At1g11950 putative DNA-binding protein
Length = 879
Score = 414 bits (1065), Expect = e-115
Identities = 223/516 (43%), Positives = 294/516 (56%), Gaps = 43/516 (8%)
Query: 800 CHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKD 859
CHQC + + IC C+ + YC+ CI KWYP + ++I CPFC CNC CL
Sbjct: 193 CHQCSKGERRYLFICTFCEVRLYCFPCIKKWYPHLSTDDILEKCPFCRGTCNCCTCLHSS 252
Query: 860 ISVMTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEVEASMHGSPLMEEDIQ-- 917
+ T + D L +L+ LP L+ + + Q E+E EA + S + DI
Sbjct: 253 GLIETSKRKLDKYERFYHLRFLIVAMLPFLKKLCKAQDQEIETEAKVQDSMASQVDISES 312
Query: 918 ---------FDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNG---------------- 952
++C TSIV+ HRSCP C Y+LCL CC E+R G
Sbjct: 313 LCSNEERVFCNHCATSIVDLHRSCPK--CSYELCLNCCQEIRGGWLSDRPECQLQFEYRG 370
Query: 953 ---LHYEDIPASGNEETIDEPPITSA-WRAEINGRIPCPPKARGGCGTSILSLRRLFEAN 1008
+H E S + + DE S W A+ NG I C PK GGCG S+L L+R+
Sbjct: 371 TRYIHGEAAEPSSSSVSEDETKTPSIKWNADENGSIRCAPKELGGCGDSVLELKRILPVT 430
Query: 1009 WVNKLVRNAEELTIQYHPPSVDLLVGCLQCHRFVVDLAQNSVRKAASRETNHDNFLYCPD 1068
W++ L + AE Y S+ + +C + + RKAASR+ + DN+LY PD
Sbjct: 431 WMSDLEQKAETFLASY---SIKPPMSYCRCSSDMSSMK----RKAASRDGSSDNYLYSPD 483
Query: 1069 AVD-MGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAF-RGANKILKEEPTT 1126
++D + E HFQ HW +GEPVIVRN +GLSW PMVMWRA + + +
Sbjct: 484 SLDVLKQEELLHFQEHWSKGEPVIVRNALNNTAGLSWEPMVMWRALCENVDSAISSNMSD 543
Query: 1127 FKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAEFI 1186
KAIDCL CEV+IN FF+GY +GR Y N WPEMLKLKDWPPS+ FE PRH EFI
Sbjct: 544 VKAIDCLANCEVKINTLCFFEGYSKGRTYENFWPEMLKLKDWPPSDKFENLLPRHCDEFI 603
Query: 1187 AMLPFSDYTHPKFGILNLATKLP-AVLKPDLGPKTYIAYGSLEELSRGDSVTKLHCDISD 1245
+ LPF +Y+ P+ GILN+ATKLP +LKPDLGPKTY+AYG+ +EL RGDSVTKLHCD+SD
Sbjct: 604 SALPFQEYSDPRSGILNIATKLPEGLLKPDLGPKTYVAYGTSDELGRGDSVTKLHCDMSD 663
Query: 1246 AVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRDL 1281
AVNIL HT EV Q I L++K++ ++ ++L
Sbjct: 664 AVNILMHTAEVTLSEEQRSAIADLKQKHKQQNEKEL 699
Score = 128 bits (321), Expect = 3e-29
Identities = 64/122 (52%), Positives = 80/122 (65%), Gaps = 4/122 (3%)
Query: 1423 KIQIDKTVPVKNDISSNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHA 1482
K Q +K + +N + ++ + + A+WDIF+R+DVPKL EYL+KH EFRH
Sbjct: 692 KQQNEKELQEQNGLEEEEVVSDEIVVYDETSGALWDIFKREDVPKLEEYLRKHCIEFRHT 751
Query: 1483 NNLPVDSVTHPIHDQILYLNEKHKRQLKKEYG----IEPWTFEQHLGEAVFIPAGCPHQV 1538
V V HPIHDQ +L +HKR+LK E+G IEPWTF Q LGEAVFIPAGCPHQV
Sbjct: 752 YCSRVTKVYHPIHDQSYFLTVEHKRKLKAEFGMVTWIEPWTFVQKLGEAVFIPAGCPHQV 811
Query: 1539 RN 1540
RN
Sbjct: 812 RN 813
>At3g07610 hypothetical protein
Length = 1027
Score = 347 bits (890), Expect = 3e-95
Identities = 244/715 (34%), Positives = 336/715 (46%), Gaps = 149/715 (20%)
Query: 681 GSKNKMKSIANKARNKFGKVRNMRGRPKGSLRKKNETAYCLDSQNERNSL--DGRTSTEA 738
G K K K N+A K K K +KNE +D N L + R +T+
Sbjct: 46 GPKRKGKRGGNRAPKKTPK--------KDEEMQKNE----IDEANRVTGLVKEKRAATKI 93
Query: 739 AYRNDVDLHRGHCSQEELLRMLSVEHKNIQGVGVEETIDYGLRSSGLMGDTERKK----- 793
R D + G S K ++G+ + G R + G+ K
Sbjct: 94 LNRKDSIIEVGEASGSM--------PKEVKGIRI------GKRKGEIDGEIPTKPGKKPK 139
Query: 794 ---ETRIL------RCHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACP 844
+ RI+ CHQC ++ V C C K+YC+ C+ WYP +E++ C
Sbjct: 140 TTVDPRIIGYRPDNMCHQCQKSDRI-VERCQTCNSKRYCHPCLDTWYPLIAKEDVAKKCM 198
Query: 845 FCLRNCNCRLCLKKDISV--MTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEV 902
FC CNCR CL+ D + + + +Q ++L LP L+ I EQ++E EV
Sbjct: 199 FCSSTCNCRACLRLDTKLKGINSNLIVSEEEKVQASKFILQSLLPHLKGINDEQVAEKEV 258
Query: 903 EASMHGSPLME-----------EDIQFDNCNTSIVNFHRSCPNPNCRYDL----CLT--- 944
EA ++G E E + D C TSI + HR+C + C +D+ CL
Sbjct: 259 EAKIYGLKFEEVRPQDAKAFPDERLYCDICKTSIYDLHRNCKS--CSFDICLSCCLEIRN 316
Query: 945 ----CCME------LRNGLHYE--------DIPASGNEETI------------------- 967
C E + GL YE + PA+ ++ +
Sbjct: 317 GKALACKEDVSWNYINRGLEYEHGQEGKVIEKPANKLDDKLKDKLDGKPDDKPKGKPKGR 376
Query: 968 -------------------------DEPPIT-------SAWRAEINGRIPCPPKARGGCG 995
DE P+ S W+A G I C CG
Sbjct: 377 PKGKPDDKPKGKLKGKQDDKPDDKPDEKPVNTDHMKYPSLWKANEAGIITCC------CG 430
Query: 996 TSILSLRRLFEANWVNKLVRNAEELT----IQYHPPSVDLLVGCLQCHRFVVDLAQNSVR 1051
L L+RL W+++LV E+ + P +V C R + D+ ++
Sbjct: 431 AGELVLKRLLPDGWISELVNRVEKTAEAGELLNLPETVLERCPCSNSDRHI-DIDSCNLL 489
Query: 1052 KAASRETNHDNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWR 1111
KAA RE + DN+LY P D+ + +HFQ HW++GEPVIVRNV E SGLSW PMVM R
Sbjct: 490 KAACREGSEDNYLYSPSVWDVQQDDLKHFQHHWVKGEPVIVRNVLEATSGLSWEPMVMHR 549
Query: 1112 AFRGANKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPS 1171
A R + + A+DCLD+CEV++N+ +FF GY +GR R GWP +LKLKDWPP+
Sbjct: 550 ACRQISHVQHGSLKDVVAVDCLDFCEVKVNLHEFFTGYTDGRYDRMGWPLVLKLKDWPPA 609
Query: 1172 NSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPA-VLKPDLGPKTYIAYGSLEEL 1230
F++ PRH EF+ LP YTHP G LNLA KLP LKPD+GPKTY+A G +EL
Sbjct: 610 KVFKDNLPRHAEEFLCSLPLKHYTHPVNGPLNLAVKLPQNCLKPDMGPKTYVASGFAQEL 669
Query: 1231 SRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRDLYGRI 1285
RGDSVTKLHCD+SDAVNILTH EV P QP I L+KK+ +D+++LY +
Sbjct: 670 GRGDSVTKLHCDMSDAVNILTHISEV--PNMQPG-IGNLKKKHAEQDLKELYSSV 721
Score = 136 bits (342), Expect = 1e-31
Identities = 63/101 (62%), Positives = 73/101 (71%), Gaps = 1/101 (0%)
Query: 1440 NFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQIL 1499
N Q ++ET G A+WDIFRR+D+PKL Y++KHHKEFRH PV V HPIHDQ
Sbjct: 733 NSRQQVQNVETDDG-ALWDIFRREDIPKLESYIEKHHKEFRHLYCCPVSQVVHPIHDQNF 791
Query: 1500 YLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRN 1540
YL H +LK+EYGIEPWTF Q LG+AV IP GCPHQVRN
Sbjct: 792 YLTRYHIMKLKEEYGIEPWTFNQKLGDAVLIPVGCPHQVRN 832
>At1g09060 hypothetical protein
Length = 911
Score = 297 bits (760), Expect = 4e-80
Identities = 163/477 (34%), Positives = 248/477 (51%), Gaps = 37/477 (7%)
Query: 800 CHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKD 859
CHQC R ++ C KC ++ +C+ C++ Y + EE+E CP C C+C+ CL+ D
Sbjct: 220 CHQCQRKDRERIISCLKCNQRAFCHNCLSARYSEISLEEVEKVCPACRGLCDCKSCLRSD 279
Query: 860 ISVMTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEVEASMHGSPL-------- 911
++ E LQ L LL+ LP+++ I EQ E+E+E + +
Sbjct: 280 NTIKVRIREIPVLDKLQYLYRLLSAVLPVIKQIHLEQCMEVELEKRLREVEIDLVRARLK 339
Query: 912 MEEDIQFDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNGLHYEDIPASGNEETIDE-- 969
+E + + C +V+++R CPN C YDLCL CC +LR + SG + + +
Sbjct: 340 ADEQMCCNVCRIPVVDYYRHCPN--CSYDLCLRCCQDLREE---SSVTISGTNQNVQDRK 394
Query: 970 --PPIT-------SAWRAEINGRIPCPPKARGGCGTSILSLRRLFEANWVNKLVRNAEEL 1020
P + W A +G IPCPPK GGCG+ L+L R+F+ NWV KLV+NAEE+
Sbjct: 395 GAPKLKLNFSYKFPEWEANGDGSIPCPPKEYGGCGSHSLNLARIFKMNWVAKLVKNAEEI 454
Query: 1021 TIQYHPPSVDLLVGCLQCHRFVVDLAQNSVRKAASRETNHDNFLYCPDAVDMGDTEYEHF 1080
+ GC D+ + K A RE + DN++Y P + F
Sbjct: 455 -----------VSGCKLSDLLNPDMCDSRFCKFAEREESGDNYVYSPSLETIKTDGVAKF 503
Query: 1081 QRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAFRG-ANKILKEEPTTFKAIDCLDWCEVQ 1139
++ W G V V+ V + +S W P +WR +++ L+E KAI+CLD EV
Sbjct: 504 EQQWAEGRLVTVKMVLDDSSCSRWDPETIWRDIDELSDEKLREHDPFLKAINCLDGLEVD 563
Query: 1140 INIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAEFIAMLPFSDYTHPKF 1199
+ + +F + Y +G+ G P + KLKDWP ++ EE EFI PF +Y HP+
Sbjct: 564 VRLGEFTRAYKDGKNQETGLPLLWKLKDWPSPSASEEFIFYQRPEFIRSFPFLEYIHPRL 623
Query: 1200 GILNLATKLPAV-LKPDLGPKTYIAYGSLEELSRGDSVTKLHCDISDAVNILTHTEE 1255
G+LN+A KLP L+ D GPK Y++ G+ +E+S GDS+T +H ++ D V +L HT E
Sbjct: 624 GLLNVAAKLPHYSLQNDSGPKIYVSCGTYQEISAGDSLTGIHYNMRDMVYLLVHTSE 680
Score = 119 bits (297), Expect = 2e-26
Identities = 56/110 (50%), Positives = 78/110 (70%), Gaps = 9/110 (8%)
Query: 1436 ISSNNFFQNDDHMETQF-----GSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSV 1490
++ N +N D+ME+ G A WD+FRRQDVPKL+ YL++ F+ +N+ D V
Sbjct: 740 VNPENLTENGDNMESSCTSSCAGGAQWDVFRRQDVPKLSGYLQR---TFQKPDNIQTDFV 796
Query: 1491 THPIHDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRN 1540
+ P+++ L+LNE HKRQL+ E+G+EPWTFEQH GEA+FIPAGCP Q+ N
Sbjct: 797 SRPLYEG-LFLNEHHKRQLRDEFGVEPWTFEQHRGEAIFIPAGCPFQITN 845
Score = 42.4 bits (98), Expect = 0.002
Identities = 22/66 (33%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query: 10 RCGRKAPPGWRCTERALSGKSVCERHFLYNQKKTERWKEGASGITPKRRSGRRKPVDNSE 69
RC R WRCT +++ K+VCE+H++ +K+ A+ KRRS + D
Sbjct: 43 RCKRSDGKQWRCTAMSMADKTVCEKHYIQAKKRAANSAFRANQKKAKRRSSLGE-TDTYS 101
Query: 70 NGVVDD 75
G +DD
Sbjct: 102 EGKMDD 107
>At4g00990 unknown protein
Length = 454
Score = 279 bits (713), Expect = 1e-74
Identities = 158/310 (50%), Positives = 201/310 (63%), Gaps = 18/310 (5%)
Query: 1085 IRGEPVIVRNVFEKASGLSWHPMVMWRAFRGAN---KILKEEPTTFKAIDCLDWCEVQIN 1141
++ EPVIVRNV EK SGLSW PMVMWRA R + K +EE T KA+DCLDWCEV+IN
Sbjct: 1 MKAEPVIVRNVLEKTSGLSWEPMVMWRACREMDPKRKGTEEETTKVKALDCLDWCEVEIN 60
Query: 1142 IFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAEFIAMLPFSDYTHPKFGI 1201
+ QFF+GYLEGR ++NGWPEMLKLKDWPPS+ FE+ PRH AEFIA LPF DYT PK GI
Sbjct: 61 LHQFFEGYLEGRMHKNGWPEMLKLKDWPPSDLFEKRLPRHNAEFIAALPFFDYTDPKSGI 120
Query: 1202 LNLATKLP-AVLKPDLGPKTYIAYGSLEELSRGDSVTKLHCDISDAVNILTHTEEVKAPL 1260
LNLAT+ P LKPDLGPKTYIAYG EEL+RGDSVTKLHCDISDAVN+LTHT +V+ P
Sbjct: 121 LNLATRFPEGSLKPDLGPKTYIAYGFHEELNRGDSVTKLHCDISDAVNVLTHTAKVEIPP 180
Query: 1261 WQPKIIKKLQKKY-EAEDMRDLYGRINKTVVSHRSKHKKCRTGISMDPK-IPENDDTMGR 1318
+ + IK QKKY EA + Y K +K K D K N++
Sbjct: 181 VKYQNIKVHQKKYAEAMLQKQQYSGQVKEASELENKSMKEVDESKKDLKDKAANEEQSNN 240
Query: 1319 NSNLRGSQSNEEIVVNE--------LSTRSSSLGESRSDSAACVQGFSESSESKSVLNAG 1370
+S GS E++++++ +ST S+ E + D+ G ++E ++ G
Sbjct: 241 SSRPSGSGEAEKVIISKEDNPTQPAVSTSVESIQEQKLDAPKETDG--NTNERSKAVHGG 298
Query: 1371 EQAILNMYKR 1380
A+ ++++R
Sbjct: 299 --AVWDIFRR 306
Score = 155 bits (393), Expect = 1e-37
Identities = 69/126 (54%), Positives = 88/126 (69%)
Query: 1416 SPVMPCSKIQIDKTVPVKNDISSNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKH 1475
+P P ++ K D ++ + G AVWDIFRR+DVPKL ++LK+H
Sbjct: 260 NPTQPAVSTSVESIQEQKLDAPKETDGNTNERSKAVHGGAVWDIFRREDVPKLIQFLKRH 319
Query: 1476 HKEFRHANNLPVDSVTHPIHDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCP 1535
EFRH NN P++SV HPIHDQ ++L++ K+QLK+E+ IEPWTFEQHLGEAVFIPAGCP
Sbjct: 320 EHEFRHFNNEPLESVIHPIHDQTMFLSDSQKKQLKEEFDIEPWTFEQHLGEAVFIPAGCP 379
Query: 1536 HQVRNR 1541
HQVRNR
Sbjct: 380 HQVRNR 385
>At4g21430 unknown protein
Length = 927
Score = 223 bits (567), Expect = 9e-58
Identities = 200/741 (26%), Positives = 318/741 (41%), Gaps = 101/741 (13%)
Query: 562 REGLENKMLSNLCQEHIEYTQPVVRGGRPKGSRNKKIKLAFQDMVDEVRFANKESDKATC 621
R LE K +C+ H ++Q ++ + K + + K+ + + DEV + E +++
Sbjct: 25 RRALEGK---KMCESH--HSQQSLKRSKQKVAESSKLVRSRRGGGDEVASSEIEPNESRI 79
Query: 622 AVGEEQKDHGSDIGKPIGLDNDKATL--ASDRDQETPNQTLAQDEVQND------KSSVK 673
K +G K + A D+ L + ++Q D K V+
Sbjct: 80 R------------SKRLGKSKRKRVMGEAEAMDEAVKKMKLKRGDLQLDLIRMVLKREVE 127
Query: 674 PKRGRPKGSKNKMKSIANKARNKFGKVRNMRGRPKGSLRKKNETAYCLDSQNERNSLDGR 733
KR R S NK KS N ++F R P G + ++
Sbjct: 128 -KRKRLPNSNNKKKS--NGGFSEFVGEELTRVLPNGIM-----------------AISPP 167
Query: 734 TSTEAAYRNDVDLHRGHCSQEELLRMLS--VEHKNIQGVGVEETIDYGLRSSGLMGDTER 791
+ T + + D+ G EE + M+ KNI+ + + + ++ GD
Sbjct: 168 SPTTSNVSSPCDVKVG----EEPISMIKRRFRSKNIEPLPIGK-----MQVVPFKGDLVN 218
Query: 792 KKETRILRCHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCN 851
++ + +RCH C + ++ C C+R+ +C +CI K G ++EE+E CP C +C
Sbjct: 219 GRKEKKMRCHWCGTRGFGDLISCLSCEREFFCIDCIEKRNKG-SKEEVEKKCPVCRGSCR 277
Query: 852 CRLCLKKDISVM----TGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEME------ 901
C++C + V + S +D +L L Y + LP+L+ I E E+E
Sbjct: 278 CKVCSVTNSGVTECKDSQSVRSDIDRVLH-LHYAVCMLLPVLKEINAEHKVEVENDAEKK 336
Query: 902 ----VEASMHGSPLMEEDIQ--FDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNGLHY 955
E +H S L +D Q + + ++V+ R C + L
Sbjct: 337 EGNPAEPQIHSSELTSDDRQPCSNGRDFAVVDLQRMCTRSSSVLRL-------------- 382
Query: 956 EDIPASGNEETIDEPPITSAWRAEINGRIPCPPKARGGCGTSILSLRRLFEANWVNKLVR 1015
+ ++E++ + I C K GC ++ LF +KL
Sbjct: 383 -NSDQDQSQESLSRKVGSVKCSNGIKSPKVCKRKEVKGCSNNLFL--SLFPLELTSKLEI 439
Query: 1016 NAEELTIQYHPPSV-DLLVGCLQCHRFVVDLAQNSVRKAASRETNHD---NFLYCPDAVD 1071
+AEE+ Y P + D GC C + + + +T D NFLY P +D
Sbjct: 440 SAEEVVSCYELPEILDKYSGCPFCIGMETQSSSSDSHLKEASKTREDGTGNFLYYPTVLD 499
Query: 1072 MGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAFRGANKILKEEPTTFKAID 1131
EHFQ HW +G PVIVR+V + S L+W P+ ++ + ++ T D
Sbjct: 500 FHQNNLEHFQTHWSKGHPVIVRSVIKSGSSLNWDPVALF-----CHYLMNRNNKTGNTTD 554
Query: 1132 CLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAEFIAMLPF 1191
C+DW EV+I + QFF G L G+ N E LKL+ W S+ F+E FP H AE + +LP
Sbjct: 555 CMDWFEVEIGVKQFFLGSLRGKAETNTCQERLKLEGWLSSSLFKEQFPNHYAEILNILPI 614
Query: 1192 SDYTHPKFGILNLATKLP-AVLKPDLGPKTYIAYGSLEELSRGDSVTKLHCDISDAVNIL 1250
S Y PK G+LN+A LP V PD GP I+Y S EE ++ DSV KL + D V+IL
Sbjct: 615 SHYMDPKRGLLNIAANLPDTVQPPDFGPCLNISYRSGEEYAQPDSVKKLGFETCDMVDIL 674
Query: 1251 THTEEVKAPLWQPKIIKKLQK 1271
+ E Q I+KL K
Sbjct: 675 LYVTETPVSTNQICRIRKLMK 695
Score = 99.8 bits (247), Expect = 1e-20
Identities = 45/102 (44%), Positives = 70/102 (68%), Gaps = 2/102 (1%)
Query: 1438 SNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQ 1497
S N+ ++ + +G A WD+F++QDV KL EY+K H E ++ V+HP+ +Q
Sbjct: 753 SCNYSCEEESLSNTYG-AQWDVFQKQDVSKLLEYIKNHSLELESMDSSK-KKVSHPLLEQ 810
Query: 1498 ILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVR 1539
YL+E HK +LK+E+ +EPW+F+Q +GEAV +PAGCP+Q+R
Sbjct: 811 SYYLDEYHKARLKEEFDVEPWSFDQCVGEAVILPAGCPYQIR 852
Score = 42.0 bits (97), Expect = 0.003
Identities = 22/60 (36%), Positives = 26/60 (42%)
Query: 10 RCGRKAPPGWRCTERALSGKSVCERHFLYNQKKTERWKEGASGITPKRRSGRRKPVDNSE 69
RC R WRC RAL GK +CE H K + K S + R G V +SE
Sbjct: 12 RCNRSDGKQWRCKRRALEGKKMCESHHSQQSLKRSKQKVAESSKLVRSRRGGGDEVASSE 71
>At4g37110 unknown protein
Length = 417
Score = 57.0 bits (136), Expect = 8e-08
Identities = 61/243 (25%), Positives = 96/243 (39%), Gaps = 44/243 (18%)
Query: 685 KMKSIANKARNKFGKVRNMRGRPKG----SLRKKNETAYCLDSQNERNSLDGRTSTEAAY 740
K+KS A+ ++G GR S+ + + + R S + +T +Y
Sbjct: 57 KLKSEIRPAKRRYGNSNANPGRETSPIQLSVSSRRSSRLKQEPPVSRRSSRLKNATPVSY 116
Query: 741 RNDVDLHRGHCSQEELLRMLSVEHKNIQGVGVEETIDYGLRSSGLMGDTERKKE------ 794
+ +L +G S+EE++ + G GV I Y L+G+TER E
Sbjct: 117 AEEPELKKGKVSKEEIVLWV--------GEGVRPEI-YTEEHEKLLGNTERTWELFVDGC 167
Query: 795 ----------TRILRCHQCWRNSWSGVVICAKCK---RKQYCYECITKWYPGKTREEIEI 841
R CHQC + + C++C R Q+C +C+ Y E +E
Sbjct: 168 DKNGKRIYDPVRGKCCHQCRQKTLGYHTQCSQCNHSVRGQFCGDCLYMRYGEHVLEALEN 227
Query: 842 A---CPFCLRNCNCRLCLKKDISVMTGSGEADTGVILQKLLYLLNKTLP--LLQHIQREQ 896
CP C CNC C T G TG +K+ L K++ L+Q Q+ +
Sbjct: 228 PDWICPVCRDICNCSFC-------RTKKGWLPTGAAYRKIHKLGYKSVAHYLIQTNQQSE 280
Query: 897 ISE 899
SE
Sbjct: 281 TSE 283
>At3g28770 hypothetical protein
Length = 2081
Score = 47.0 bits (110), Expect = 9e-05
Identities = 94/472 (19%), Positives = 188/472 (38%), Gaps = 46/472 (9%)
Query: 290 NEEARCEALRPLSKRGRPKGSKNENDNKQLST---ALDGQSVGGDDNAGTIGMSSVTDLG 346
N++ + R +K +G+K E++ + + D +SV DN S T+
Sbjct: 758 NKKTKTNENRVRNKEENVQGNKKESEKVEKGEKKESKDAKSVETKDNKKL----SSTENR 813
Query: 347 IEIAVLSGEKDKSS-DEVADLGETARAEKSGRPKVSKNKIRRVEHVGNVVAVKIVGPKKH 405
E SGE +K +E D EK+ V N VGN K + K
Sbjct: 814 DEAKERSGEDNKEDKEESKDYQSVEAKEKNENGGVDTN-------VGNKEDSKDL---KD 863
Query: 406 GRPKSSKCRKKNIMEAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGAGEIVRPK 465
R K K+ M+ E DK + + N +D + + G+GE V+ K
Sbjct: 864 DRSVEVKANKEESMKKKREEVQR--NDKSSTK---EVRDFANNMDIDVQ-KGSGESVKYK 917
Query: 466 KRGRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSA 525
K + G+K + + + S K G K + K SKN N+ + ++
Sbjct: 918 KDEKKEGNKEENKDTINTSSKQK--------GKDKKKKKKESKNS--NMKKKEEDKKEYV 967
Query: 526 DCEIAGPKKCGRPKGSMKKRKSLVCASILEGAGGITREGLENKMLSNLCQEHIEYTQPVV 585
+ E+ + + KK + S L+ +E E++ ++ +E EY +
Sbjct: 968 NNEL-------KKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEKKS 1020
Query: 586 RGGRPKGSRNKKIKLAFQDMVD-EVRFANKESDKATCAVGEEQKDHGSDIGKPIGLDNDK 644
+ KK + ++ D E R + KE +++ +++++ + + +N K
Sbjct: 1021 KTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKES---ENHK 1077
Query: 645 ATLASDRDQETPNQTLAQDEVQNDKSSVKPKRGRPKGSKNK-MKSIANKARNKFGKVRNM 703
+ D+ + N+++ ++E + +K + + R K K M+ + ++ NK + +N
Sbjct: 1078 SKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNE 1137
Query: 704 RGRPKGSLRKKNETAYCLDSQNERNSLDGRTSTEAAYRNDVDLHRGHCSQEE 755
+ + + K E+ +NE S + + +N+VD S+++
Sbjct: 1138 KKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQ 1189
Score = 43.1 bits (100), Expect = 0.001
Identities = 85/470 (18%), Positives = 175/470 (37%), Gaps = 45/470 (9%)
Query: 289 TNEEARCEALRPLSKRGRPKGSKNENDNKQLSTALDGQSVGGDDNAGTIGMSSVTDLGIE 348
+ E+ + E + KG + + +NK + D + + + SV D E
Sbjct: 675 SKEDTKSEVEVKKNDGSSEKGEEGKENNKD--SMEDKKLENKESQTDSKDDKSVDDKQEE 732
Query: 349 IAVLSGEKDKSSDEVADLGETARAEKSGRPKVSKNKIRRVEHVGNVVAVKIVGPKKHGRP 408
+ GE K V G+ ++++ + K ++N++R E + G KK
Sbjct: 733 AQIYGGES-KDDKSVEAKGKKKESKENKKTKTNENRVRNKEE-------NVQGNKK---- 780
Query: 409 KSSKCRKKNIMEAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGAG---EIVRPK 465
+S K K E+ D + E +KKL + + + + N E + V K
Sbjct: 781 ESEKVEKGEKKESKDAKSVETKDNKKLSSTENRDEAKERSGEDNKEDKEESKDYQSVEAK 840
Query: 466 KRGRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEV---A 522
++ G V N E SK ++ K+ K + Q+N ++EV A
Sbjct: 841 EKNENGGVDTNVGN-KEDSKDLKDDRSVEVKANKEESMKKKREEVQRNDKSSTKEVRDFA 899
Query: 523 GSADCEIAGP-------KKCGRPKGSMKKRKSLVCASILEGAGGITREGLENKMLSNLCQ 575
+ D ++ KK + +G+ ++ K + S + ++ E+K SN+ +
Sbjct: 900 NNMDIDVQKGSGESVKYKKDEKKEGNKEENKDTINTSSKQKGKDKKKKKKESKN-SNMKK 958
Query: 576 EHIEYTQPVVRGGRPKGSRNKKIKLAFQDMVDEVRFANKESDKATCAVG--------EEQ 627
+ + + V + + K+ + + E NKE ++ + EE+
Sbjct: 959 KEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEK 1018
Query: 628 KDHGSDIGKPIGL--------DNDKATLASDRDQETPNQTLAQDEVQNDKSSVKPKRGRP 679
K + K + D S +++E A+ + + K + + +
Sbjct: 1019 KSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKS 1078
Query: 680 KGSKNKMKSIANKARNKFGKVRNMRGRPKGSLRKKNETAYCLDSQNERNS 729
K ++K + NK+ K + + + RKK E ++ ++NS
Sbjct: 1079 KKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNS 1128
Score = 35.8 bits (81), Expect = 0.20
Identities = 92/445 (20%), Positives = 155/445 (34%), Gaps = 72/445 (16%)
Query: 314 NDNKQLSTALDGQSVGGDDNAGTIGMSSVTDLGIEIAVLSGEKDKSSDEVADLGETARAE 373
N ST G V G N G + M + L G K + E ++ E +
Sbjct: 214 NGENSESTQEKGDGVEGS-NGGDVSMEN----------LQGNKVEDLKEGNNVVENGETK 262
Query: 374 KSGRPKVSKNKIRRVEHVGNVVAVKIVGPKKHGRPKSSKCRKKNIMEAGDEAAGEIGGDK 433
++ V N + VE G + +G + SK K+ +EA + +
Sbjct: 263 ENNGENVESNNEKEVEGQG-----ESIGDSAIEKNLESKEDVKSEVEAAKNDGSSM--TE 315
Query: 434 KLGRPKGSLNKLKNTVDCNNEGSGAGEIVRPKKRGRPRGSKNKVNNIMEVSKKAASGGDC 493
LG +G N +T+D E G GE + + SK V + +E +K A S
Sbjct: 316 NLGEAQG--NNGVSTIDNEKEVEGQGESIEDSDIEKNLESKEDVKSEVEAAKNAGSSMTG 373
Query: 494 KIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSADCEIAGPKKCGRPKGSMKKRKSLVCASI 553
K+ ++ R G + ++ E GS E K +K +
Sbjct: 374 KL---EEAQRNNGVSTNE----TMNSENKGSG--ESTNDKMVNATTNDEDHKKENKEETH 424
Query: 554 LEGAGGITREGLENKMLSNLCQEHIEYTQPVVRGGRPKGSRNKKIKLAFQDMVDEVR--- 610
+ E LENK N E + V KG+ + + K + +E R
Sbjct: 425 ENNGESVKGENLENK-AGNEESMKGENLENKVGNEELKGNASVEAKTNNESSKEEKREES 483
Query: 611 ------FANKESDKATC------AVGEEQKDHG----SDIGKPIGLDNDKATLASDRDQE 654
+ NKE+ K ++G+ KD+ D+ + + +R QE
Sbjct: 484 QRSNEVYMNKETTKGENVNIQGESIGDSTKDNSLENKEDVKPKVDANESDGNSTKERHQE 543
Query: 655 --------TPNQTL----AQDEVQNDKS---------SVKPKRGRPKGSKNKMKSIANKA 693
T ++ L A ++ +NDKS K KR +G N +S+ N+
Sbjct: 544 AQVNNGVSTEDKNLDNIGADEQKKNDKSVEVTTNDGDHTKEKREETQG--NNGESVKNEN 601
Query: 694 RNKFGKVRNMRGRPKGSLRKKNETA 718
+ ++ + NET+
Sbjct: 602 LENKEDKKELKDDESVGAKTNNETS 626
Score = 35.4 bits (80), Expect = 0.26
Identities = 60/249 (24%), Positives = 97/249 (38%), Gaps = 48/249 (19%)
Query: 243 EVGGFVENPCFEGENDSNKEGPGSNYK-MSALGFEEEIGLLLSRGGTTNEEARCEALRPL 301
++G +E+ +G+ D+ E N K + G EE S+ G TNE E
Sbjct: 1578 KIGESLEDNKVKGKEDNGDEVGKENSKTIEVKGRHEE-----SKDGKTNENGGKEV---- 1628
Query: 302 SKRGRPKGSKNENDNKQLSTALDGQSVGGDDNAGTIGMSSVTDLGIEIAVLSGEKDKSSD 361
+GSK+ N ++ D G +D G + + G E++ G KD +
Sbjct: 1629 ---STEEGSKDSNIVERNGGKEDSIKEGSED-----GKTVEINGGEELSTEEGSKDGKIE 1680
Query: 362 EVADLGETARAEKSGRPKVSKNKIRRVEHVGNVVAVKIVGPKKHGRPKSSKCRKKNIMEA 421
E + E + E S K+ + + K++ +SSK K N
Sbjct: 1681 EGKEGKENSTKEGSKDDKIEEG----------------MEGKENSTKESSKDGKIN---- 1720
Query: 422 GDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGAGEIVRPK----KRGRPRGSKNKV 477
EI GDK+ +GS + N+ +++ S + EI K K G N++
Sbjct: 1721 ------EIHGDKEATMEEGSKDGGTNSTGKDSKDSKSVEINGVKDDSLKDDSKNGDINEI 1774
Query: 478 NNIMEVSKK 486
NN E S K
Sbjct: 1775 NNGKEDSVK 1783
>At5g38690 unknown protein
Length = 572
Score = 43.1 bits (100), Expect = 0.001
Identities = 26/85 (30%), Positives = 39/85 (45%), Gaps = 17/85 (20%)
Query: 800 CHQCWRNSWSGVVICAKCKRKQ-----YCYECITKWYPGKTREEIEIA----CPFCLRNC 850
CHQC + V C K+ + C +CI Y G+ +E+ + CP C NC
Sbjct: 32 CHQCRQKRTDLVGSCVTKKKDKTCPIKLCTKCILNRY-GENAQEVALKKDWICPKCRGNC 90
Query: 851 NCRLCLKKDISVMTGSGEADTGVIL 875
NC C+KK G+ TG+++
Sbjct: 91 NCSYCMKK-------RGQKPTGILV 108
>At1g67780 putative protein
Length = 512
Score = 43.1 bits (100), Expect = 0.001
Identities = 26/87 (29%), Positives = 39/87 (43%), Gaps = 17/87 (19%)
Query: 800 CHQCWRNSWSGVVICAKCKRKQ-----YCYECITKWYPGKTREEI----EIACPFCLRNC 850
CHQC + + V C K+ + +C++C+ Y G+ EE+ + CP C C
Sbjct: 35 CHQCRQKTMDFVASCKAMKKDKQCTINFCHKCLINRY-GENAEEVAKLDDWICPQCRGIC 93
Query: 851 NCRLCLKKDISVMTGSGEADTGVILQK 877
NC C KK G TG++ K
Sbjct: 94 NCSFCRKK-------RGLNPTGILAHK 113
>At1g67270 unknown protein
Length = 495
Score = 40.4 bits (93), Expect = 0.008
Identities = 21/67 (31%), Positives = 33/67 (48%), Gaps = 10/67 (14%)
Query: 800 CHQCWRNSWSGVVICAKCKRK-----QYCYECITKWYPGKTREEI----EIACPFCLRNC 850
CHQC + + C +RK ++CY+C++ Y G+ EE+ + CP C C
Sbjct: 27 CHQCRQKTLDFAAPCKAMRRKKLCPIKFCYKCLSIRY-GENAEEVAKLDDWICPLCRGIC 85
Query: 851 NCRLCLK 857
C +C K
Sbjct: 86 ICSVCRK 92
>At1g56460 unknown protein
Length = 502
Score = 38.5 bits (88), Expect = 0.030
Identities = 73/348 (20%), Positives = 131/348 (36%), Gaps = 45/348 (12%)
Query: 430 GGDKKLGRPKGSL--NKLKNTVDCNNEGSGAGEIVRPKKRGRPRGSKNKVNNIMEVSKKA 487
GG +G L +K K+T+D +G E + K + + GS +K NI+ S A
Sbjct: 78 GGGHSTASTEGFLVPSKKKDTIDTTERRAGFDENIIKKVKLKLGGS-SKTINIISASDGA 136
Query: 488 ASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSADCEIAGPK--KCGRPKGSMKKR 545
+ G C + + + I Q QE++ ++ G + R
Sbjct: 137 SDIGLCST-------KSSHASDDALGIGQTYQEISNERSTKLRGTPLDTASKSDSCNDTR 189
Query: 546 KSLVCASILEGAGGITREGLENKMLSNLCQ--EHIEYTQPVVRGGRPKGSRNKKIKLAFQ 603
S + + + I++ + ++ L +L E I++ + R K ++ ++
Sbjct: 190 DSKANTNPIRKSNRISKRRVLDEELDSLDDDDEEIQFLR------RMKMAKVVAVEEDVD 243
Query: 604 DMVDEVRFANKESD------KATCAVGEEQKDHGSDIGKPIGLDNDKATLASDRDQETPN 657
D D R K S + VG +K D +G D A D D+E
Sbjct: 244 DDEDRTRKHKKLSKVMKQNVEFPRGVGTSEKSAKKD---KMGKAFDDADYVKDDDEEEEE 300
Query: 658 QTLAQDEVQNDKSSVKPKRGRPKGSKNKMKSIANKARNKFGKVRNM----RGRPKGSLRK 713
A +V+ + S + +RG +G + R + G N+ RG P RK
Sbjct: 301 ---AVSDVELENKSARTRRGAEEGQSEVKTEMTVTTRRRSGHSGNLIEFPRGLPPAPPRK 357
Query: 714 KNETAYCLDSQNERNSLDGRTSTEAAYRNDVDLHR-GHCSQEELLRML 760
+ E +D Q ++ EAA R + + + S+ E +R +
Sbjct: 358 RKENGLEVDQQLKK--------AEAAQRRKLQVEKAARESEAEAIRKI 397
>At1g56660 hypothetical protein
Length = 522
Score = 37.7 bits (86), Expect = 0.052
Identities = 62/252 (24%), Positives = 94/252 (36%), Gaps = 41/252 (16%)
Query: 303 KRGRPKGSKNENDNKQLSTALDGQSVGGDDNAGTIGMSSVTDLGIEIAVLSGEKDKSSDE 362
K+ +P K E D ST + + + G G D G + + + DE
Sbjct: 263 KKKKPDKEKKEKDE---STEKEDKKLKGKKGKGE--KPEKEDEGKKTKEHDATEQEMDDE 317
Query: 363 VADLGETARAEKSGRPKVSKNKIRRVEHVGNVVAVKIVGPK--KHGRPKSSKCRKKNIME 420
AD K G+ K +K+K ++ E V + V K K G K K +KK
Sbjct: 318 AAD-------HKEGKKKKNKDKAKKKETVIDEVCEKETKDKDDDEGETKQKKNKKK---- 366
Query: 421 AGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGAGEIVRPKKRGRPRGSKNKVNNI 480
E E G+K + K N L+ V + R K P K + ++
Sbjct: 367 ---EKKSE-KGEKDVKEDKKKENPLETEV-----------MSRDIKLEEPEAEKKEEDDT 411
Query: 481 MEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQ------VSQEVAGSADCEIAGPKK 534
E K GG+ + G KK + K KNK+K+ + ++ S D +I G K
Sbjct: 412 EEKKKSKVEGGESE-EGKKKKKKDK-KKNKKKDTKEPKMTEDEEEKKDDSKDVKIEGSKA 469
Query: 535 CGRPKGSMKKRK 546
K K+K
Sbjct: 470 KEEKKDKDVKKK 481
>At1g26210 hypothetical protein
Length = 168
Score = 37.0 bits (84), Expect = 0.089
Identities = 25/105 (23%), Positives = 44/105 (41%), Gaps = 5/105 (4%)
Query: 642 NDKATLASDRDQETPNQTLAQDEVQNDKSSVKPKRGRPKGSKNKMKSIANKARNKFGK-V 700
ND++++ D D + A D +ND G + + ++ N+ K +
Sbjct: 33 NDQSSVVVDDDDDDTQVKEADDGYENDDGDTSDDGGDEESDDSMASDASSGPSNQLPKHI 92
Query: 701 RNMRGRPKGS----LRKKNETAYCLDSQNERNSLDGRTSTEAAYR 741
R GS L+K+ T + ++ E++ L RT T AA R
Sbjct: 93 NKHAARKNGSKQVYLQKRQHTEKTISNEGEKSDLKARTRTSAASR 137
>At5g26910 putative protein
Length = 900
Score = 36.6 bits (83), Expect = 0.12
Identities = 45/227 (19%), Positives = 89/227 (38%), Gaps = 31/227 (13%)
Query: 598 IKLAFQDMVDEVRFANKESDKATCAVGEEQKDHGSDIGKPIGLDNDKATLASDRDQETPN 657
+ + QD+ +++ A K S + + + ++ P G N+K S T +
Sbjct: 281 VPMRIQDLREKLEAAQKVSSR-------QNSNDTFNLKYPSGKHNEKRITTSLTTPST-S 332
Query: 658 QTLAQDEVQNDKSSVKPK--RGRPKGSKNKMKSIANKARNK--------FGKVRN-MRGR 706
+ + + K VKP + K + N A K K +N +RG
Sbjct: 333 KFMGKSSTDGLKGKVKPSYVSAQAKAGTTPLSVTRNSANQKEKADAKKCVVKSQNALRGA 392
Query: 707 P----KGSLRKKNETAYCLDSQNERNSLDGRTSTEAAYR--NDVDLHRGHCSQEELLRML 760
P K ++ N+ C D+Q S+ + S++ + N V + G S++ L
Sbjct: 393 PISMGKNMFKQNNQKQNCRDNQPSMTSVLNQKSSKVNNKVVNKVPVESGSISKQLGLSTA 452
Query: 761 SVEHKNIQGVGVEETIDY------GLRSSGLMGDTERKKETRILRCH 801
S E + ++T+ G++ SG+ D K+ +++C+
Sbjct: 453 SAEKNTSLSLSRKKTLPRSKKLPNGMQKSGISDDKRTKRSENMIKCN 499
>At2g45480 unknown protein
Length = 431
Score = 36.6 bits (83), Expect = 0.12
Identities = 16/40 (40%), Positives = 21/40 (52%)
Query: 3 ETGEECRRCGRKAPPGWRCTERALSGKSVCERHFLYNQKK 42
ET E RC R WRC++ LSG+ C++H KK
Sbjct: 305 ETDNEPGRCRRTDGKKWRCSKDVLSGQKYCDKHMHRGMKK 344
Score = 33.9 bits (76), Expect = 0.75
Identities = 16/53 (30%), Positives = 25/53 (46%)
Query: 1 MDETGEECRRCGRKAPPGWRCTERALSGKSVCERHFLYNQKKTERWKEGASGI 53
+D E RC R WRC+ L + CERH +K++ + E +S +
Sbjct: 85 IDTLETEPTRCRRTDGKKWRCSNTVLLFEKYCERHMHRGRKRSRKLVESSSEV 137
>At2g42040 unknown protein
Length = 269
Score = 36.6 bits (83), Expect = 0.12
Identities = 15/30 (50%), Positives = 18/30 (60%)
Query: 6 EECRRCGRKAPPGWRCTERALSGKSVCERH 35
EE RC R GWRC ++ L G S+CE H
Sbjct: 164 EEGSRCSRVNGRGWRCCQQTLVGYSLCEHH 193
>At5g55660 putative protein
Length = 759
Score = 36.2 bits (82), Expect = 0.15
Identities = 31/154 (20%), Positives = 70/154 (45%), Gaps = 12/154 (7%)
Query: 592 GSRNKKIKLAFQDMVDEVRFANKESDKATCAVGEEQKDHGSDIGKPIGLDNDKAT----L 647
GS + K Q +E NK+S + EE+K+ + K ++ ++ +
Sbjct: 478 GSSSSKRSAKSQKKTEEATRTNKKSVAHSDDESEEEKEDDEEEEKEQEVEEEEEENENGI 537
Query: 648 ASDRDQETPNQTLAQDEVQNDKSSVKPKRGRPKGSK--NKMKSIANKARNKFGKVRNMRG 705
+ E P + +++ V++++ S + + + +GS+ + K A K+R+K V
Sbjct: 538 PDKSEDEAPQLSESEENVESEEESEEETKKKKRGSRTSSDKKESAGKSRSKKTAVPTKSS 597
Query: 706 RPKGSLRKKNETAYCLDSQNERNSLDGRTSTEAA 739
PK + +K++ + +++ D TS +A+
Sbjct: 598 PPKKATQKRSA------GKRKKSDDDSDTSPKAS 625
>At5g03450 unknown protein
Length = 630
Score = 36.2 bits (82), Expect = 0.15
Identities = 25/76 (32%), Positives = 32/76 (41%), Gaps = 8/76 (10%)
Query: 782 SSGLMGDTERKK-ETRILRCHQC---WRNSWSGVVICAKCKRKQYCYECITKWYPGKTRE 837
SSG D E K+ +T L C C W + V C C Y Y CI KW+ +
Sbjct: 101 SSGSQEDVEWKQGDTEGLSCSICMEVWTSGGQHQVCCLPCGHL-YGYSCINKWFQQRRSG 159
Query: 838 EIEIACPFCLRNCNCR 853
CP C + C+ R
Sbjct: 160 G---KCPLCNKICSLR 172
>At5g01030 unknown protein (At5g01030)
Length = 744
Score = 35.4 bits (80), Expect = 0.26
Identities = 67/321 (20%), Positives = 114/321 (34%), Gaps = 25/321 (7%)
Query: 390 HVGNVVAVKIVGPK-----KHGRPKSSKCRKKNIMEAGDEAAGEIGGDKKLGRPKGSLNK 444
H NV+ V ++ + KHGR K + ++ + A G + P S N+
Sbjct: 63 HQSNVLNVGVLDWESLQRWKHGRAKGGEISGRSERKVSTIATTSTSG---VVVPNDSANR 119
Query: 445 LK---NTVDCNNEGSGAGEIVRPKKRGRPRGSKNKVNN--IMEVSKKAASGGDCKIAGPK 499
K C+N G S++ +N I S K+ SG D K P+
Sbjct: 120 CKIDDQVHTCSNLGKVKASRDLQYSLEPQLASRDSLNKQEIATCSYKS-SGRDHKGVEPR 178
Query: 500 KCGRPKGSKNKQKNIVQVSQEVAGSADCEIAGPKKCGRPKGSMKKRKSLVCASILEGAG- 558
K R ++ + AGS + K+ G + ++ C L+G
Sbjct: 179 KSRRTHSNRESTTGLSSEMGNSAGSLFRDKETQKRAGEIHAKEARERAKECVEKLDGDEK 238
Query: 559 --GITREGL--ENKMLSNLCQEHIEYTQPVVRGGRPKGSRNKKIKLAFQDMVDEVRFANK 614
G + GL E + SN+ G P+ SR L F D ++ F +
Sbjct: 239 IIGDSEAGLTSEKQEFSNIFLLRSRKQSRSTLSGEPQISREVNRSLDFSDGINS-SFGLR 297
Query: 615 ESDKATCAVGEEQKDHGSDIGKPIGLDNDKATLASDRDQETPNQTLAQDEVQNDKSSVKP 674
++C + + + D+ P+G D L+ R + ++T ++ +
Sbjct: 298 SQIPSSCPLSFDLERDSEDMMLPLGTD-----LSGKRGGKRHSKTTSRIFDREFPEDESR 352
Query: 675 KRGRPKGSKNKMKSIANKARN 695
K P SK S +RN
Sbjct: 353 KERHPSPSKRFSFSFGRLSRN 373
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,998,162
Number of Sequences: 26719
Number of extensions: 1830019
Number of successful extensions: 4084
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 3955
Number of HSP's gapped (non-prelim): 141
length of query: 1541
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1429
effective length of database: 8,326,068
effective search space: 11897951172
effective search space used: 11897951172
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0103.12


