

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0101.4
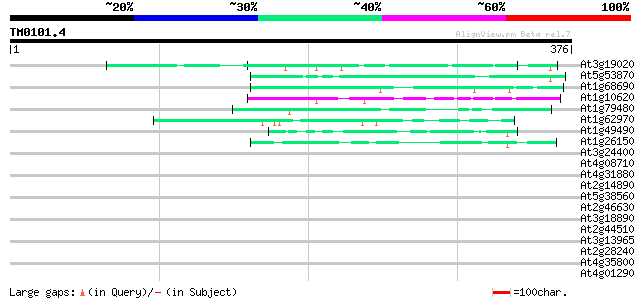
(376 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g19020 hypothetical protein 56 4e-08
At5g53870 predicted GPI-anchored protein 50 3e-06
At1g68690 protein kinase, putative 44 1e-04
At1g10620 putative serine/threonine protein kinase emb|CAA18823.1 43 2e-04
At1g79480 hypothetical protein 42 7e-04
At1g62970 unknown protein 42 7e-04
At1g49490 hypothetical protein 42 7e-04
At1g26150 Pto kinase interactor, putative 42 7e-04
At3g24400 protein kinase, putative 41 0.001
At4g08710 putative protein 41 0.001
At4g31880 unknown protein 39 0.006
At2g14890 arabinogalactan-protein AGP9 39 0.006
At5g38560 putative protein 38 0.008
At2g46630 putative extensin 38 0.010
At3g18890 unknown protein 37 0.023
At2g44510 unknown protein 37 0.023
At3g13965 unknown protein 36 0.030
At2g28240 putative proline/hydroxyproline-rich glycoprotein 36 0.030
At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain 36 0.039
At4g01290 hypothetical protein 36 0.039
>At3g19020 hypothetical protein
Length = 951
Score = 55.8 bits (133), Expect = 4e-08
Identities = 68/282 (24%), Positives = 106/282 (37%), Gaps = 34/282 (12%)
Query: 66 DPPDFESPRKRKQVEASEEKDPSDGDDDDDDEPPPQKKGKKVRIATQVTHLQPTQELART 125
+PP+ E P K K E+ + + PS + EP K+ K + P E +
Sbjct: 420 NPPNLEEPSKPKPEESPKPQQPSPKPETPSHEPSNPKEPKPESPKQE----SPKTEQPKP 475
Query: 126 APSTRITRSVVRASSKFVVLLDDDLNLLDAIYDETNPEPTPVDQPTSHPSPPRASFFQ-- 183
P + S + + K ++ P+P Q +S PP+
Sbjct: 476 KPESPKQESPKQEAPK---------------PEQPKPKPESPKQESSKQEPPKPEESPKP 520
Query: 184 --PSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVS---ETQAQEIPNPEKSNSEP 238
P EE P + + P SP P +T +P S + QE P PE+S P
Sbjct: 521 EPPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEES---P 577
Query: 239 QPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEASRQFFQLARERVSEILEHFLT 298
+P E+P + E +P L + + + +D S ++ Q E +
Sbjct: 578 KPQPPKQEQPPKTE--APKMGSPPLESPVPNDPYDASPIKKRRPQPPSPSTEETKT--TS 633
Query: 299 VLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPPRSPLP 340
SP +S P P P+ P + + P M PPPP SP P
Sbjct: 634 PQSPPVHSPPPPPPVHSPPPPVFSPP-PPMHSPPPPVYSPPP 674
Score = 44.7 bits (104), Expect = 8e-05
Identities = 54/214 (25%), Positives = 78/214 (36%), Gaps = 41/214 (19%)
Query: 160 TNPEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSAST----SPIILPYNPQT 215
+NP P P PT P+ P++EE K ++ KP + +P P NP+
Sbjct: 400 SNPSPKPT--PTPKAPEPKKEINPPNLEEPSKPKPEESPKPQQPSPKPETPSHEPSNPKE 457
Query: 216 SEPIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKS 275
+P E+ QE P E+ +P+P E P + P K
Sbjct: 458 PKP---ESPKQESPKTEQP--KPKPESPKQESPKQEAPKPEQPKPK--------PESPKQ 504
Query: 276 EASRQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPP 335
E+S+Q E SP + P+PE P Q P P PP
Sbjct: 505 ESSKQEPPKPEE-------------SP-KPEPPKPEESPKPQPPKQETPKPEESPKPQPP 550
Query: 336 RSPLPENNASVSNHSSDKSQNPETET--PHQNPK 367
+ P+ S K Q P+ ET P ++PK
Sbjct: 551 KQETPK------PEESPKPQPPKQETPKPEESPK 578
Score = 38.5 bits (88), Expect = 0.006
Identities = 49/206 (23%), Positives = 67/206 (31%), Gaps = 45/206 (21%)
Query: 162 PEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVS 221
P P P H PP P + P + + P + P ++P PI S
Sbjct: 688 PPPVHSPPPPVHSPPPPVHSPPPPVHSPP--PPVHSPPPPVQSPPPPPVFSPPPPAPIYS 745
Query: 222 ETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEASRQF 281
P +S P P HS P V SP P V + S
Sbjct: 746 PPPPPVHSPPPPVHSPPPPPVHSPPPP----VHSPPPPVHSPPPPVHSPP---------- 791
Query: 282 FQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPPRSPLPE 341
P +S P P P+ P P+ S PPP P +PLP
Sbjct: 792 --------------------PPVHSPPPPSPIYSPP-----PPVFS---PPPKPVTPLPP 823
Query: 342 NNASVSNHSS-DKSQNPETETPHQNP 366
+ ++N + S++ E TP Q P
Sbjct: 824 ATSPMANAPTPSSSESGEISTPVQAP 849
Score = 31.2 bits (69), Expect = 0.97
Identities = 21/64 (32%), Positives = 28/64 (42%), Gaps = 13/64 (20%)
Query: 306 SGPRPEPLVDPDFLIQAVPLASMP-HPPPPPRSPLPENNASVSNHSSDKSQNPETETPHQ 364
S P+PE P Q P P H P P+ P PE S K ++P+TE P
Sbjct: 428 SKPKPEESPKPQ---QPSPKPETPSHEPSNPKEPKPE---------SPKQESPKTEQPKP 475
Query: 365 NPKT 368
P++
Sbjct: 476 KPES 479
Score = 29.6 bits (65), Expect = 2.8
Identities = 18/62 (29%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query: 308 PRPEPLVDPDFLIQAVPLASMPHPPPPPRSPLPENNASVSNHSSDKSQNPETETPHQ-NP 366
P P+ ++P L + P P P P+ P P+ +H + P+ E+P Q +P
Sbjct: 413 PEPKKEINPPNLEE--PSKPKPEESPKPQQPSPK--PETPSHEPSNPKEPKPESPKQESP 468
Query: 367 KT 368
KT
Sbjct: 469 KT 470
Score = 29.3 bits (64), Expect = 3.7
Identities = 14/48 (29%), Positives = 21/48 (43%), Gaps = 2/48 (4%)
Query: 329 PHPPPPPRSPLPENNASVSNHSSDKSQNPETETPHQ--NPKTNTSAHQ 374
P P P P++P P+ + N PE Q +PK T +H+
Sbjct: 404 PKPTPTPKAPEPKKEINPPNLEEPSKPKPEESPKPQQPSPKPETPSHE 451
>At5g53870 predicted GPI-anchored protein
Length = 370
Score = 49.7 bits (117), Expect = 3e-06
Identities = 57/221 (25%), Positives = 82/221 (36%), Gaps = 25/221 (11%)
Query: 162 PEPTPVDQPTSH-PSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIV 220
P +P P SH P P A PS + P + A +P S+SPI + P S
Sbjct: 143 PSVSPTQPPKSHSPVSPVAPASAPSKSQPPRSSVSPA-QPPKSSSPI--SHTPALSP--- 196
Query: 221 SETQAQEIPNPEKSNSEPQPSDHS-NEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEASR 279
S + P S P P HS + P+ SPA T S S S + +
Sbjct: 197 SHATSHSPATPSPSPKSPSPVSHSPSHSPAHTPSHSPAHTPSHSPAHAPSHSPAHAPSHS 256
Query: 280 QFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPPRSPL 339
+ T SP+ S P P A P P P P SP
Sbjct: 257 PAHAPSHSPAHSPSHSPATPKSPSPSSSPAQSP---------ATPSPMTPQSPSPVSSPS 307
Query: 340 PENNASVSNHSSDKSQNPETET--------PHQNPKTNTSA 372
P+ +A+ S+ S+ + +P T P +P+TN+++
Sbjct: 308 PDQSAAPSDQSTPLAPSPSETTPTADNITAPAPSPRTNSAS 348
Score = 34.7 bits (78), Expect = 0.088
Identities = 44/182 (24%), Positives = 62/182 (33%), Gaps = 7/182 (3%)
Query: 196 QAGKPSASTSPIILPYNPQTSEPIVSETQAQEIPN----PEKSNSEPQPSDHSNEEPSER 251
Q P+ S P + P P S VS P+ P S S QP S+ S
Sbjct: 133 QPSAPAHSPVPSVSPTQPPKSHSPVSPVAPASAPSKSQPPRSSVSPAQP-PKSSSPISHT 191
Query: 252 EVLSPAPTVSFLTNVLFSSSFDKSEASRQFFQLARERVSEILEHFLTVLSPNRYS-GPRP 310
LSP+ S S S S S H + + S P
Sbjct: 192 PALSPSHATSHSPATPSPSPKSPSPVSHSPSHSPAHTPSHSPAHTPSHSPAHAPSHSPAH 251
Query: 311 EPLVDPDFLIQAVPLASMPHPPPPPRSPLPENNASVSNHSSDKSQNPETETPHQNPKTNT 370
P P P S H P P+SP P ++ + S ++ P++ +P +P +
Sbjct: 252 APSHSPAHAPSHSPAHSPSHSPATPKSPSPSSSPAQS-PATPSPMTPQSPSPVSSPSPDQ 310
Query: 371 SA 372
SA
Sbjct: 311 SA 312
Score = 33.1 bits (74), Expect = 0.26
Identities = 32/128 (25%), Positives = 51/128 (39%), Gaps = 19/128 (14%)
Query: 161 NPEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSP-----IILPYNPQT 215
+P P P PS A S P PS S+SP P PQ+
Sbjct: 240 SPAHAPSHSPAHAPSHSPAHAPSHSPAHSPSHSPATPKSPSPSSSPAQSPATPSPMTPQS 299
Query: 216 SEPIVSETQAQE-IPNPEKSNSEPQPSDHSNEEPSEREVLSPAPT----------VSFLT 264
P+ S + Q P+ + + P PS+ + P+ + +PAP+ V+ +
Sbjct: 300 PSPVSSPSPDQSAAPSDQSTPLAPSPSETT---PTADNITAPAPSPRTNSASGLAVTSVM 356
Query: 265 NVLFSSSF 272
+ LFS++F
Sbjct: 357 STLFSATF 364
>At1g68690 protein kinase, putative
Length = 708
Score = 43.9 bits (102), Expect = 1e-04
Identities = 61/221 (27%), Positives = 75/221 (33%), Gaps = 26/221 (11%)
Query: 162 PEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVS 221
P P P +S P P P P ++ + PSAS P ++P P + P S
Sbjct: 69 PPPLPKPPESSSPPPQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPPPPAS 128
Query: 222 ETQAQEIPNPEKSNSEPQPSDHSNEE----PSEREVLSPAPTVSFLTNVLFSSSFDKSEA 277
+ P+P P PS + PS+R SP P S
Sbjct: 129 VPPPRPSPSPPILVRSPPPSVRPIQSPPPPPSDRPTQSPPP------------PSPPSPP 176
Query: 278 SRQFFQLARERVSEILEHFLTVLSPNRYSGPRP--EPLVDPDFLIQAVPLASMPHPPP-- 333
S + Q SE SP RP P P+ P S PPP
Sbjct: 177 SERPTQSPPSPPSERPTQSPPPPSPPSPPSDRPSQSPPPPPEDTKPQPPRRSPNSPPPTF 236
Query: 334 --PPRSPLPENNASVSNHSSDKSQNPETETPHQNP-KTNTS 371
PPRSP PE SN+ S NP P P TN S
Sbjct: 237 SSPPRSP-PEILVPGSNNPS--QNNPTLRPPLDAPNSTNNS 274
Score = 40.0 bits (92), Expect = 0.002
Identities = 29/101 (28%), Positives = 37/101 (35%), Gaps = 17/101 (16%)
Query: 161 NPEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIV 220
+P P P D+PT P PP S P + Q+ S P P P P
Sbjct: 154 SPPPPPSDRPTQSPPPP-------SPPSPPSERPTQSPPSPPSERPTQSPPPPSPPSP-- 204
Query: 221 SETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVS 261
P+ S S P P + + +P R SP PT S
Sbjct: 205 --------PSDRPSQSPPPPPEDTKPQPPRRSPNSPPPTFS 237
Score = 35.4 bits (80), Expect = 0.051
Identities = 37/109 (33%), Positives = 44/109 (39%), Gaps = 14/109 (12%)
Query: 160 TNPEPTPV-DQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTS-E 217
T+P P PV P SPP A P + P P S SP IL +P S
Sbjct: 94 TSPPPQPVIPSPPPSASPPPA--LVPPLPSSPP-PPASVPPPRPSPSPPILVRSPPPSVR 150
Query: 218 PIVS--------ETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAP 258
PI S TQ+ P+P SE +P+ PSER SP P
Sbjct: 151 PIQSPPPPPSDRPTQSPPPPSPPSPPSE-RPTQSPPSPPSERPTQSPPP 198
Score = 28.9 bits (63), Expect = 4.8
Identities = 16/50 (32%), Positives = 20/50 (40%), Gaps = 4/50 (8%)
Query: 302 PNRYSGPRPEPLVDPDFLIQAVPLASMPHPP----PPPRSPLPENNASVS 347
PNR P P P + P +P PP PPP+ +P S S
Sbjct: 46 PNRAPPPPPPVTTSPPPVANGAPPPPLPKPPESSSPPPQPVIPSPPPSTS 95
>At1g10620 putative serine/threonine protein kinase emb|CAA18823.1
Length = 718
Score = 43.1 bits (100), Expect = 2e-04
Identities = 57/216 (26%), Positives = 88/216 (40%), Gaps = 28/216 (12%)
Query: 160 TNPEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSAST--SPIILPY-NPQTS 216
T +P P P + P P S PSI P Q S T SP+++P+ PQ
Sbjct: 64 TAAQPPPNQPPNTTPPPTPPSSPPPSITPPPSPPQPQPPPQSTPTGDSPVVIPFPKPQLP 123
Query: 217 EPIVSETQAQEIPNPEKSNS--EPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDK 274
P + P P N +P+P+D++ EP + P+P + FS +
Sbjct: 124 PPSL-------FPPPSLVNQLPDPRPNDNNILEPINNPISLPSP-----PSTPFSPPSQE 171
Query: 275 SEASRQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDP-DFLIQAVPLASMPHPPP 333
+ S+ +S +L L L+PN P +PL P VP +S P P
Sbjct: 172 NSGSQ-----GSPPLSSLLPPMLP-LNPNSPGNPL-QPLDSPLGGESNRVP-SSSSSPSP 223
Query: 334 PPRSPLPENNASVSNHSSDKSQNPETETPHQNPKTN 369
P S NN S ++ + + N + T Q+ ++N
Sbjct: 224 PSLS--GSNNHSGGSNRHNANSNGDGGTSQQSNESN 257
>At1g79480 hypothetical protein
Length = 356
Score = 41.6 bits (96), Expect = 7e-04
Identities = 52/217 (23%), Positives = 81/217 (36%), Gaps = 36/217 (16%)
Query: 150 LNLLDAIYDETNPEPTPVDQPTSHPSPPRASFFQPSI--EEEPLCKMLQAGKPSASTSPI 207
LNL + ++ P T+ PP S S+ P C PS+S +
Sbjct: 18 LNLAQPLDYSSSSNTQPYGVSTTLTLPPYVSLPPLSVPGNAPPFCINPPNTPPSSSYPGL 77
Query: 208 ILPYNPQT-SEPIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNV 266
P P T P S + PNP +S+S P P D S+ S +P P V+
Sbjct: 78 SPPPGPITLPNPPDSSSNPNSNPNPPESSSNPNPPDSSSNPNS-----NPNPPVTVPNPP 132
Query: 267 LFSSSFDKSEASRQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLA 326
SS+ + ++S +PN P PE +P+ P
Sbjct: 133 ESSSNPNPPDSSS---------------------NPNSNPNP-PESSSNPN------PPV 164
Query: 327 SMPHPPPPPRSPLPENNASVSNHSSDKSQNPETETPH 363
++P+PP +P P ++S N PE+ +P+
Sbjct: 165 TVPNPPESSSNPNPPESSSNPNPPITIPYPPESSSPN 201
Score = 41.2 bits (95), Expect = 0.001
Identities = 47/183 (25%), Positives = 70/183 (37%), Gaps = 31/183 (16%)
Query: 162 PEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVS 221
P +P P + P+PP +S P+ P P +S++P P P T
Sbjct: 75 PGLSPPPGPITLPNPPDSSS-NPNSNPNPPESSSNPNPPDSSSNPNSNPNPPVT------ 127
Query: 222 ETQAQEIPNPEKSNSEPQPSDHS---NEEPSEREVLS-PAPTVSFLTNVLFSSSFDKSEA 277
+PNP +S+S P P D S N P+ E S P P V+ SS+ + E+
Sbjct: 128 ------VPNPPESSSNPNPPDSSSNPNSNPNPPESSSNPNPPVTVPNPPESSSNPNPPES 181
Query: 278 SRQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLV--DPDFLIQAVPLASMPHPPPPP 335
S +T+ P S P P +V P+ P+ P+ P P
Sbjct: 182 SSN------------PNPPITIPYPPESSSPNPPEIVPSPPESGYTPGPVLGPPYSEPGP 229
Query: 336 RSP 338
+P
Sbjct: 230 STP 232
>At1g62970 unknown protein
Length = 825
Score = 41.6 bits (96), Expect = 7e-04
Identities = 61/264 (23%), Positives = 95/264 (35%), Gaps = 53/264 (20%)
Query: 97 EPPPQKKGKKVRIATQVTHLQPTQELARTAPSTRITRSVVRASSKFVVLLDDDLNLLDAI 156
+PP K + ++Q + P + ++ + +++S R+ V + N
Sbjct: 397 QPPSNSKPFPMSQSSQNSKPFPVSQSSQKSKPLLVSQSSQRSKPLPVSQSLQNSNPFPVS 456
Query: 157 YDETNPEPTPVD--QPTSHPSP---PRA-----SFFQPSIEEEPLCKMLQAGKPSASTSP 206
+N +P PV QP S+P P PR S QPS P A +P A++
Sbjct: 457 QPSSNSKPFPVSQPQPASNPFPVSQPRPNSQPFSMSQPSSTARP----FPASQPPAASKS 512
Query: 207 IILPYNPQTSEPIVSETQAQEIPNPEKSN---------SEPQPSDHS---NEEPSEREVL 254
+ P TS+P VS+ P P S+P P+ S ++ P+ L
Sbjct: 513 FPISQPPTTSKPFVSQPPNTSKPMPVSQPPTTSKPLPVSQPPPTFQSTCPSQPPAASSSL 572
Query: 255 SPAPTVSFLTNVLFSSSFDKSEASRQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLV 314
SP P V T SF S + T+ SP P P P+
Sbjct: 573 SPLPPVFNST-----QSFQSPPVS---------TTPSAVPEASTIPSP-----PAPAPVA 613
Query: 315 DPDFLIQAVPLASMPHPPPPPRSP 338
P + PPP ++P
Sbjct: 614 QPTHVFN--------QTPPPEQTP 629
Score = 39.7 bits (91), Expect = 0.003
Identities = 57/234 (24%), Positives = 93/234 (39%), Gaps = 32/234 (13%)
Query: 163 EPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGK----PSASTSPIILPYNPQTSEP 218
+PTPV +P H + +P+ + + + +A + PS S S
Sbjct: 307 KPTPVSEPVRHSELVPWQYSEPARQYQLSSRSSEAAQLSLLPSVSDSSHASQPTRSNQSH 366
Query: 219 IVSETQAQEIPNPEKSNSEPQPSDHS---NEEPSEREVLSPAPTVSFLTNVLFSSSFDKS 275
VS+ Q P+P S+P P+ + ++ PS + P P N S F S
Sbjct: 367 AVSKPQPVSKPHPPFPMSQPPPTSNPFPLSQPPSNSK---PFPMSQSSQN---SKPFPVS 420
Query: 276 EASRQFFQLARER---------VSEILEHF--LTVLSPNRYSGP----RPEPLVDPDFLI 320
++S++ L + VS+ L++ V P+ S P +P+P +P +
Sbjct: 421 QSSQKSKPLLVSQSSQRSKPLPVSQSLQNSNPFPVSQPSSNSKPFPVSQPQPASNPFPVS 480
Query: 321 QAVPLA---SMPHPPPPPRSPLPENNASVSNHSSDKSQNPETETPHQNPKTNTS 371
Q P + SM P R P P + ++ S SQ P T P + NTS
Sbjct: 481 QPRPNSQPFSMSQPSSTAR-PFPASQPPAASKSFPISQPPTTSKPFVSQPPNTS 533
Score = 38.9 bits (89), Expect = 0.005
Identities = 50/218 (22%), Positives = 73/218 (32%), Gaps = 40/218 (18%)
Query: 163 EPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPI-VS 221
+P P P PP S P + K + S + P+++ + Q S+P+ VS
Sbjct: 385 QPPPTSNPFPLSQPPSNSKPFPMSQSSQNSKPFPVSQSSQKSKPLLVSQSSQRSKPLPVS 444
Query: 222 ETQAQEIPNP---EKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEAS 278
++ P P SNS+P P + V P P S F S+ S
Sbjct: 445 QSLQNSNPFPVSQPSSNSKPFPVSQPQPASNPFPVSQPRPN---------SQPFSMSQPS 495
Query: 279 RQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPPRS- 337
+ P S P P F I P S P PP +
Sbjct: 496 -------------------STARPFPASQP---PAASKSFPISQPPTTSKPFVSQPPNTS 533
Query: 338 -PLPENNASVSNHSSDKSQNPET---ETPHQNPKTNTS 371
P+P + ++ SQ P T P Q P ++S
Sbjct: 534 KPMPVSQPPTTSKPLPVSQPPPTFQSTCPSQPPAASSS 571
>At1g49490 hypothetical protein
Length = 847
Score = 41.6 bits (96), Expect = 7e-04
Identities = 49/177 (27%), Positives = 72/177 (39%), Gaps = 41/177 (23%)
Query: 174 PSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVSETQAQEIPNPEK 233
P+PPR S +PS + EP+ KPS S+ P P+T E Q P P K
Sbjct: 396 PNPPRTSEPKPS-KPEPVMP-----KPSDSSK----PETPKTPE------QPSPKPQPPK 439
Query: 234 SNS-EPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEASRQFFQLARERVSEI 292
S +P+ ++ +E P ++E P P+ K E + + + +I
Sbjct: 440 HESPKPEEPENKHELPKQKESPKPQPSK--------PEDSPKPEQPKP-EESPKPEQPQI 490
Query: 293 LEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPP---------PPPRSPLP 340
E V PN GP P+ DP A P+ + PP PPP+ P+P
Sbjct: 491 PEPTKPVSPPNEAQGPTPD---DP---YDASPVKNRRSPPPPKVEDTRVPPPQPPMP 541
Score = 40.8 bits (94), Expect = 0.001
Identities = 64/332 (19%), Positives = 106/332 (31%), Gaps = 42/332 (12%)
Query: 67 PPDFESPR-----------KRKQVEASEEKDPSDGDDDDDDEPPPQKKGKKVRIATQVTH 115
PP ESP+ K+K+ + P D + +P K ++ +I
Sbjct: 437 PPKHESPKPEEPENKHELPKQKESPKPQPSKPEDSPKPEQPKPEESPKPEQPQIPEPTKP 496
Query: 116 LQPTQELARTAPSTRITRSVVRASSKFVVLLDDDLNL------------LDAIYDETNPE 163
+ P E P S V+ +D + IY P
Sbjct: 497 VSPPNEAQGPTPDDPYDASPVKNRRSPPPPKVEDTRVPPPQPPMPSPSPPSPIYSPPPPV 556
Query: 164 PTPVDQPTSHPSPPRASFFQPSIEEEPLCK---MLQAGKPSASTSPIILPYNPQTSEPIV 220
+P S P PP P + P + + P SP ++P P+
Sbjct: 557 HSPPPPVYSSPPPPHVYSPPPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFSPPPPSPVY 616
Query: 221 SETQAQEIP-----NPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKS 275
S P +P P P+ ++N+ P + APT S T + + S +
Sbjct: 617 SPPPPSHSPPPPVYSPPPPTFSPPPTHNTNQPPMGAPTPTQAPTPSSETTQVPTPSSESD 676
Query: 276 EASRQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPP 335
++ ++ ++ V S S P P QA L+ + P P
Sbjct: 677 QS----------QILSPVQAPTPVQSSTPSSEPTQVPTPSSSESYQAPNLSPVQAPTPVQ 726
Query: 336 RSPLPENNASVSNHSSDKSQNP-ETETPHQNP 366
+ V SS+ +Q+P + TP P
Sbjct: 727 APTTSSETSQVPTPSSESNQSPSQAPTPILEP 758
Score = 39.7 bits (91), Expect = 0.003
Identities = 55/231 (23%), Positives = 80/231 (33%), Gaps = 23/231 (9%)
Query: 162 PEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVS 221
P P+P S P PP S P P + PS S P + P T P +
Sbjct: 583 PPPSPPPPVHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPPPTFSPPPT 642
Query: 222 ETQAQEIPNPEKSNSEPQPSDHSNEEPS------EREVLSP--APT-------VSFLTNV 266
Q P PS + + P+ + ++LSP APT S T V
Sbjct: 643 HNTNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQILSPVQAPTPVQSSTPSSEPTQV 702
Query: 267 LFSSSFDKSEASRQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLA 326
SS + +A A V T P S P P +++ P+
Sbjct: 703 PTPSSSESYQAPNLSPVQAPTPVQAPTTSSETSQVPTPSSESNQSPSQAPTPILE--PVH 760
Query: 327 SMPHPPPPPRSPLPENNASVSNHSSDKSQNPE------TETPHQNPKTNTS 371
+ P +SP P + S S++ + PE + P +P T+TS
Sbjct: 761 APTPNSKPVQSPTPSSEPVSSPEQSEEVEAPEPTPVNPSSVPSSSPSTDTS 811
Score = 38.5 bits (88), Expect = 0.006
Identities = 49/184 (26%), Positives = 72/184 (38%), Gaps = 18/184 (9%)
Query: 158 DETNPE-PTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTS 216
D + PE P +QP+ P PP+ +P EEP K P SP PQ S
Sbjct: 418 DSSKPETPKTPEQPSPKPQPPKHESPKP---EEPENKH---ELPKQKESP-----KPQPS 466
Query: 217 EPIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSE 276
+P ++ E P PE+S P+P EP+ + V P + + +S K+
Sbjct: 467 KP--EDSPKPEQPKPEES---PKPEQPQIPEPT-KPVSPPNEAQGPTPDDPYDASPVKNR 520
Query: 277 ASRQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPPR 336
S ++ RV + P+ P P P + + P + PPPP
Sbjct: 521 RSPPPPKVEDTRVPPPQPPMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVA 580
Query: 337 SPLP 340
SP P
Sbjct: 581 SPPP 584
Score = 35.0 bits (79), Expect = 0.067
Identities = 24/105 (22%), Positives = 43/105 (40%), Gaps = 4/105 (3%)
Query: 159 ETNPEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSE- 217
ET+ PTP + PS +P P K +Q+ P+ S+ P+ P + E
Sbjct: 733 ETSQVPTPSSESNQSPSQAPTPILEPVHAPTPNSKPVQS--PTPSSEPVSSPEQSEEVEA 790
Query: 218 PIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSF 262
P + +P+ S P +N++ + + + P P + F
Sbjct: 791 PEPTPVNPSSVPSSSPSTDTSIPPPENNDDDDDGDFVLP-PHIGF 834
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 41.6 bits (96), Expect = 7e-04
Identities = 49/209 (23%), Positives = 73/209 (34%), Gaps = 56/209 (26%)
Query: 162 PEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVS 221
P TP+ P PSPP PS+ P + + P S P + P + P+ S
Sbjct: 63 PPETPLSSPPPEPSPP-----SPSLTGPPPTTIPVSPPPEPSPPPPLPTEAPPPANPVSS 117
Query: 222 ETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEASRQF 281
P PE S P P+ E P + SP+P +
Sbjct: 118 -------PPPESSPPPPPPT----EAPPTTPITSPSPPTN-------------------- 146
Query: 282 FQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPP----PPPRS 337
E ++ +P+ S P P P + P P +P PP PPP
Sbjct: 147 -------PPPPPESPPSLPAPDPPSNPLPPPKLVPP---SHSPPRHLPSPPASEIPPPPR 196
Query: 338 PLPENNASVSNHSSDKSQNPETETPHQNP 366
LP S +S++ P +++ H +P
Sbjct: 197 HLP------SPPASERPSTPPSDSEHPSP 219
Score = 37.4 bits (85), Expect = 0.014
Identities = 54/204 (26%), Positives = 68/204 (32%), Gaps = 37/204 (18%)
Query: 67 PPDFESPRKRKQVEASEEKDPSDGDDDDDDEPPPQKKGKKVRIATQVTHLQPTQELARTA 126
PP SP EA +P + PPP T PT + +
Sbjct: 94 PPPEPSPPPPLPTEAPPPANPVSSPPPESSPPPPPP-----------TEAPPTTPITSPS 142
Query: 127 PSTRITRSVVRASSKFVVLLDDDLNLLDAIYDETNPEPTPVDQPTSHPSPPRASFFQPSI 186
P T S L A +NP P P P SH SPPR P+
Sbjct: 143 PPTNPPPPPESPPS------------LPAPDPPSNPLPPPKLVPPSH-SPPRHLPSPPAS 189
Query: 187 EEEPLCKMLQAGKPSASTSPIILPYNPQ-TSEPIVSETQAQEIPNPEKSN----SEPQPS 241
E P + L + P AS P P + + S P + +E P P S S P PS
Sbjct: 190 EIPPPPRHLPS--PPASERPSTPPSDSEHPSPPPPGHPKRREQPPPPGSKRPTPSPPSPS 247
Query: 242 DH------SNEEPSEREVLSPAPT 259
D S P E + P P+
Sbjct: 248 DSKRPVHPSPPSPPEETLPPPKPS 271
Score = 32.0 bits (71), Expect = 0.57
Identities = 23/74 (31%), Positives = 27/74 (36%), Gaps = 8/74 (10%)
Query: 306 SGPRPEPLVD--------PDFLIQAVPLASMPHPPPPPRSPLPENNASVSNHSSDKSQNP 357
S P PEP P + + P P PP P +P P N S S P
Sbjct: 69 SSPPPEPSPPSPSLTGPPPTTIPVSPPPEPSPPPPLPTEAPPPANPVSSPPPESSPPPPP 128
Query: 358 ETETPHQNPKTNTS 371
TE P P T+ S
Sbjct: 129 PTEAPPTTPITSPS 142
Score = 31.2 bits (69), Expect = 0.97
Identities = 25/87 (28%), Positives = 35/87 (39%), Gaps = 11/87 (12%)
Query: 162 PEPTPVDQPT--SHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTS-PIILPYNPQTSEP 218
P P+ +P S PSPP + P +PL PS S+S P +LP + S P
Sbjct: 244 PSPSDSKRPVHPSPPSPPEETLPPPKPSPDPL--------PSNSSSPPTLLPPSSVVSPP 295
Query: 219 IVSETQAQEIPNPEKSNSEPQPSDHSN 245
NP +N P + S+
Sbjct: 296 SPPRKSVSGPDNPSPNNPTPVTDNSSS 322
>At3g24400 protein kinase, putative
Length = 694
Score = 41.2 bits (95), Expect = 0.001
Identities = 54/212 (25%), Positives = 72/212 (33%), Gaps = 9/212 (4%)
Query: 164 PTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVSET 223
P+P QP P PP+ P L L P + P + P P T+ P + +
Sbjct: 11 PSPPPQPLPIPPPPQPLPVTPPPPPTALPPALPPPPPPTALPPALPPPPPPTTVPPIPPS 70
Query: 224 QAQEIPNPEKSNSEPQPS------DHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEA 277
P S P P+ S PS SP P ++ + L S S
Sbjct: 71 TPSPPPPLTPSPLPPSPTTPSPPLTPSPTTPSPPLTPSPPPAIT-PSPPLTPSPLPPSPT 129
Query: 278 SRQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPP--PP 335
+ + S L SP R S P P P ++ P S P PPP
Sbjct: 130 TPSPPPPSPSIPSPPLTPSPPPSSPLRPSSPPPPSPATPSTPPRSPPPPSTPTPPPRVGS 189
Query: 336 RSPLPENNASVSNHSSDKSQNPETETPHQNPK 367
SP P + S S S P + P Q+ K
Sbjct: 190 LSPPPPASPSGGRSPSTPSTTPGSSPPAQSSK 221
Score = 37.4 bits (85), Expect = 0.014
Identities = 49/206 (23%), Positives = 70/206 (33%), Gaps = 34/206 (16%)
Query: 162 PEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVS 221
P P P T P PP PSI PL PS S + P +P P
Sbjct: 121 PSPLPPSPTTPSPPPP-----SPSIPSPPL-------TPSPPPSSPLRPSSPPPPSPATP 168
Query: 222 ETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEASRQF 281
T + P P P+ S P+ SP+ S T S +++S++
Sbjct: 169 STPPRSPPPPSTPTPPPRVGSLSPPPPA-----SPSGGRSPSTPSTTPGSSPPAQSSKEL 223
Query: 282 FQLARE----------RVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHP 331
+ A V+ L FL R + P P+V P P + P
Sbjct: 224 SKGAMVGIAIGGGFVLLVALALIFFLCKKKRRRDNEAPPAPIVPP-------PKSPSSAP 276
Query: 332 PPPPRSPLPENNASVSNHSSDKSQNP 357
P PP ++ ++ SD+S P
Sbjct: 277 PRPPHFMSSGSSGDYDSNYSDQSVLP 302
>At4g08710 putative protein
Length = 715
Score = 40.8 bits (94), Expect = 0.001
Identities = 30/116 (25%), Positives = 55/116 (46%), Gaps = 8/116 (6%)
Query: 65 QDPPDF---ESPRKRKQVEASEEKDPSDGDDDDDDEPPPQKKGKKVRIATQVTHLQPTQE 121
QDP + ES + K+ E EE++ +G++++++E ++K ++ + + + T+
Sbjct: 77 QDPENVEEEESEEEEKEEEEKEEEEEEEGEEEEEEEEEEEEKEEEENVGGEESSDDSTRS 136
Query: 122 LARTAPSTRITRSVVRASSKFVVLLDDDLNLLDAIYDETNPEPTPVDQPTSHPSPP 177
L + P RI + KF L + + LL I +ET P P S PP
Sbjct: 137 LGKRPPPMRIWMRRHQLQWKFRRLCNMNECLLQEIDEETEAVP-----PLSMYFPP 187
>At4g31880 unknown protein
Length = 873
Score = 38.5 bits (88), Expect = 0.006
Identities = 55/251 (21%), Positives = 101/251 (39%), Gaps = 25/251 (9%)
Query: 8 ETKVEVKIADARPRRRIHLKDYPLWSKADDPEAIR----IYLDDLKQQDLEIVVDEFFRN 63
ET+VE + P R KD S + A + + D +K+QD DE
Sbjct: 278 ETEVEKAAEISTPERTDAPKDESGKSGVSNGVAQQNDSSVDTDSMKKQDDTGAKDE---- 333
Query: 64 LQDPPDFESPRKRKQVEASEEK-DPSDGDDDDDDEPPPQK-----KGKKVRIATQVTHLQ 117
P ++PR +EEK D ++ ++E K K ++ T+ L
Sbjct: 334 ---PQQLDNPRNTDLNNTTEEKPDVEHQIEEKENESSSVKQADLSKDSDIKEETEPAELL 390
Query: 118 PTQELARTAPSTRITRSVVRASSKFVVLLDDDLNLLDAIYDETNPEPTP-VDQPTSHPSP 176
++++ + P + SV A+S ++ + + +T+ + T V P+
Sbjct: 391 DSKDVLTSPP---VDSSVTAATSSE----NEKNKSVQILPSKTSGDETANVSSPSMAEEL 443
Query: 177 PRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVSETQAQEIPNPEKSNS 236
P S + + ++ + KPSAS + + P TSEP V++ +++ + K+
Sbjct: 444 PEQSVPKKTANQKKKESSTEEVKPSASIATEEVSEEPNTSEPQVTKKSGKKVASSSKTKP 503
Query: 237 EPQPSDHSNEE 247
PS S E
Sbjct: 504 TVPPSKKSTSE 514
Score = 34.7 bits (78), Expect = 0.088
Identities = 60/261 (22%), Positives = 93/261 (34%), Gaps = 53/261 (20%)
Query: 36 DDPEAIRIYLDDLKQQDLEIVVDEFFRNLQDPPDFESPRKRKQVEASEEKDPS---DGDD 92
D+P+ LD+ + DL +E PD E + K+ E+S K D D
Sbjct: 332 DEPQQ----LDNPRNTDLNNTTEE-------KPDVEHQIEEKENESSSVKQADLSKDSDI 380
Query: 93 DDDDEPPPQKKGKKVRIATQVTHLQPTQELARTAPSTRITRSVVRASSKFVVLLDDDLNL 152
++ EP K V + V + A T+ +SV SK D+ N+
Sbjct: 381 KEETEPAELLDSKDVLTSPPVD----SSVTAATSSENEKNKSVQILPSK--TSGDETANV 434
Query: 153 LDAIYDETNPEPTPVDQPTSHPSP---------PRASFFQPSIEEEPLCKMLQAGKPS-- 201
E PE + V + T++ P AS + EEP Q K S
Sbjct: 435 SSPSMAEELPEQS-VPKKTANQKKKESSTEEVKPSASIATEEVSEEPNTSEPQVTKKSGK 493
Query: 202 -----ASTSPIILPYNPQTSE---------PIVSETQAQEIPNPEKSNSEP-------QP 240
+ T P + P TSE +V AQE P++ +P +
Sbjct: 494 KVASSSKTKPTVPPSKKSTSETKVAKQSEKKVVGSDNAQESTKPKEEKKKPGRGKAIDEE 553
Query: 241 SDHSNEEPSEREVLSPAPTVS 261
S H++ +E+ +S S
Sbjct: 554 SLHTSSGDNEKPAVSSGKLAS 574
>At2g14890 arabinogalactan-protein AGP9
Length = 191
Score = 38.5 bits (88), Expect = 0.006
Identities = 30/103 (29%), Positives = 43/103 (41%), Gaps = 5/103 (4%)
Query: 160 TNPEPTPVDQPTSHP---SPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTS 216
T P P P S P SPP A+ + P+ A P +T P +P
Sbjct: 63 TAPPPANPPPPVSSPPPASPPPATPPPVASPPPPVASPPPATPPPVATPPPAPLASPPAQ 122
Query: 217 EPIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPT 259
P + T + P+P S+S P PS S+ + +SPAP+
Sbjct: 123 VPAPAPTTKPDSPSPSPSSSPPLPS--SDAPGPSTDSISPAPS 163
Score = 31.6 bits (70), Expect = 0.74
Identities = 46/203 (22%), Positives = 62/203 (29%), Gaps = 57/203 (28%)
Query: 166 PVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVSETQA 225
P PT+ P+PP + P+A+ P+ P TS P V T A
Sbjct: 23 PTSPPTATPAPPTPT----------------TPPPAATPPPVSAPPPVTTSPPPV--TTA 64
Query: 226 QEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEASRQFFQLA 285
NP S P P+ P V SP P V+
Sbjct: 65 PPPANPPPPVSSPPPASPPPATPP--PVASPPPPVASPPPA------------------- 103
Query: 286 RERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPPRS---PLPEN 342
+P + P P PL P + A + P P P S PLP +
Sbjct: 104 ---------------TPPPVATPPPAPLASPPAQVPAPAPTTKPDSPSPSPSSSPPLPSS 148
Query: 343 NASVSNHSSDKSQNPETETPHQN 365
+A + S T+ QN
Sbjct: 149 DAPGPSTDSISPAPSPTDVNDQN 171
>At5g38560 putative protein
Length = 681
Score = 38.1 bits (87), Expect = 0.008
Identities = 55/210 (26%), Positives = 74/210 (35%), Gaps = 47/210 (22%)
Query: 164 PTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVSET 223
P P+ P S S A P ++ +P PSA P PQ+ P+VS +
Sbjct: 6 PLPILSPPSSNSSTTAP---PPLQTQPTT-------PSAPPPVTPPPSPPQSPPPVVSSS 55
Query: 224 QAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTV--SFLTNVLFSSSFDKSEASRQF 281
P S P PS S+ PS + SP PTV S V+ +S + A+
Sbjct: 56 P------PPPVVSSPPPS--SSPPPSPPVITSPPPTVASSPPPPVVIASPPPSTPAT--- 104
Query: 282 FQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPPRSPLPE 341
T +P + P P P P + P + +PPP P SP P
Sbjct: 105 ----------------TPPAPPQTVSPPPPPDASP-----SPPAPTTTNPPPKP-SPSPP 142
Query: 342 NNASVSNHSSDKSQNPETETPHQNPKTNTS 371
+ P TP P T TS
Sbjct: 143 GETPSPPGETPSPPKPSPSTP--TPTTTTS 170
Score = 36.6 bits (83), Expect = 0.023
Identities = 52/221 (23%), Positives = 80/221 (35%), Gaps = 48/221 (21%)
Query: 162 PEPTPVDQP----TSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTS---PIIL----P 210
P P+P P +S P PP S PS P ++ + P+ ++S P+++ P
Sbjct: 40 PPPSPPQSPPPVVSSSPPPPVVSSPPPSSSPPPSPPVITSPPPTVASSPPPPVVIASPPP 99
Query: 211 YNPQTSEPIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSS 270
P T+ P +T + P P+ S S P P+ +N P SP+P + +
Sbjct: 100 STPATTPPAPPQTVSPP-PPPDASPSPPAPTT-TNPPPKP----SPSPPGETPSPPGETP 153
Query: 271 SFDKSEASRQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPH 330
S K S +P + P P P A P +S P
Sbjct: 154 SPPKPSPSTP--------------------TPTTTTSPPPPPAT------SASPPSSNPT 187
Query: 331 PP---PPPRSPLP--ENNASVSNHSSDKSQNPETETPHQNP 366
P PP +PLP ++ + S N P +P
Sbjct: 188 DPSTLAPPPTPLPVVPREKPIAKPTGPASNNGNNTLPSSSP 228
>At2g46630 putative extensin
Length = 394
Score = 37.7 bits (86), Expect = 0.010
Identities = 32/99 (32%), Positives = 43/99 (43%), Gaps = 10/99 (10%)
Query: 165 TPVDQPTSHPSPP--RASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVSE 222
TP Q SPP R+ + P P M P A+T P P + TS P S
Sbjct: 75 TPPRQKAPPTSPPQERSPYHSP-----PSRHMSPPTPPKAATPPPPPPRSSYTSPP--SP 127
Query: 223 TQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVS 261
+ QE P K NS P P+ HS+ + V + +P+ S
Sbjct: 128 KEVQEALPPRKPNSPPSPA-HSSRSTTSESVKTRSPSES 165
Score = 35.4 bits (80), Expect = 0.051
Identities = 28/103 (27%), Positives = 47/103 (45%), Gaps = 9/103 (8%)
Query: 164 PTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPI-----ILPYNPQTSEP 218
PTP T P PPR+S+ P +E + + L KP++ SP + +T P
Sbjct: 104 PTPPKAATPPPPPPRSSYTSPPSPKE-VQEALPPRKPNSPPSPAHSSRSTTSESVKTRSP 162
Query: 219 IVSETQAQEIPNPEKSNSEPQPSD--HSNEEPSEREVLSPAPT 259
SE ++ P+P + P+ HS E +++ +L+ T
Sbjct: 163 SESENH-RKAPSPRVLSPYSLPASLLHSERETTQKNILTAEKT 204
>At3g18890 unknown protein
Length = 641
Score = 36.6 bits (83), Expect = 0.023
Identities = 30/111 (27%), Positives = 42/111 (37%), Gaps = 8/111 (7%)
Query: 61 FRNLQDPPDFESPRKRKQVEASEEKDPSDGDDDDDDEPPPQKKGKKVRIATQVTHLQPTQ 120
+ NL+ PP SP S+ P D D D K + +AT VT
Sbjct: 451 YENLK-PPSSPSPTA-SSTRKSDSLSPGPTDSDTDKSSTVAKTVTETAVATSVTETSVAT 508
Query: 121 ELARTAPSTRITRSVVRASSKFVVLLDDDLNLLDAIYDETNPEPTPVDQPT 171
+ TA +T +T + A+SK L AIY + P +P T
Sbjct: 509 SVPETAVATSVTETAAPATSKMRPLSP------YAIYADLKPPTSPTPAST 553
>At2g44510 unknown protein
Length = 326
Score = 36.6 bits (83), Expect = 0.023
Identities = 23/73 (31%), Positives = 34/73 (46%), Gaps = 6/73 (8%)
Query: 210 PYNPQTSEPIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREV------LSPAPTVSFL 263
P + Q S+ +S + Q+IPN + E Q SD S+EE S+ +V P PT
Sbjct: 39 PEDCQCSDEDISFDEKQKIPNLPRKGKEEQVSDSSDEEDSQEDVQADFEFFDPKPTDFHG 98
Query: 264 TNVLFSSSFDKSE 276
+L + D E
Sbjct: 99 VKILLQNYLDDKE 111
>At3g13965 unknown protein
Length = 209
Score = 36.2 bits (82), Expect = 0.030
Identities = 37/143 (25%), Positives = 54/143 (36%), Gaps = 21/143 (14%)
Query: 219 IVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSE-- 276
++ ++Q Q +P P+ + P PS N LSP P F+ + F ++
Sbjct: 18 LLVQSQQQVLPQPQLQSPPPPPSPLQN--------LSPPPQQQFMQSPPFPPESGNTQPP 69
Query: 277 --ASRQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPP 334
QF S I V+SP P P P P P S P PPP
Sbjct: 70 PLTPPQFNTPPPPSKSTI-----NVMSPPPPPLPSPPPPPPPPTPSPPPPPISKPPPPPR 124
Query: 335 PRSPLPENNASVSNHSSDKSQNP 357
++ P +N N S ++Q P
Sbjct: 125 AQASPPHSN----NRESHRNQPP 143
>At2g28240 putative proline/hydroxyproline-rich glycoprotein
Length = 660
Score = 36.2 bits (82), Expect = 0.030
Identities = 46/238 (19%), Positives = 83/238 (34%), Gaps = 48/238 (20%)
Query: 163 EPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEP---- 218
E P+ QP S+ S PR Q + +PS + +L + P ++ P
Sbjct: 402 EARPLHQPHSNTSQPRPIPQQALAQSNTNITSTALPRPSITAEARLL-HQPHSNTPQPRP 460
Query: 219 --------IVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSS 270
++ + +P P + P S + P + + S P V T+++ S+
Sbjct: 461 IPQKALVQANTDINSTALPRPLVTAEAPPLHQSSCKAPQPKPI-SQQPAVQSKTDIINST 519
Query: 271 SFDKSEASRQFFQLARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPH 330
+ + + + L + R P+P+P+ P + S PH
Sbjct: 520 ALPRPSVTTEARPLHQPRSKT----------------PQPKPVSQPPAKQSNTEINSTPH 563
Query: 331 P------------PPPPRSPLPENNASVSNHSSDKSQNPETETPHQNPKTNTSAHQTL 376
P PP +P P +SNH+ P + P P + A +T+
Sbjct: 564 PRPSVTSKAISLQSPPCNTPQPRPPPLISNHT------PTSYQPASAPPVHGIARRTM 615
>At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain
Length = 1840
Score = 35.8 bits (81), Expect = 0.039
Identities = 25/91 (27%), Positives = 37/91 (40%), Gaps = 3/91 (3%)
Query: 157 YDETNPEPTPVDQ---PTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNP 213
Y T+P +P PTS P+++ + PSI P L P + TSP P +P
Sbjct: 1706 YSPTSPSYSPTSPSYGPTSPSYNPQSAKYSPSIAYSPSNARLSPASPYSPTSPNYSPTSP 1765
Query: 214 QTSEPIVSETQAQEIPNPEKSNSEPQPSDHS 244
S S + + +P S D+S
Sbjct: 1766 SYSPTSPSYSPSSPTYSPSSPYSSGASPDYS 1796
>At4g01290 hypothetical protein
Length = 736
Score = 35.8 bits (81), Expect = 0.039
Identities = 30/116 (25%), Positives = 51/116 (43%), Gaps = 14/116 (12%)
Query: 128 STRITRSVVRASSKFVVLLDDDL--NLLDAIYDETNPEPTPVDQPTSHPSPPRASFFQPS 185
ST T+SV VL +DL ++L + D +P P PVDQ TS PS + S
Sbjct: 226 STSTTKSVTAVPP---VLTCEDLEQSILSEVGDSYHPPPPPVDQDTSVPSVKMTKQRKTS 282
Query: 186 IEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVSETQAQEIPNPEKSNSEPQPS 241
++++ +L + S+ +P++ + + + P P + P PS
Sbjct: 283 VDDQASQHLLSLLQRSS---------DPKSQDTQLLSATERRPPPPSMKTTTPPPS 329
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.129 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,135,095
Number of Sequences: 26719
Number of extensions: 543908
Number of successful extensions: 6837
Number of sequences better than 10.0: 343
Number of HSP's better than 10.0 without gapping: 135
Number of HSP's successfully gapped in prelim test: 213
Number of HSP's that attempted gapping in prelim test: 4808
Number of HSP's gapped (non-prelim): 1329
length of query: 376
length of database: 11,318,596
effective HSP length: 101
effective length of query: 275
effective length of database: 8,619,977
effective search space: 2370493675
effective search space used: 2370493675
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0101.4


