

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0101.14
(1733 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
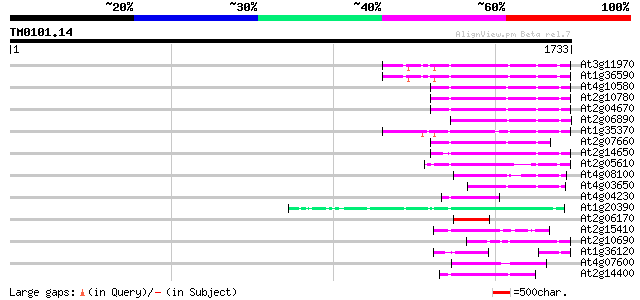
Score E
Sequences producing significant alignments: (bits) Value
At3g11970 hypothetical protein 216 1e-55
At1g36590 hypothetical protein 216 1e-55
At4g10580 putative reverse-transcriptase -like protein 207 6e-53
At2g10780 pseudogene 204 5e-52
At2g04670 putative retroelement pol polyprotein 202 1e-51
At2g06890 putative retroelement integrase 202 1e-51
At1g35370 hypothetical protein 187 4e-47
At2g07660 putative retroelement pol polyprotein 186 8e-47
At2g14650 putative retroelement pol polyprotein 179 1e-44
At2g05610 putative retroelement pol polyprotein 176 8e-44
At4g08100 putative polyprotein 159 1e-38
At4g03650 putative reverse transcriptase 139 1e-32
At4g04230 putative transposon protein 123 8e-28
At1g20390 hypothetical protein 117 6e-26
At2g06170 putative Ty3-gypsy-like retroelement pol polyprotein 113 1e-24
At2g15410 putative retroelement pol polyprotein 100 6e-21
At2g10690 putative retroelement pol polyprotein 100 6e-21
At1g36120 putative reverse transcriptase gb|AAD22339.1 100 6e-21
At4g07600 100 1e-20
At2g14400 putative retroelement pol polyprotein 92 3e-18
>At3g11970 hypothetical protein
Length = 1499
Score = 216 bits (549), Expect = 1e-55
Identities = 163/604 (26%), Positives = 287/604 (46%), Gaps = 49/604 (8%)
Query: 1151 KLSAAEGSKLIIKYKIPSAIIKNDSLEIETPFLLVRNLTHKVIIGTPFIKKLFPYNTDEK 1210
++S A+G KL ++ K+ K + ++ LL+ +++G +++ L + + K
Sbjct: 433 RVSVADGRKLRVEGKVTDFSWKLQTTTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFK 492
Query: 1211 GITVQHLGKPILFKFSKPPFS------------KTLNIISYKEKQINFLK---EEISHKS 1255
+ ++ FKF+ K + +E Q+ +E+S +
Sbjct: 493 KLEMR-------FKFNNQKVLLHGLTSGSVREVKAQKLQKLQEDQVQLAMLCVQEVSEST 545
Query: 1256 IEVQLQQPSVKTRIGNILENIQSSICSDLPNAFWERKSHMVELPYEKDFSDKQI------ 1309
++ + +G E++ + ++ P+ F E + LP ++ + +I
Sbjct: 546 EGELCTINALTSELGE--ESVVEEVLNEYPDIFIEPTA----LPPFREKHNHKIKLLEGS 599
Query: 1310 -PTKARPIQMNEELLQFCQKEINDLLQKKLIRRSKSPWSCATFYVNKQAEIERGTPRLVI 1368
P RP + + K + DLL ++ S SP++ V K+ GT RL +
Sbjct: 600 NPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSPYASPVVLVKKKD----GTWRLCV 655
Query: 1369 NYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPF 1428
+Y+ LN +PIP +DL+ L A IFSK D+++G+ Q+++ D KTAF
Sbjct: 656 DYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHS 715
Query: 1429 GQYEWNVMPFGLKNAPSEFQRIMNEIFNPY-SNFTIVYIDDVLIFSQSIDQHFKHLNTFI 1487
G +E+ VMPFGL NAP+ FQ +MN IF P+ F +V+ DD+L++S S+++H +HL
Sbjct: 716 GHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVF 775
Query: 1488 SVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRF 1547
V++ N L +K + K+ +LGH I I I+ ++P K QL+ F
Sbjct: 776 EVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLK-QLRGF 834
Query: 1548 LGCLNYVADFCPQLSTIIKLLHDRLKKDPPPWSDVHTNVVKQIKLRIKNLPCLYLPNPQA 1607
LG Y F I LH K D W+ V + +K + P L LP
Sbjct: 835 LGLAGYYRRFVRSFGVIAGPLHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLFDK 894
Query: 1608 FKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISKFQ 1667
+VETDA G G +L Q+ +A+ S+ Q + S +KE+LA++ ++ K++
Sbjct: 895 QFVVETDACGQGIGAVLMQE----GHPLAYISRQLKGKQLHLSIYEKELLAVIFAVRKWR 950
Query: 1668 YDLINQTFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPD 1727
+ L+ F+++ D +S K +L++ + + I +W L FD+EI+Y +G N + D
Sbjct: 951 HYLLQSHFIIKTDQRSLKYLLEQRL----NTPIQQQWLPKLLEFDYEIQYRQGKENVVAD 1006
Query: 1728 FLTR 1731
L+R
Sbjct: 1007 ALSR 1010
>At1g36590 hypothetical protein
Length = 1499
Score = 216 bits (549), Expect = 1e-55
Identities = 163/604 (26%), Positives = 287/604 (46%), Gaps = 49/604 (8%)
Query: 1151 KLSAAEGSKLIIKYKIPSAIIKNDSLEIETPFLLVRNLTHKVIIGTPFIKKLFPYNTDEK 1210
++S A+G KL ++ K+ K + ++ LL+ +++G +++ L + + K
Sbjct: 433 RVSVADGRKLRVEGKVTDFSWKLQTTTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFK 492
Query: 1211 GITVQHLGKPILFKFSKPPFS------------KTLNIISYKEKQINFLK---EEISHKS 1255
+ ++ FKF+ K + +E Q+ +E+S +
Sbjct: 493 KLEMR-------FKFNNQKVLLHGLTSGSVREVKAQKLQKLQEDQVQLAMLCVQEVSEST 545
Query: 1256 IEVQLQQPSVKTRIGNILENIQSSICSDLPNAFWERKSHMVELPYEKDFSDKQI------ 1309
++ + +G E++ + ++ P+ F E + LP ++ + +I
Sbjct: 546 EGELCTINALTSELGE--ESVVEEVLNEYPDIFIEPTA----LPPFREKHNHKIKLLEGS 599
Query: 1310 -PTKARPIQMNEELLQFCQKEINDLLQKKLIRRSKSPWSCATFYVNKQAEIERGTPRLVI 1368
P RP + + K + DLL ++ S SP++ V K+ GT RL +
Sbjct: 600 NPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSPYASPVVLVKKKD----GTWRLCV 655
Query: 1369 NYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPF 1428
+Y+ LN +PIP +DL+ L A IFSK D+++G+ Q+++ D KTAF
Sbjct: 656 DYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHS 715
Query: 1429 GQYEWNVMPFGLKNAPSEFQRIMNEIFNPY-SNFTIVYIDDVLIFSQSIDQHFKHLNTFI 1487
G +E+ VMPFGL NAP+ FQ +MN IF P+ F +V+ DD+L++S S+++H +HL
Sbjct: 716 GHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVF 775
Query: 1488 SVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRF 1547
V++ N L +K + K+ +LGH I I I+ ++P K QL+ F
Sbjct: 776 EVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLK-QLRGF 834
Query: 1548 LGCLNYVADFCPQLSTIIKLLHDRLKKDPPPWSDVHTNVVKQIKLRIKNLPCLYLPNPQA 1607
LG Y F I LH K D W+ V + +K + P L LP
Sbjct: 835 LGLAGYYRRFVRSFGVIAGPLHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLFDK 894
Query: 1608 FKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISKFQ 1667
+VETDA G G +L Q+ +A+ S+ Q + S +KE+LA++ ++ K++
Sbjct: 895 QFVVETDACGQGIGAVLMQE----GHPLAYISRQLKGKQLHLSIYEKELLAVIFAVRKWR 950
Query: 1668 YDLINQTFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPD 1727
+ L+ F+++ D +S K +L++ + + I +W L FD+EI+Y +G N + D
Sbjct: 951 HYLLQSHFIIKTDQRSLKYLLEQRL----NTPIQQQWLPKLLEFDYEIQYRQGKENVVAD 1006
Query: 1728 FLTR 1731
L+R
Sbjct: 1007 ALSR 1010
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 207 bits (526), Expect = 6e-53
Identities = 136/436 (31%), Positives = 219/436 (50%), Gaps = 15/436 (3%)
Query: 1299 PYEKDFSDKQIPTKARPIQMNEELLQFCQKEINDLLQKKLIRRSKSPWSCATFYVNKQAE 1358
P+ + P P +M + +K++ DLL K IR S SPW +V K+
Sbjct: 480 PFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSPWGAPVLFVKKKD- 538
Query: 1359 IERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKD 1418
G+ RL I+Y+ LN+ RYP+P +LL +L A FSK D+ SG+ QI + E D
Sbjct: 539 ---GSFRLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEAD 595
Query: 1419 RYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPY-SNFTIVYIDDVLIFSQSID 1477
KTAF +G +E+ VMPFGL NAP+ F R+MN +F + F I++IDD+L++S+S +
Sbjct: 596 VRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPE 655
Query: 1478 QHFKHLNTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQ 1537
+ HL + +++ L +K S +Q ++ FLGH + + IE +P +
Sbjct: 656 EQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWP-R 714
Query: 1538 IIDKTQLQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPP-PWSDVHTNVVKQIKLRIKN 1596
+ T+++ FLG Y F +++ + + KD P WS +K + +
Sbjct: 715 PTNATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTS 774
Query: 1597 LPCLYLPNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEV 1656
P L LP +V TDAS +G G +L Q + ++IA+ S+ + NY T E+
Sbjct: 775 TPVLALPEHGQPYMVYTDASRVGLGCVLMQ----HGKVIAYASRQLMKHEGNYPTHDLEM 830
Query: 1657 LAIVLSISKFQYDLINQTFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIE 1716
A++ ++ ++ L V D KS K I + NL + RW +++ +D EI
Sbjct: 831 AAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQR----RWMELVADYDLEIA 886
Query: 1717 YIKGSTNSLPDFLTRE 1732
Y G N + D L+R+
Sbjct: 887 YHPGKANVVVDALSRK 902
>At2g10780 pseudogene
Length = 1611
Score = 204 bits (518), Expect = 5e-52
Identities = 131/435 (30%), Positives = 218/435 (50%), Gaps = 15/435 (3%)
Query: 1299 PYEKDFSDKQIPTKARPIQMNEELLQFCQKEINDLLQKKLIRRSKSPWSCATFYVNKQAE 1358
P+ + P P +M + +K++ +LL K IR S SPW +V K+
Sbjct: 648 PFTIELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSPWGAPVLFVKKKD- 706
Query: 1359 IERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKD 1418
G+ RL I+Y+ LN+ +YP+P +L+ +L A+ FSK D+ SG+ QI ++ D
Sbjct: 707 ---GSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTD 763
Query: 1419 RYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPY-SNFTIVYIDDVLIFSQSID 1477
KTAF + +E+ VMPFGL NAP+ F ++MN +F + F I++I+D+L++S+S +
Sbjct: 764 VRKTAFRTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWE 823
Query: 1478 QHFKHLNTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQ 1537
H +HL + ++++ L +K S +Q + FLGH I + I ++P +
Sbjct: 824 AHQEHLRAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWP-R 882
Query: 1538 IIDKTQLQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPP-PWSDVHTNVVKQIKLRIKN 1596
+ T+++ FLG Y F +++ + L KD WSD ++K + N
Sbjct: 883 PRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTN 942
Query: 1597 LPCLYLPNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEV 1656
P L LP V TDAS +G G +L QK +IA+ S+ ++NY T E+
Sbjct: 943 APVLVLPEEGEPYTVYTDASIVGLGCVLMQK----GSVIAYASRQLRKHEKNYPTHDLEM 998
Query: 1657 LAIVLSISKFQYDLINQTFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIE 1716
A+V + ++ L + D KS K I + NL + RW +++ ++ +I
Sbjct: 999 AAVVFFLKIWRSYLYGAKVQIYTDHKSLKYIFTQPELNLRQR----RWMELVADYNLDIA 1054
Query: 1717 YIKGSTNSLPDFLTR 1731
Y G N + D L+R
Sbjct: 1055 YHPGKANQVADALSR 1069
Score = 35.0 bits (79), Expect = 0.38
Identities = 26/91 (28%), Positives = 42/91 (45%), Gaps = 11/91 (12%)
Query: 876 SKPHSTQAAKNPPENRPSQGKNV-----TCYNCGKPGHISRYCRLKRRISELHLEPEIED 930
SK H+ Q+ P+ P Q K + TC+ CGK GH+ R C +++ + ++
Sbjct: 404 SKDHAIQSC---PKMEPGQSKVLGEETRTCFYCGKTGHLKREC---PKLTAEKQAGQRDN 457
Query: 931 KINNLLIQTSDEEESASSDSEVSEDLNQIQN 961
+ N L + AS E+SE+ N N
Sbjct: 458 RGGNGLPPPPKRQAVASRVYELSEEANDAGN 488
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 202 bits (515), Expect = 1e-51
Identities = 134/436 (30%), Positives = 215/436 (48%), Gaps = 15/436 (3%)
Query: 1299 PYEKDFSDKQIPTKARPIQMNEELLQFCQKEINDLLQKKLIRRSKSPWSCATFYVNKQAE 1358
P+ + P P +M + +K++ DLL K IR S SPW +V K+
Sbjct: 506 PFTIELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKKKD- 564
Query: 1359 IERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKD 1418
G+ RL I+Y+ LN +YP+P +LL +L A FSK D+ SG+ QI + E D
Sbjct: 565 ---GSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEAD 621
Query: 1419 RYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPY-SNFTIVYIDDVLIFSQSID 1477
KTAF +G +E+ VMPF L NAP+ F R+MN +F + F I++IDD+L++S+S +
Sbjct: 622 VRKTAFRTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPE 681
Query: 1478 QHFKHLNTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQ 1537
+H HL + +++ L +K S +Q +I FLGH + + IE +P +
Sbjct: 682 EHEVHLRRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWP-R 740
Query: 1538 IIDKTQLQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPP-PWSDVHTNVVKQIKLRIKN 1596
+ T+++ FL Y F +++ + + KD P WS +K + +
Sbjct: 741 PTNATEIRSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTS 800
Query: 1597 LPCLYLPNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEV 1656
P L LP +V TDAS +G G +L Q+ ++IA+ S+ + NY T E+
Sbjct: 801 TPVLALPEHGQPYMVYTDASRVGLGCVLMQR----GKVIAYASRQLRKHEGNYPTHDLEM 856
Query: 1657 LAIVLSISKFQYDLINQTFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIE 1716
++ ++ ++ L V D KS K I + NL RW +++ +D EI
Sbjct: 857 AVVIFALKIWRSYLYGGKVQVFTDHKSLKYIFNQPELNLRQ----MRWMELVADYDLEIA 912
Query: 1717 YIKGSTNSLPDFLTRE 1732
Y G N + D L+R+
Sbjct: 913 YHPGKANVVADALSRK 928
>At2g06890 putative retroelement integrase
Length = 1215
Score = 202 bits (514), Expect = 1e-51
Identities = 123/376 (32%), Positives = 199/376 (52%), Gaps = 15/376 (3%)
Query: 1362 GTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYK 1421
G+ R+ + + +N +PIP D+L LH + IFSK D+KSG+ QI++ E D +K
Sbjct: 467 GSWRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIFSKIDLKSGYHQIRMNEGDEWK 526
Query: 1422 TAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPYSN-FTIVYIDDVLIFSQSIDQHF 1480
TAF G YEW VMPFGL +APS F R+MN + + F IVY DD+L++S+S+ +H
Sbjct: 527 TAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFIGIFVIVYFDDILVYSESLREHI 586
Query: 1481 KHLNTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIID 1540
+HL++ ++V++K L + K + + FLG + + ++ +P
Sbjct: 587 EHLDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVSADGVKVDEEKVKAIRDWPS---P 643
Query: 1541 KT--QLQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPP-PWSDVHTNVVKQIKLRIKNL 1597
KT +++ F G + F STI+ L + +KKD W + +K ++ N
Sbjct: 644 KTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVMKKDVGFKWEKAQEEAFQSLKDKLTNA 703
Query: 1598 PCLYLPNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEVL 1657
P L L +E DAS IG G +L Q ++++IAF S+ A NY T KE+
Sbjct: 704 PVLILSEFLKTFEIECDASGIGIGAVLMQ----DQKLIAFFSEKLGGATLNYPTYDKELY 759
Query: 1658 AIVLSISKFQYDLINQTFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEY 1717
A+V ++ ++Q+ L + F++ D +S K + K + L +H ARW + F + I+Y
Sbjct: 760 ALVRALQRWQHYLWPKVFVIHTDHESLKHL--KGQQKLNKRH--ARWVEFIETFAYVIKY 815
Query: 1718 IKGSTNSLPDFLTREY 1733
KG N + D L++ Y
Sbjct: 816 KKGKDNVVADALSQRY 831
Score = 43.1 bits (100), Expect = 0.001
Identities = 42/156 (26%), Positives = 64/156 (40%), Gaps = 24/156 (15%)
Query: 817 QQLTKEKSQNRRDLGTFCEQFGIQGCPKKPKPRKHDPPPKQQWRRNSSRNHDHRKPKPRS 876
+Q K KS +R G+ G KP ++ + R +S N P+ S
Sbjct: 90 EQQVKRKSSSRSSYGS--------GTIAKPTYQREE-------RTSSYHNKPIVSPRAES 134
Query: 877 KPHST-QAAKNPPENRPSQGKNVTCYNCGKPGHISRYCRLKRRI-----SELHLEPEIED 930
KP++ Q K E S+ ++V CY C GH + C KR + E+ E EI D
Sbjct: 135 KPYAAVQDHKGKAEISTSRVRDVRCYKCQGKGHYANECPNKRVMILLDNGEIEPEEEIPD 194
Query: 931 KINNLLIQTSDEEESASSDSEVSEDLNQIQNDDDPQ 966
++L +EE A + V+ +Q D Q
Sbjct: 195 SPSSL---KENEELPAQGELLVARRTLSVQTKTDEQ 227
>At1g35370 hypothetical protein
Length = 1447
Score = 187 bits (476), Expect = 4e-47
Identities = 159/597 (26%), Positives = 275/597 (45%), Gaps = 43/597 (7%)
Query: 1151 KLSAAEGSKLIIKYKIPSAIIKNDSLEIETPFLLVRNLTHKVIIGTPFIKKLFPYNTDEK 1210
K++ A+G KL + +I K S ++ LL+ +++G +++ L + + K
Sbjct: 412 KVAVADGRKLNVDGQIKGFTWKLQSTTFQSDILLIPLQGVDMVLGVQWLETLGRISWEFK 471
Query: 1211 GITVQHLGKPILFKFSKPPFSKTLNIISYKEKQINFLKEEISHKSIEVQLQQPSVKTRIG 1270
+ +Q K +I ++K ++ ++I + V+ + IG
Sbjct: 472 KLEMQFFYKNQRVWLHGIITGSVRDIKAHKLQKTQ--ADQIQLAMVCVREVVSDEEQEIG 529
Query: 1271 NIL--------ENIQSSICSDLPNAFWERKSHMVELPYEKDFSDKQI-------PTKARP 1315
+I E++ +I + P+ F E +LP ++ D +I P RP
Sbjct: 530 SISALTSDVVEESVVQNIVEEFPDVFAE----PTDLPPFREKHDHKIKLLEGANPVNQRP 585
Query: 1316 IQMNEELLQFCQKEINDLLQKKLIRRSKSPWSCATFYVNKQAEIERGTPRLVINYKPLNQ 1375
+ K + D+++ I+ S SP++ V K + GT RL ++Y LN
Sbjct: 586 YRYVVHQKDEIDKIVQDMIKSGTIQVSSSPFASPVVLVKK----KDGTWRLCVDYTELNG 641
Query: 1376 ALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNV 1435
R+ IP +DL+ L + +FSK D+++G+ Q+++ D KTAF G +E+ V
Sbjct: 642 MTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHFEYLV 701
Query: 1436 MPFGLKNAPSEFQRIMNEIFNPY-SNFTIVYIDDVLIFSQSIDQHFKHLNTFISVIKKNG 1494
M FGL NAP+ FQ +MN +F + F +V+ DD+LI+S SI++H +HL V++ +
Sbjct: 702 MLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVMRLHK 761
Query: 1495 LAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYV 1554
L +K LGH I I I+ ++P K Q++ FLG Y
Sbjct: 762 LFAKGSK--------EHLGHFISAREIETDPAKIQAVKEWPTPTTVK-QVRGFLGFAGYY 812
Query: 1555 ADFCPQLSTIIKLLHDRLKKDPPPWSDVHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETD 1614
F I LH K D WS + +K + N P L LP +VETD
Sbjct: 813 RRFVRNFGVIAGPLHALTKTDGFCWSLEAQSAFDTLKAVLCNAPVLALPVFDKQFMVETD 872
Query: 1615 ASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISKFQYDLINQT 1674
A G +L QK +A+ S+ Q + S +KE+LA + ++ K+++ L+
Sbjct: 873 ACGQGIRAVLMQK----GHPLAYISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYLLPSH 928
Query: 1675 FLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTR 1731
F+++ D +S K +L++ + + +W L FD+EI+Y +G N + D L+R
Sbjct: 929 FIIKTDQRSLKYLLEQRLNTPVQQ----QWLPKLLEFDYEIQYRQGKENLVADALSR 981
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 186 bits (473), Expect = 8e-47
Identities = 118/375 (31%), Positives = 193/375 (51%), Gaps = 11/375 (2%)
Query: 1299 PYEKDFSDKQIPTKARPIQMNEELLQFCQKEINDLLQKKLIRRSKSPWSCATFYVNKQAE 1358
P+ + P P +M + +K++ +LL K IR S SPW +V K+
Sbjct: 143 PFTIELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGAPVLFVKKKD- 201
Query: 1359 IERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKD 1418
G+ RL I+Y+ LN+ +YP+P +L+ +L A+ FSK D+ SG+ QI ++ D
Sbjct: 202 ---GSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTD 258
Query: 1419 RYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPY-SNFTIVYIDDVLIFSQSID 1477
KTAF +G +E+ VMPFGL NAP+ F ++MN +F + F I++IDD+L+ S+S +
Sbjct: 259 VRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWE 318
Query: 1478 QHFKHLNTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQ 1537
H +HL + ++++ L +K S +Q + FLGH I + I ++P +
Sbjct: 319 AHQEHLRAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWP-R 377
Query: 1538 IIDKTQLQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPP-PWSDVHTNVVKQIKLRIKN 1596
+ T+++ FLG Y F +++ + L KD WSD ++K + N
Sbjct: 378 PRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLIN 437
Query: 1597 LPCLYLPNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEV 1656
P L LP V TDAS +G G +L QK +IA+ S+ ++NY T E+
Sbjct: 438 APVLVLPEEGEPYTVYTDASIVGLGCVLMQK----GSVIAYASRQLRKHEKNYPTHDLEM 493
Query: 1657 LAIVLSISKFQYDLI 1671
A+V ++ ++ LI
Sbjct: 494 AAVVFALKIWRSYLI 508
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 179 bits (454), Expect = 1e-44
Identities = 125/437 (28%), Positives = 211/437 (47%), Gaps = 29/437 (6%)
Query: 1299 PYEKDFSDKQIPTKARPIQMNEELLQFCQKEINDLLQKKLIRRSKSPWSCATFYVNKQAE 1358
P+ + P P +M + +K++ DLL ++ +V K+
Sbjct: 467 PFTIELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGAPVL------------FVKKKD- 513
Query: 1359 IERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKD 1418
G+ RL I+Y+ LN +YP+P +LL +L A FSK D+ SG+ I + E D
Sbjct: 514 ---GSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEAD 570
Query: 1419 RYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-YSNFTIVYIDDVLIFSQSID 1477
KTAF +G +E+ VMPFGL NAP+ F R+MN +F F I++IDD+L++S+S++
Sbjct: 571 VRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLE 630
Query: 1478 QHFKHLNTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIH-QGTIIPINRAIEFTDKFPD 1536
+H HL + +++ L +K S +Q ++ FLGH + +G + + D
Sbjct: 631 EHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTP 690
Query: 1537 QIIDKTQLQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPP-PWSDVHTNVVKQIKLRIK 1595
+ T+++ FLG Y F +++ + + KD P WS +K +
Sbjct: 691 --TNATEIRSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLT 748
Query: 1596 NLPCLYLPNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKE 1655
+ P L LP +V TDAS +G G +L Q+ ++IA+ S+ + NY T E
Sbjct: 749 STPVLALPEHGEPYMVYTDASGVGLGCVLMQR----GKVIAYASRQLRKHEGNYPTHDLE 804
Query: 1656 VLAIVLSISKFQYDLINQTFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEI 1715
+ A++ ++ ++ L V D KS K I + NL + +W +++ +D EI
Sbjct: 805 MAAVIFALKIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQR----QWMELVADYDLEI 860
Query: 1716 EYIKGSTNSLPDFLTRE 1732
Y G N + D L+ +
Sbjct: 861 AYHPGKANVVADALSHK 877
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 176 bits (447), Expect = 8e-44
Identities = 128/451 (28%), Positives = 210/451 (46%), Gaps = 72/451 (15%)
Query: 1282 SDLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFCQKEINDLLQKKLIRR 1341
+DLP F H +EL + + P RP + K ++D+L I+
Sbjct: 22 TDLP-PFRAPHDHKIELLEDSN------PVNQRPYRYVVHQKNEIDKIVDDMLASGTIQA 74
Query: 1342 SKSPWSCATFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFS 1401
S SP++ V K+ GT RL ++Y+ LN R+PIP +DL+ L + ++S
Sbjct: 75 SSSPYASPVVLVKKKD----GTWRLCVDYRELNGMTVKDRFPIPLIEDLMDELGGSNVYS 130
Query: 1402 KFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPY-SN 1460
K D+++G+ Q+++ D +KTAF G YE+ VMPFGL NAP+ FQ +MN F P+
Sbjct: 131 KIDLRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMNSFFKPFLRK 190
Query: 1461 FTIVYIDDVLIFSQSIDQHFKHLNTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGT 1520
F +V+ DD+LI+S S+++H KHL V++ + L +K + ++ +LGH I
Sbjct: 191 FVLVFFDDILIYSTSMEEHKKHLEAVFEVMRVHHLFAKMSKCAFAVPRVEYLGHFISGEG 250
Query: 1521 IIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPPPWS 1580
I I+ +P ++ QL FLG Y F
Sbjct: 251 IATDPAKIKAVQDWPVP-VNLKQLCGFLGLTGYYRRFF---------------------- 287
Query: 1581 DVHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSK 1640
+VE DA G G +L Q+ +AF S+
Sbjct: 288 -----------------------------VVEMDACGHGIGAVLMQE----GHPLAFISR 314
Query: 1641 HWNPAQQNYSTVKKEVLAIVLSISKFQYDLINQTFLVRVDCKSAKDILQKDVKNLASKHI 1700
Q + S +KE+LA++ + K+++ LI+ F+++ D +S K +L++ + + I
Sbjct: 315 QLKGKQLHLSIYEKELLAVIFVVRKWRHYLISSHFIIKTDQRSLKYLLEQRL----NTPI 370
Query: 1701 FARWQAILSVFDFEIEYIKGSTNSLPDFLTR 1731
+W L FD+EI+Y +G N + D L+R
Sbjct: 371 QQQWLPKLLEFDYEIQYKQGKENLVADALSR 401
>At4g08100 putative polyprotein
Length = 1054
Score = 159 bits (402), Expect = 1e-38
Identities = 103/350 (29%), Positives = 168/350 (47%), Gaps = 39/350 (11%)
Query: 1371 KPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQ 1430
+ +N R+PIP D L +LH + IFSK D+KSG+ Q +++E D +KTA
Sbjct: 529 RAINNITVKYRHPIPRLDDTLDKLHGSSIFSKIDLKSGYHQTRMKEGDEWKTAIKTKQRL 588
Query: 1431 YEWNVMPFGLKNAPSEFQRIMNEIFNPYSN-FTIVYIDDVLIFSQSIDQHFKHLNTFISV 1489
YEW VMPFGL NAP+ F R+MN + + F IVY DD+L++S++++ H HL + +
Sbjct: 589 YEWLVMPFGLTNAPNTFMRLMNHVLRKHIGVFVIVYFDDILVYSKNLEYHVMHLKLVLDL 648
Query: 1490 IKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLG 1549
++K L + K + + FLG + I ++ ++P+ K ++ +G
Sbjct: 649 LRKEKLYANLKKCTFCTDNLVFLGFVVSADGIKVDEEKVKAIREWPN---PKNVSEKDIG 705
Query: 1550 CLNYVADFCPQLSTIIKLLHDRLKKDPPPWSDVHTNVVKQIKLRIKNLPCLYLPNPQAFK 1609
W D N + +K ++ N LPN
Sbjct: 706 F---------------------------KWEDAQENAFQALKEKLTNSSVPILPNFMKSF 738
Query: 1610 IVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISKFQYD 1669
+E DAS +G G +L Q + + IA+ S+ A NY T KE+ A+V ++ +Q+
Sbjct: 739 EIECDASGLGIGAVLMQ----DHKPIAYFSEKLGGATLNYPTYDKELYALVRALQTWQHY 794
Query: 1670 LINQTFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIK 1719
L + F++ D +S K + K + L +H ARW + F + I+Y K
Sbjct: 795 LWPKKFVIHTDHESLKHL--KGQQKLNKRH--ARWVEFIETFPYVIKYKK 840
>At4g03650 putative reverse transcriptase
Length = 839
Score = 139 bits (350), Expect = 1e-32
Identities = 93/306 (30%), Positives = 153/306 (49%), Gaps = 11/306 (3%)
Query: 1414 LQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPY-SNFTIVYIDDVLIF 1472
L E D KTAF +G +E+ VMPFGL NAP+ F R+MN +F + F I++IDD+L++
Sbjct: 516 LAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVY 575
Query: 1473 SQSIDQHFKHLNTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTD 1532
S+S ++H HL + +++ L +K S +Q ++ FLGH + + IE
Sbjct: 576 SKSPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIR 635
Query: 1533 KFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPP-PWSDVHTNVVKQIK 1591
+P + + T+++ FLG Y F +++ + + KD P WS +K
Sbjct: 636 DWP-RPTNATEIRSFLGLAGYYRRFIKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLK 694
Query: 1592 LRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYST 1651
+ + P L LP +V TDAS +G G L Q+ ++IA+ S+ + NY T
Sbjct: 695 EMLTSTPVLALPEHGEPYMVYTDASGVGLGCALMQR----GKVIAYASRQLRKHEGNYPT 750
Query: 1652 VKKEVLAIVLSISKFQYDLINQTFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVF 1711
E+ A++ ++ ++ L V D KS K I + NL + RW +++ +
Sbjct: 751 HDLEMAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQR----RWMELVADY 806
Query: 1712 DFEIEY 1717
D EI Y
Sbjct: 807 DLEIAY 812
>At4g04230 putative transposon protein
Length = 315
Score = 123 bits (309), Expect = 8e-28
Identities = 67/179 (37%), Positives = 100/179 (55%), Gaps = 5/179 (2%)
Query: 1334 LQKKLIRRSKSPWSCATFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLAR 1393
+ K IR S SP + V K+ GT R+ ++ + +N R+PIP D+L
Sbjct: 125 IHKGYIRESLSPCAVPVLLVPKKD----GTWRMCLDCRAINNITIKYRHPIPRLYDMLDE 180
Query: 1394 LHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNE 1453
L A IFSK D++SG+ Q++++E D +KTAF G YE VMPFGL NAPS F R+MN+
Sbjct: 181 LSGAIIFSKVDLRSGYHQVRMREGDEWKTAFKTKQGLYECLVMPFGLTNAPSTFMRLMNQ 240
Query: 1454 IFNPY-SNFTIVYIDDVLIFSQSIDQHFKHLNTFISVIKKNGLAVSKTKVSLFQTKIRF 1511
+ + F +VY DD+LI+++S H + L + ++K GL + K + K F
Sbjct: 241 VLRSFIGKFVVVYFDDILIYNKSYSDHIQPLELILKTLRKEGLYANLKKCTFCSDKFFF 299
>At1g20390 hypothetical protein
Length = 1791
Score = 117 bits (293), Expect = 6e-26
Identities = 187/875 (21%), Positives = 344/875 (38%), Gaps = 85/875 (9%)
Query: 862 NSSRNHDHRKPKPRSKPHSTQAAKNPPENRPSQGKNVTCYNCGKPGHISRYCRLKRRISE 921
N R H R P KP S + P+ +P N + P YC + S
Sbjct: 363 NEPRQHLDRNPSAGRKPTSFLVSTETPDAKPW---NKYIRDADSPAAGPMYCEYHK--SR 417
Query: 922 LHLEPEIEDKINNLLIQTSDEEESASSDSEVSEDLNQIQNDDDPQSSSSINVLTNEQDLL 981
H N +Q + S + D I N + ++ ++ N+Q
Sbjct: 418 AHSTE------NCRFLQGLLMAKYKSGGITIECDRPPINNKNQRRNETTARQYLNDQ--- 468
Query: 982 FRAINSIPDPDEKKIYLERLKFTLEDKPPKNPITTNKFNLRDTFRRLEKSTIKPVTIQDL 1041
P P E+ I T D P K +PV ++ +
Sbjct: 469 ----TKPPTPAEQGI------ITSADDPAAKRQRNGK-----------AIAAEPVVVRQV 507
Query: 1042 QSEVHTLQA---EVKSLKQIQISQQLILDKLTEENSEESSSSSSTPNSASNNNVGDFLEI 1098
+ LQ V+S+KQ + ++++ S + + S P S ++ ++
Sbjct: 508 HVIMGGLQNCSDSVRSIKQYRKKAEMVV-AWPSSTSTTRNPNQSAPISFTDVDLEGLDTP 566
Query: 1099 INNVIIQKFYINIKIIIGDFILETPALFDTGANSSCISEGL-----IPTRYFEKTTEKLS 1153
N+ ++ +++II D + T L DTG++ I + + I R + ++ L+
Sbjct: 567 HNDPLV------VELIISDSRV-TRVLIDTGSSVDLIFKDVLTAMNITDRQIKPVSKPLA 619
Query: 1154 AAEGSKLIIKYKIPSAIIKNDSLEIETPFLLVRNLTHKVIIGTPFIKKL--FPYNTDEKG 1211
+G ++ I I + ++ + + VI+GTP+I ++ P +
Sbjct: 620 GFDGDFVMTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQCV 679
Query: 1212 ITVQHLGKPILFKFSKPPFSKTLNIISYKEKQINF-----LKEEISHKSIEVQLQ-QPSV 1265
H G +F P +KT + SY+E ++ + E + + V + PS+
Sbjct: 680 KFPTHNG---IFTLRAPKEAKTPSR-SYEESELCRTEMVNIDESDPTRCVGVGAEISPSI 735
Query: 1266 KTRIGNILENIQSSIC---SDLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEEL 1322
+ + +L+ + D+ +H EL + F P K + ++ E
Sbjct: 736 RLELIALLKRNSKTFAWSIEDMKGIDPAITAH--ELNVDPTFK----PVKQKRRKLGPER 789
Query: 1323 LQFCQKEINDLLQKKLIRRSKSP-WSCATFYVNKQAEIERGTPRLVINYKPLNQALCWIR 1381
+ +E+ LL+ I K P W V K+ G R+ ++Y LN+A
Sbjct: 790 ARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKK----NGKWRVCVDYTDLNKACPKDS 845
Query: 1382 YPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLK 1441
YP+P+ L+ + S D SG+ QI + + D+ KT+F G Y + VM FGLK
Sbjct: 846 YPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGTYCYKVMSFGLK 905
Query: 1442 NAPSEFQRIMNEIFNPYSNFTI-VYIDDVLIFSQSIDQHFKHLNTFISVIKKNGLAVSKT 1500
NA + +QR +N++ T+ VYIDD+L+ S + H +HL+ V+ G+ ++ T
Sbjct: 906 NAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDVLNTYGMKLNPT 965
Query: 1501 KVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQ 1560
K + T FLG+ + + I + I + P + ++QR G + + F +
Sbjct: 966 KCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSP-RNAREVQRLTGRIAALNRFISR 1024
Query: 1561 LSTIIKLLHDRLKKDPP-PWSDVHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIG 1619
+ ++ LK+ W +++K + P L P + SD
Sbjct: 1025 STDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVGETLYLYIAVSDHA 1084
Query: 1620 FGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISKFQYDLINQTFLVRV 1679
+L ++ ++ I +TSK A+ Y ++K LA+V S K + + T V
Sbjct: 1085 VSSVLVREDRGEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSARKLRPYFQSHTIAVLT 1144
Query: 1680 DCKSAKDILQKDVKNLASKHIFARWQAILSVFDFE 1714
D L+ + + + +W LS +D +
Sbjct: 1145 D-----QPLRVALHSPSQSGRMTKWAVELSEYDID 1174
>At2g06170 putative Ty3-gypsy-like retroelement pol polyprotein
Length = 587
Score = 113 bits (282), Expect = 1e-24
Identities = 50/113 (44%), Positives = 78/113 (68%), Gaps = 1/113 (0%)
Query: 1371 KPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQ 1430
K +N+ R+PIP D+L L +K+FSK D++SG+ QI+++ D +KT F G
Sbjct: 401 KAINKITIKYRFPIPQLDDMLDELSGSKVFSKIDLRSGYHQIRIRPGDEWKTDFKSKDGL 460
Query: 1431 YEWNVMPFGLKNAPSEFQRIMNEIFNPYS-NFTIVYIDDVLIFSQSIDQHFKH 1482
YEW VMPFG+ NAPS F R+MN+I P++ +F +VY DD+LI+S++ +++ +H
Sbjct: 461 YEWQVMPFGMSNAPSTFMRLMNQILRPFTGSFVVVYFDDILIYSKTKEEYLEH 513
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 100 bits (250), Expect = 6e-21
Identities = 92/366 (25%), Positives = 164/366 (44%), Gaps = 38/366 (10%)
Query: 1310 PTKARPIQMNEELLQFCQKEINDLLQKKLIRRSKSP-WSCATFYVNKQAEIERGTPRLVI 1368
P K + ++ + + +E+ LL I + P W V K+ G R+ I
Sbjct: 782 PLKQKRRKLGPDRTKDVNEEVKKLLDAGSIVEVRYPDWLRNPVVVKKK----NGKWRVCI 837
Query: 1369 NYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPF 1428
++ LN+A +P+P+ L+ ++ S D SG+ QI + + DR KT F
Sbjct: 838 DFTDLNKACPKDSFPLPHIDRLVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQ 897
Query: 1429 GQYEWNVMPFGLKNAPSEFQRIMNEIFNPYSNFTI-VYIDDVLIFSQSIDQHFKHLNTFI 1487
G Y + VMPFGLKNA + + R++N++F + ++ VYIDD+L+ S ++H HL
Sbjct: 898 GTYCYKVMPFGLKNAGATYPRLVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCF 957
Query: 1488 SVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIID------K 1541
V+ + + ++ +K + T FLG+ + R IE K IID
Sbjct: 958 QVLNRYNMKLNPSKCTFGVTSGEFLGY-------LVTRRGIEANPKQISAIIDLPSPRNT 1010
Query: 1542 TQLQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPPPWSDVHTNVVKQIKLRIKNLPCLY 1601
++QR +G +++ + + + K P + + N + + + LY
Sbjct: 1011 REVQRLIG----------RIAALNRFISRSTDKCLPFYQLLRANKRFEWDEKCEEGETLY 1060
Query: 1602 LPNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEVLAIVL 1661
L + V T A G+L ++ + I + SK + A+ Y T++K A+V+
Sbjct: 1061 L-----YIAVSTSA----VSGVLVREDRGEQHPIFYVSKTLDGAELRYPTLEKLAFAVVI 1111
Query: 1662 SISKFQ 1667
S K +
Sbjct: 1112 SARKLR 1117
>At2g10690 putative retroelement pol polyprotein
Length = 622
Score = 100 bits (250), Expect = 6e-21
Identities = 93/328 (28%), Positives = 148/328 (44%), Gaps = 19/328 (5%)
Query: 1410 WQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPY-SNFTIVYIDD 1468
+++Q + KT FT P G + + M FGL NAP FQR M IF+ + V++DD
Sbjct: 154 FKLQFTLMIKKKTTFTCPNGTFAYKRMSFGLCNAPGTFQRSMTSIFSDFIEEIMEVFMDD 213
Query: 1469 VLIFSQSIDQHFKHLNTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRA- 1527
FS + +L + ++ L ++ K + LGH I G I +++A
Sbjct: 214 FSGFSSCL----LNLCRVLERCEETNLVLNWEKCHFMVHEGIVLGHKI-SGKGIEVDKAK 268
Query: 1528 --IEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPP-PWSDVHT 1584
+ + P + D ++ FLG + F S I + L L K+ + +
Sbjct: 269 IDVMIQLQPPKTVKD---IRSFLGHAEFYRRFIKDFSKIARPLTRLLCKETEFNFDEDCL 325
Query: 1585 NVVKQIKLRIKNLPCLYLPN-PQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWN 1643
IK + + P PN F+I+ DA D G +L QKI D +I + S+ +
Sbjct: 326 KAFHLIKETLVSAPIFQAPNWDHPFEIM-CDAFDYAVGAVLDQKIDDKLHVIYYASRTMD 384
Query: 1644 PAQQNYSTVKKEVLAIVLSISKFQYDLINQTFLVRVDCKSAKDILQKDVKNLASKHIFAR 1703
AQ Y+T++KE+LA+V + K + L+ V +D + + I K +K R
Sbjct: 385 EAQTRYATIEKELLAVVFAFEKIRSYLVGFKVKVYIDHAALRHIYAKK----ETKSRLLR 440
Query: 1704 WQAILSVFDFEIEYIKGSTNSLPDFLTR 1731
W +L FD EI KG N + D L+R
Sbjct: 441 WILLLQEFDMEIIDKKGVENGVADHLSR 468
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 100 bits (250), Expect = 6e-21
Identities = 59/171 (34%), Positives = 87/171 (50%), Gaps = 23/171 (13%)
Query: 1310 PTKARPIQMNEELLQFCQKEINDLLQKKLIRRSKSPWSCATFYVNKQAEIERGTPRLVIN 1369
P P +M + +K++ DLL K IR S S W G P L
Sbjct: 535 PLSKTPYRMVPAEIAELKKQLEDLLGKGFIRPSTSRW---------------GAPGL--- 576
Query: 1370 YKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFG 1429
N+ +YP+P +LL +L A FSK D+ G+ Q + E D KTAF +G
Sbjct: 577 ----NRVTLKNKYPLPRIDELLDQLRGATCFSKIDLTPGYHQFPIAEADVRKTAFRTRYG 632
Query: 1430 QYEWNVMPFGLKNAPSEFQRIMNEIFNPY-SNFTIVYIDDVLIFSQSIDQH 1479
+E+ VMPFGL NAP+ R+MN +F + F I++IDD+L++ +S ++H
Sbjct: 633 HFEFVVMPFGLTNAPTALMRLMNSVFQEFLDEFVIIFIDDILVYFKSPEEH 683
Score = 48.5 bits (114), Expect = 3e-05
Identities = 30/100 (30%), Positives = 49/100 (49%), Gaps = 4/100 (4%)
Query: 1633 QIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISKFQYDLINQTFLVRVDCKSAKDILQKDV 1692
++I + S+ + NY T E+ AI+ ++ + L V D KS KDI +
Sbjct: 723 KVIVYASRQLRKHEGNYPTHDLEMAAIIFALKIWGSYLYGGKVQVFTDHKSLKDIFTQPE 782
Query: 1693 KNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTRE 1732
NL + RW +++ +D EI Y G TN + D L+R+
Sbjct: 783 LNLRQR----RWMELVADYDLEIAYHPGKTNVVADALSRK 818
>At4g07600
Length = 630
Score = 100 bits (248), Expect = 1e-20
Identities = 82/294 (27%), Positives = 129/294 (42%), Gaps = 21/294 (7%)
Query: 1365 RLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAF 1424
R+ I+Y+ LN A +P+P +L RL + + D SGF+QI + D KT F
Sbjct: 356 RMCIDYRNLNAASRNDHFPLPFTDQMLERLANHPYYCFLDGYSGFFQIPIHPNDHEKTTF 415
Query: 1425 TVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-YSNFTIVYIDDVLIFSQSIDQHFKHL 1483
T P+G + + MPFGL NAP+ FQR M IF+ V++DD ++ S +L
Sbjct: 416 TCPYGTFAYERMPFGLCNAPATFQRCMTSIFSDIIEEMVEVFMDDFSVYGPSFSSCLLNL 475
Query: 1484 NTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQ 1543
++ ++ L ++ K + LGH I + + IE +DK
Sbjct: 476 GRVLTRCEETNLVLNWEKCHFMVKEGIMLGHKISE-------KGIE---------VDK-- 517
Query: 1544 LQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPP-PWSDVHTNVVKQIKLRIKNLPCLYL 1602
FLG + F S I + L L K+ + + IK + + P +
Sbjct: 518 -GCFLGHAGFYRRFIKDFSKIARPLTRLLCKETEFEFDEDCPKSFHTIKEALVSAPVVRA 576
Query: 1603 PNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEV 1656
N + DA D G +L QKI +I + S+ + AQ Y+T +KE+
Sbjct: 577 TNWDYPFEIMCDALDYAVGAVLGQKIDKKLHVIYYASRTLDEAQGRYATTEKEL 630
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 91.7 bits (226), Expect = 3e-18
Identities = 78/299 (26%), Positives = 137/299 (45%), Gaps = 8/299 (2%)
Query: 1329 EINDLLQKKLIRRSKSP-WSCATFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 1387
E++ LL+ IR + P W T V K+ G R+ I++ LN+A +P+P+
Sbjct: 483 EVDKLLKIGSIREVQYPDWLANTVVVKKK----NGKDRVCIDFTDLNKACPKDSFPLPHI 538
Query: 1388 KDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEF 1447
L+ ++ S D SG+ QI + +D+ KT F G Y + VMPFGL+NA + +
Sbjct: 539 DRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPFGLRNAGATY 598
Query: 1448 QRIMNEIFNPYSNFTI-VYIDDVLIFSQSIDQHFKHLNTFISVIKKNGLAVSKTKVSLFQ 1506
R++N++F+ + T+ VYIDD+LI S + H KHL +++ + + ++ K +
Sbjct: 599 PRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMKLNPAKCTFGV 658
Query: 1507 TKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIK 1566
FLG+ + + I I P K ++QR G + + F + +
Sbjct: 659 PSGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFK-EVQRLTGRIAALNRFISRSTDKSL 717
Query: 1567 LLHDRLKKDPP-PWSDVHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGIL 1624
+ LK + W + Q+K + + P L+ P + S+ G+L
Sbjct: 718 PFYQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVSNHAVSGVL 776
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.339 0.149 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,672,059
Number of Sequences: 26719
Number of extensions: 1644048
Number of successful extensions: 10325
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 79
Number of HSP's that attempted gapping in prelim test: 9870
Number of HSP's gapped (non-prelim): 414
length of query: 1733
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1620
effective length of database: 8,299,349
effective search space: 13444945380
effective search space used: 13444945380
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0101.14


