

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0101.13
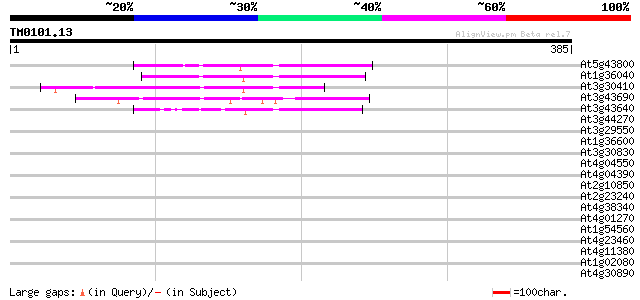
(385 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g43800 putative protein 75 6e-14
At1g36040 putative envelope-like protein gb|AAD20656 69 3e-12
At3g30410 hypothetical protein 61 9e-10
At3g43690 unknown protein 53 3e-07
At3g43640 putative protein 52 5e-07
At3g44270 putative protein 40 0.003
At3g29550 hypothetical protein 38 0.008
At1g36600 hypothetical protein 38 0.011
At3g30830 hypothetical protein 36 0.041
At4g04550 putative protein 33 0.20
At4g04390 hypothetical protein 33 0.34
At2g10850 pseudogene 33 0.34
At2g23240 metallothionein-like protein 31 1.0
At4g38340 putative protein 30 1.7
At4g01270 putative RING zinc finger protein 30 2.2
At1g54560 30 2.2
At4g23460 beta adaptin - like protein 29 5.0
At4g11380 beta-adaptin - like protein 29 5.0
At1g02080 unknown protein 29 5.0
At4g30890 ubiquitin-specific protease 24 (UBP24) 28 6.5
>At5g43800 putative protein
Length = 515
Score = 75.1 bits (183), Expect = 6e-14
Identities = 54/170 (31%), Positives = 82/170 (47%), Gaps = 12/170 (7%)
Query: 86 KEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSP 145
++++ +I L TV I +++ EF N N T KVFVRGK FSP
Sbjct: 203 RDMMKVIEDNHLLDTVAEIDPFVKEVIWEFYANF-HIFNEETKKS--KVFVRGKMYEFSP 259
Query: 146 AIINQTLERSVV------EFVEEELSLDTVAEELTAGRVKKWPAKKLLSTGVLSVKYAIL 199
+IN+T + EF +S +A+ L+ G+V W K L T V+ A L
Sbjct: 260 QMINKTFRSPAIKWHAKAEFERVTMSKHALAKYLSDGKVVTW---KDLRTPVMPTHLAGL 316
Query: 200 NRIGVVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLKHAET 249
I +NW+PS + S V+ AK++Y + I ++FG V+ Q K A +
Sbjct: 317 YLICCINWIPSKNNSNVTVDRAKMLYLLNERIPFNFGKLVYDQVEKAARS 366
>At1g36040 putative envelope-like protein gb|AAD20656
Length = 444
Score = 69.3 bits (168), Expect = 3e-12
Identities = 48/161 (29%), Positives = 76/161 (46%), Gaps = 12/161 (7%)
Query: 91 LIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSPAIINQ 150
L+ GL TV I ++V EF NL + VFVR FS AIINQ
Sbjct: 201 LLFNGGLLPTVTEIDSYVHEVVMEFYANLPDGEEGDNLAY--SVFVRRNMYEFSLAIINQ 258
Query: 151 TLERSVVEF-------VEEELSLDTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNRIG 203
+ + ++ S+D VA L+ G+ W K+L++ +LS + A+LN+I
Sbjct: 259 MFQLPNPSYPLDGMSEIQVPESMDEVAIALSNGKANSW---KMLTSRLLSPELALLNKIC 315
Query: 204 VVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTL 244
NW P+ + SV+ L+Y + + ++FG +F + L
Sbjct: 316 CHNWSPTVNRSVLKPERMTLLYMVAKALPFNFGKLIFDENL 356
>At3g30410 hypothetical protein
Length = 421
Score = 61.2 bits (147), Expect = 9e-10
Identities = 57/208 (27%), Positives = 89/208 (42%), Gaps = 19/208 (9%)
Query: 22 EGDVRDITA-------SEKKMFSGKRIPSDVPNVPLDNVSIHAPENAFKWKYVYQRRVAK 74
+GDV I E + R P +N + A E ++K + R
Sbjct: 106 DGDVTQIAPPLFLSGYQENRRLLSARDPQVSSETSYNNCFLTA-EGLGRYKLIGGRYFND 164
Query: 75 DREIDSDVLACKEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKV 134
R + D + L++ AGL TV I ++V EF NL + F V
Sbjct: 165 MRFLSLDGNNTASTLQLLSTAGLLLTVTEIDSYVQEVVMEFYANLPDGEEGDQLAYF--V 222
Query: 135 FVRGKCVNFSPAIINQTLERSVVEF------VEEELSLDTVAEELTAGRVKKWPAKKLLS 188
FVRG FSPAIINQ + + ++ S+D V + + W K+L+
Sbjct: 223 FVRGNMYEFSPAIINQIFQLPNPPYPLNMSKIKVPESMDEVDVARSNSKANSW---KMLT 279
Query: 189 TGVLSVKYAILNRIGVVNWVPSNHTSVV 216
+ +LS + A+LN+I NW+P+ + SV+
Sbjct: 280 SQLLSPEIALLNKIYCHNWMPTVNRSVL 307
>At3g43690 unknown protein
Length = 539
Score = 52.8 bits (125), Expect = 3e-07
Identities = 52/215 (24%), Positives = 100/215 (46%), Gaps = 25/215 (11%)
Query: 46 PNVPLDNVSIHAPENAFKWKYVYQRRVA--KDREIDSDVLACKEIIALIAKAGLRKTVMN 103
P VP + A ++K++ +R K +D +VL + + +G+ +TV+
Sbjct: 233 PLVPPHTKRFLSAAAAERYKHIAKRDFIFQKTLPLDPEVLTATKYF--LEHSGMAQTVVA 290
Query: 104 IGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSPAIINQ--TLERSVVEFVE 161
+ + ++V+EF NL E + V+VRGK FSPA+IN +++ S ++ E
Sbjct: 291 VEQFVPEVVREFYANLPEMEYRECGLDL--VYVRGKMYEFSPALINHMFSIDDSALD-PE 347
Query: 162 EELSLDTVAEE-----LTAGRVKKW----PAKKLLSTGVLSVKYAILNRIGVVNWVPSNH 212
++L T + + +T G ++W PA L + +L+++ NW P+ +
Sbjct: 348 APVTLSTASRDDLALMMTGGTTRRWLRLQPADHLDT-------MKMLHKVCCGNWFPTTN 400
Query: 213 TSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLKHA 247
TS + +LI +++ G V + T+ A
Sbjct: 401 TSTLRVDRLRLIDMGTHGKSFNLGKLVVTHTMSLA 435
>At3g43640 putative protein
Length = 428
Score = 52.0 bits (123), Expect = 5e-07
Identities = 43/161 (26%), Positives = 82/161 (50%), Gaps = 17/161 (10%)
Query: 86 KEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSP 145
+ ++A+ + G K + N +C++ + F L D NN+ S + + +F G F+
Sbjct: 164 RRLLAVRDRLGRYKVIGN--RCFNDM--RF---LPLDGNNTASTQ-QLLFNAGLLPTFTE 215
Query: 146 AIINQTLERSVVEFV----EEELSLDTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNR 201
I+ + V+EF + S+D VA L+ G+ W K+L++ +LS + A+LN+
Sbjct: 216 --IDSYVHEVVMEFYANLPDVPESMDEVAIALSNGKANSW---KMLTSRLLSPELALLNK 270
Query: 202 IGVVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQ 242
I NW P+ + SV+ L+Y + + ++FG +F +
Sbjct: 271 ICCHNWSPTVNHSVLKPERMTLLYMVAKALPFNFGKLIFDE 311
>At3g44270 putative protein
Length = 324
Score = 39.7 bits (91), Expect = 0.003
Identities = 42/156 (26%), Positives = 66/156 (41%), Gaps = 27/156 (17%)
Query: 92 IAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSPAIINQT 151
I AGL +TV + ++LV EF NL +STS +++ Q
Sbjct: 147 IYDAGLIRTVTELKPFEEELVFEFWANLPTVKVDSTSV----------------SVMQQM 190
Query: 152 LERSVVEFVEEELSLDTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNRIGVVNWVPSN 211
++E D VA+ LT G+VK K L S+ L ++ NW P++
Sbjct: 191 DMAGLLE--------DEVAQFLTDGKVK---LLKRLLLSAFSLPGRELFKLCCTNWSPTS 239
Query: 212 HTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLKHA 247
+ S AKL+Y I ++ ++F F L+ A
Sbjct: 240 NNSYAQPDRAKLVYMIMHKMPFNFVRLAFDHILQLA 275
>At3g29550 hypothetical protein
Length = 223
Score = 38.1 bits (87), Expect = 0.008
Identities = 43/152 (28%), Positives = 66/152 (43%), Gaps = 40/152 (26%)
Query: 90 ALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSPAIIN 149
+ I KAGL +TV ++ ++LV EF NL ++T+ V VR FSP IN
Sbjct: 36 SFIEKAGLIRTVTDLKPYKEELVFEFWANLPTVKVDTTNV---GVLVRNWEYEFSPENIN 92
Query: 150 QTLERSVVEFVEEELSL-----DTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNRIGV 204
+ + V+ ++ + + D VA+ LT G+VK S+K + +R
Sbjct: 93 ELFGLASVDVRQQRMDMVGLMEDEVAKFLTDGKVK-------------SLKGLLPDR--- 136
Query: 205 VNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFG 236
AKL+Y I I +DFG
Sbjct: 137 ----------------AKLVYMIMHNIPFDFG 152
>At1g36600 hypothetical protein
Length = 403
Score = 37.7 bits (86), Expect = 0.011
Identities = 26/92 (28%), Positives = 40/92 (43%), Gaps = 3/92 (3%)
Query: 167 DTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNRIGVVNWVPSNHTSVVSAMLAKLIYR 226
+ VA LT GRV+ + ++ S L +I NW P + S +L+YR
Sbjct: 173 EEVARYLTDGRVQNL---QNIAMNKFSENCKSLFKISCRNWSPQTNEGYASIDRGRLVYR 229
Query: 227 IGTEIAYDFGNFVFSQTLKHAETCAVKLPISF 258
I ++ DFG V+ L+ A K + F
Sbjct: 230 IAHKMPIDFGEMVYDHVLQLALMSVSKFFLMF 261
>At3g30830 hypothetical protein
Length = 327
Score = 35.8 bits (81), Expect = 0.041
Identities = 30/147 (20%), Positives = 51/147 (34%), Gaps = 30/147 (20%)
Query: 96 GLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSPAIINQTLERS 155
G K + I ++V EF NL VFVRG FS AI NQ
Sbjct: 130 GRYKVIGEINSYVQEVVMEFYANLPGGEEGDQLAY--SVFVRGNMYEFSLAITNQMFHFP 187
Query: 156 VVEFVEEELSLDTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNRIGVVNWVPSNHTSV 215
+ ++ + ++ LN+I NW+P + +
Sbjct: 188 NLSYL----------------------------VNMSEIQITQLNKICCHNWMPKVNRFI 219
Query: 216 VSAMLAKLIYRIGTEIAYDFGNFVFSQ 242
+ L+Y + + ++FG +F +
Sbjct: 220 LKPERVTLLYMVAKALPFNFGKLIFDE 246
>At4g04550 putative protein
Length = 307
Score = 33.5 bits (75), Expect = 0.20
Identities = 22/73 (30%), Positives = 31/73 (42%), Gaps = 3/73 (4%)
Query: 167 DTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNRIGVVNWVPSNHTSVVSAMLAKLIYR 226
D V LT G+VK + L S L + NW P+ S L+YR
Sbjct: 147 DEVTSYLTNGQVK---VLQSLPMSTFSENCRKLFKFSCRNWSPTTSEGYASIHRVLLMYR 203
Query: 227 IGTEIAYDFGNFV 239
I ++A+DFG +
Sbjct: 204 IAHKLAFDFGQML 216
>At4g04390 hypothetical protein
Length = 963
Score = 32.7 bits (73), Expect = 0.34
Identities = 19/81 (23%), Positives = 40/81 (48%), Gaps = 1/81 (1%)
Query: 165 SLDTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNRIGVVNWVPSNHTSVVSAMLAKLI 224
+L + ++T RV KK+ L ++YA+L + +P++H + A+++
Sbjct: 132 TLFELESDVTVSRVITMLKKKVTDDPSLRIRYAVLALVDGY-LLPTSHYPKIVKEHAEMV 190
Query: 225 YRIGTEIAYDFGNFVFSQTLK 245
+ + +AY +G F T+K
Sbjct: 191 ENLSSFLAYPWGRLTFEMTMK 211
>At2g10850 pseudogene
Length = 285
Score = 32.7 bits (73), Expect = 0.34
Identities = 39/152 (25%), Positives = 61/152 (39%), Gaps = 15/152 (9%)
Query: 33 KKMFSGKRIPSDVPNVPLDNVSIHAPENAFKWKYVYQRRVAKDREIDSDVL-ACKEIIAL 91
K+M GK + + D + + E ++ QR ++ E+ A +E I
Sbjct: 98 KRMKQGKGVTG---SSSFDGIRFVSTEAEKRYAQFSQRNFIEEVELSRKTNEAAREFIK- 153
Query: 92 IAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSPAIINQT 151
+AGL +TV LV EF NL KV VR + SP IN+
Sbjct: 154 --QAGLIRTVTKFNPFTQNLVFEFWANLP---TMKVDTYMVKVLVRNREYELSPGKINEM 208
Query: 152 LERSVVEFVEEELSL-----DTVAEELTAGRV 178
V+ ++ + + + VAE LT G+V
Sbjct: 209 YGLPSVDARQQRMDIAGLVDEQVAEFLTGGKV 240
>At2g23240 metallothionein-like protein
Length = 85
Score = 31.2 bits (69), Expect = 1.0
Identities = 17/60 (28%), Positives = 24/60 (39%), Gaps = 2/60 (3%)
Query: 307 DICSCLYVCLRNSQCHC*T*GHFQGTSRDHQDCCC*ETKG--GCSHPNSQGGSWSRG*AC 364
D C C C C C G ++H C C E G C+ P +Q + ++G C
Sbjct: 15 DRCGCPSPCPGGESCRCKMMSEASGGDQEHNTCPCGEHCGCNPCNCPKTQTQTSAKGCTC 74
>At4g38340 putative protein
Length = 767
Score = 30.4 bits (67), Expect = 1.7
Identities = 28/105 (26%), Positives = 51/105 (47%), Gaps = 5/105 (4%)
Query: 119 LSEDVNNSTSPEFRKVFVRGKCVNFSPAIINQTLERSVVEFVEEELSLDTVAEELTAGRV 178
++E+V ++ +P + +GK V+FS + + R E++++LDT+ + AG +
Sbjct: 454 IAEEVRDAATPPLTQEDPKGKQVSFSFSSASSLENRKRKTKAEKDITLDTLRQHF-AGSL 512
Query: 179 KKWPAKKLLSTGVLSVKYAILNRIGVVNWVPSNHTSVVSAMLAKL 223
K A K + ++K I + G+ W PS V L KL
Sbjct: 513 K--DAAKNIGVCPTTLK-RICRQNGISRW-PSRKIKKVGHSLRKL 553
>At4g01270 putative RING zinc finger protein
Length = 506
Score = 30.0 bits (66), Expect = 2.2
Identities = 29/107 (27%), Positives = 44/107 (41%), Gaps = 14/107 (13%)
Query: 85 CKEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFS 144
CK+ +L K R + G D + + +V + ED V +RG+
Sbjct: 59 CKQKCSL--KDPCRLYFQSSGNQTDSIASDKVVGIEED----------PVLLRGEVKRLE 106
Query: 145 PAIINQT--LERSVVEFVEEELSLDTVAEELTAGRVKKWPAKKLLST 189
+ N T LE E VE L E+L +VK+W A + +ST
Sbjct: 107 GKVQNLTSALEAKKKENVEVSDKLHQCNEQLKEDKVKRWEALQEIST 153
>At1g54560
Length = 1529
Score = 30.0 bits (66), Expect = 2.2
Identities = 20/89 (22%), Positives = 43/89 (47%), Gaps = 11/89 (12%)
Query: 90 ALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTS---------PEFRKVFVRGKC 140
ALI++ GL KT+ + + +D LV++ V++ +D + + F+ C
Sbjct: 396 ALISRDGLAKTIYS--RLFDWLVEKINVSIGQDATSRSLIGVLDIYGFESFKTNSFEQFC 453
Query: 141 VNFSPAIINQTLERSVVEFVEEELSLDTV 169
+NF+ + Q + V + +EE + + +
Sbjct: 454 INFTNEKLQQHFNQHVFKMEQEEYTKEAI 482
>At4g23460 beta adaptin - like protein
Length = 893
Score = 28.9 bits (63), Expect = 5.0
Identities = 16/41 (39%), Positives = 22/41 (53%)
Query: 71 RVAKDREIDSDVLACKEIIALIAKAGLRKTVMNIGKCYDKL 111
++A DR ID +L KE + +RK V IG+C KL
Sbjct: 344 KLASDRNIDQVLLEFKEYATEVDVDFVRKAVRAIGRCAIKL 384
>At4g11380 beta-adaptin - like protein
Length = 894
Score = 28.9 bits (63), Expect = 5.0
Identities = 16/41 (39%), Positives = 22/41 (53%)
Query: 71 RVAKDREIDSDVLACKEIIALIAKAGLRKTVMNIGKCYDKL 111
++A DR ID +L KE + +RK V IG+C KL
Sbjct: 344 KLASDRNIDQVLLEFKEYATEVDVDFVRKAVRAIGRCAIKL 384
>At1g02080 unknown protein
Length = 2226
Score = 28.9 bits (63), Expect = 5.0
Identities = 21/89 (23%), Positives = 45/89 (49%), Gaps = 4/89 (4%)
Query: 143 FSPAIINQTLERSVVEFVEEELSLDTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNRI 202
+S +IN + + ++ +++ + +T A+ + A ++ K+ + +L + G +Y LN I
Sbjct: 2060 YSVPLINSLVLYTGMQAIQQLQAGETQAQNVVALQMFKYLSMELDTEG----RYLFLNAI 2115
Query: 203 GVVNWVPSNHTSVVSAMLAKLIYRIGTEI 231
P+NHT S ++ L + EI
Sbjct: 2116 ANQLRYPNNHTHYFSFIMLYLFFESDQEI 2144
>At4g30890 ubiquitin-specific protease 24 (UBP24)
Length = 551
Score = 28.5 bits (62), Expect = 6.5
Identities = 25/102 (24%), Positives = 43/102 (41%), Gaps = 10/102 (9%)
Query: 82 VLACKEIIALIAKAGLR---KTVMNIGKCYDKLVKEFIVNLSEDVNNSTSP-----EFRK 133
+L+C + L+ K L+ K + + + E V S + N+ + FR
Sbjct: 215 LLSCSPFVQLLQKIQLQDIPKADSPTLAAFSEFISELDVPSSSSIRNNVTVVEAGRPFRP 274
Query: 134 VFVRGKCVNFSPAIINQTLERSVVEFVEEELS--LDTVAEEL 173
G NF+P ++N R E +E LS +D + +EL
Sbjct: 275 AMFEGVLRNFTPDVLNNMSGRPRQEDAQEFLSFIMDQMHDEL 316
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.339 0.147 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,055,168
Number of Sequences: 26719
Number of extensions: 326079
Number of successful extensions: 1236
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 1214
Number of HSP's gapped (non-prelim): 27
length of query: 385
length of database: 11,318,596
effective HSP length: 101
effective length of query: 284
effective length of database: 8,619,977
effective search space: 2448073468
effective search space used: 2448073468
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0101.13


