

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100c.1
(773 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
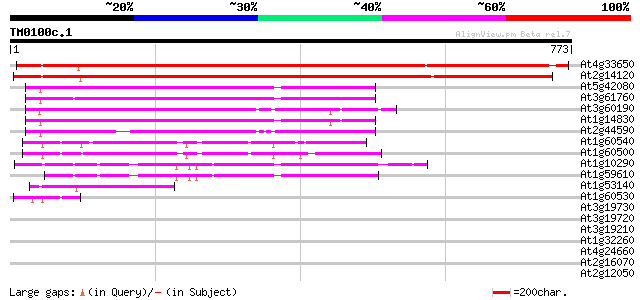
Score E
Sequences producing significant alignments: (bits) Value
At4g33650 dynamin like protein 2a (ADL2a) 1046 0.0
At2g14120 dynamin like protein 2b (ADL2b) 1025 0.0
At5g42080 dynamin-like protein (pir||S59558) 322 4e-88
At3g61760 dynamin-like protein B 312 5e-85
At3g60190 dynamin-like protein 4 (ADL4) 311 7e-85
At1g14830 dynamin-like protein 304 1e-82
At2g44590 dynamin-like protein D 273 3e-73
At1g60540 179 5e-45
At1g60500 178 9e-45
At1g10290 putative phragmoplastin 99 9e-21
At1g59610 dynamin-like protein CF1 94 4e-19
At1g53140 dynamin-like protein 75 2e-13
At1g60530 unknown protein 67 5e-11
At3g19730 dynamin-like protein 41 0.003
At3g19720 hypothetical protein 35 0.21
At3g19210 DNA repair protein, putative 33 0.61
At1g32260 hypothetical protein 33 0.79
At4g24660 unknown protein 31 2.3
At2g16070 unknown protein 31 3.0
At2g12050 hypothetical protein 31 3.0
>At4g33650 dynamin like protein 2a (ADL2a)
Length = 808
Score = 1046 bits (2705), Expect = 0.0
Identities = 541/769 (70%), Positives = 639/769 (82%), Gaps = 18/769 (2%)
Query: 10 TAASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGR 69
++++T A PLGSSVI +VN+LQDIF+++G+QSTI LPQV VVGSQSSGKSSVLE+LVGR
Sbjct: 24 SSSTTNAAPLGSSVIPIVNKLQDIFAQLGSQSTIA-LPQVVVVGSQSSGKSSVLEALVGR 82
Query: 70 DFLPRGTDICTRRPLVLQLVHTKP-----SHPEFGEFLHLPGRKFHDFTQIRHEIQAETD 124
DFLPRG DICTRRPLVLQL+ TK S E+GEF HLP +F+DF++IR EI+AET+
Sbjct: 83 DFLPRGNDICTRRPLVLQLLQTKSRANGGSDDEWGEFRHLPETRFYDFSEIRREIEAETN 142
Query: 125 REAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKI 184
R GENKGV++ QIRLKI SPNVL+ITLVDLPGITKVPVGDQPSDIEARIRTMI+SYIK
Sbjct: 143 RLVGENKGVADTQIRLKISSPNVLNITLVDLPGITKVPVGDQPSDIEARIRTMILSYIKQ 202
Query: 185 PTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIP 244
TCLILAVTPAN+DLANSDALQ+A + DPDG+RTIGVITKLDIMD+GTDAR LLLG V+P
Sbjct: 203 DTCLILAVTPANTDLANSDALQIASIVDPDGHRTIGVITKLDIMDKGTDARKLLLGNVVP 262
Query: 245 LRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILA 304
LRLGYVGVVNR QEDI +NR++K+AL+AEE+FFRS PVY GLAD GVPQLAKKLN+IL
Sbjct: 263 LRLGYVGVVNRCQEDILLNRTVKEALLAEEKFFRSHPVYHGLADRLGVPQLAKKLNQILV 322
Query: 305 QHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGK 364
QHIK +LP LK+RIS +LVA AKEH SYGE+TES+AGQGAL+LN LSKYC+A+SS+LEGK
Sbjct: 323 QHIKVLLPDLKSRISNALVATAKEHQSYGELTESRAGQGALLLNFLSKYCEAYSSLLEGK 382
Query: 365 NKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEV 424
++EMST ELSGGARIHYIFQSI+V+SLE VDPCEDLTDDDIRTA+QNATGP+SALF+P+V
Sbjct: 383 SEEMSTSELSGGARIHYIFQSIFVKSLEEVDPCEDLTDDDIRTAIQNATGPRSALFVPDV 442
Query: 425 PFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRD 484
PFE+ VRRQISRLLDPSLQCARFI++EL+KISHRCM ELQRFP LRK MDEVIG+FLR+
Sbjct: 443 PFEVLVRRQISRLLDPSLQCARFIFEELIKISHRCMMNELQRFPVLRKRMDEVIGDFLRE 502
Query: 485 GLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVS--LDALESD 542
GLEPSE MI II+MEMDYINTSHPNFIGG+KA+EAA+ Q K SR+ PV+ D +E D
Sbjct: 503 GLEPSEAMIGDIIDMEMDYINTSHPNFIGGTKAVEAAMHQVKSSRIPHPVARPKDTVEPD 562
Query: 543 KGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDNR 602
+ +S KSR + RQANG+V D GV A D EK P+ NA + W I SIF G D R
Sbjct: 563 RTSSSTSQVKSRSFLGRQANGIVTDQGVVSA-DAEKAQPAANASDTRWGIPSIFRGGDTR 621
Query: 603 VYVKENTSTKPHAEPVHNVQSS-STIHLREPPPVLRPSESNSEMEGVEITVTKLLLRSYY 661
K++ KP +E V ++ + S I+L+EPP VLRP+E++SE E VEI +TKLLLRSYY
Sbjct: 622 AVTKDSLLNKPFSEAVEDMSHNLSMIYLKEPPAVLRPTETHSEQEAVEIQITKLLLRSYY 681
Query: 662 DIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCRE 721
DI RKNIED VPKAIMHFLVN+TKRELHNVFI+KLYRE+LFEEMLQEP E+A KRKR +E
Sbjct: 682 DIVRKNIEDSVPKAIMHFLVNHTKRELHNVFIKKLYRENLFEEMLQEPDEIAVKRKRTQE 741
Query: 722 LLRAYQQAFRDLDELPLEAETVERGYGSPEATGLPRIRGLPSSSMYSSS 770
L QQA+R LDELPLEA++V + G+ + + L +SS YS+S
Sbjct: 742 TLHVLQQAYRTLDELPLEADSV--------SAGMSKHQELLTSSKYSTS 782
>At2g14120 dynamin like protein 2b (ADL2b)
Length = 780
Score = 1025 bits (2650), Expect = 0.0
Identities = 534/752 (71%), Positives = 629/752 (83%), Gaps = 14/752 (1%)
Query: 6 DEAVTAASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLES 65
D+ ++++A TPLGSSVI +VN+LQDIF+++G+QSTI LPQVAVVGSQSSGKSSVLE+
Sbjct: 4 DDLPPSSASAVTPLGSSVIPIVNKLQDIFAQLGSQSTIA-LPQVAVVGSQSSGKSSVLEA 62
Query: 66 LVGRDFLPRGTDICTRRPLVLQLVHTKPSHP-----EFGEFLHL-PGRKFHDFTQIRHEI 119
LVGRDFLPRG DICTRRPL LQLV TKPS E+GEFLH P R+ +DF++IR EI
Sbjct: 63 LVGRDFLPRGNDICTRRPLRLQLVQTKPSSDGGSDEEWGEFLHHDPVRRIYDFSEIRREI 122
Query: 120 QAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIM 179
+AET+R +GENKGVS+ I LKIFSPNVLDI+LVDLPGITKVPVGDQPSDIEARIRTMI+
Sbjct: 123 EAETNRVSGENKGVSDIPIGLKIFSPNVLDISLVDLPGITKVPVGDQPSDIEARIRTMIL 182
Query: 180 SYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLL 239
+YIK P+CLILAV+PAN+DLANSDALQ+A ADPDG+RTIGVITKLDIMDRGTDARN LL
Sbjct: 183 TYIKEPSCLILAVSPANTDLANSDALQIAGNADPDGHRTIGVITKLDIMDRGTDARNHLL 242
Query: 240 GKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKL 299
GK IPLRLGYVGVVNRSQEDI MNRSIKDALVAEE+FFRSRPVYSGL D GVPQLAKKL
Sbjct: 243 GKTIPLRLGYVGVVNRSQEDILMNRSIKDALVAEEKFFRSRPVYSGLTDRLGVPQLAKKL 302
Query: 300 NKILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSS 359
N++L QHIKA+LP LK+RI+ +L A AKE+ SYG+ITES+ GQGAL+L+ ++KYC+A+SS
Sbjct: 303 NQVLVQHIKALLPSLKSRINNALFATAKEYESYGDITESRGGQGALLLSFITKYCEAYSS 362
Query: 360 MLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSAL 419
LEGK+KEMST ELSGGARI YIFQS++V+SLE VDPCEDLT DDIRTA+QNATGP+SAL
Sbjct: 363 TLEGKSKEMSTSELSGGARILYIFQSVFVKSLEEVDPCEDLTADDIRTAIQNATGPRSAL 422
Query: 420 FIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIG 479
F+P+VPFE+ VRRQISRLLDPSLQCARFI+DEL+KISH+CM ELQRFP L+K MDEVIG
Sbjct: 423 FVPDVPFEVLVRRQISRLLDPSLQCARFIFDELVKISHQCMMKELQRFPVLQKRMDEVIG 482
Query: 480 NFLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVS--LD 537
NFLR+GLEPS+ MI +IEMEMDYINTSHPNFIGG+KA+E A+Q K SR+ PV+ D
Sbjct: 483 NFLREGLEPSQAMIRDLIEMEMDYINTSHPNFIGGTKAVEQAMQTVKSSRIPHPVARPRD 542
Query: 538 ALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWR-ISSIF 596
+E ++ +S K+R + RQANG++ D V A+D E+ AP AG +SW SSIF
Sbjct: 543 TVEPERTASSGSQIKTRSFLGRQANGIITDQAVPTAADAERPAP---AGSTSWSGFSSIF 599
Query: 597 GGRDNRVYVKENTSTKPHAEPVHNV-QSSSTIHLREPPPVLRPSESNSEMEGVEITVTKL 655
G D + K N KP +E V Q+ STI+L+EPP +L+ SE++SE E VEI +TKL
Sbjct: 600 RGSDGQAAAKNNLLNKPFSETTQEVYQNLSTIYLKEPPTILKSSETHSEQESVEIEITKL 659
Query: 656 LLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAK 715
LL+SYYDI RKN+EDLVPKAIMHFLVN TKRELHNVFIEKLYRE+L EE+L+EP E+A K
Sbjct: 660 LLKSYYDIVRKNVEDLVPKAIMHFLVNYTKRELHNVFIEKLYRENLIEELLKEPDELAIK 719
Query: 716 RKRCRELLRAYQQAFRDLDELPLEAETVERGY 747
RKR +E LR QQA R LDELPLEAE+VERGY
Sbjct: 720 RKRTQETLRILQQANRTLDELPLEAESVERGY 751
>At5g42080 dynamin-like protein (pir||S59558)
Length = 610
Score = 322 bits (826), Expect = 4e-88
Identities = 191/493 (38%), Positives = 286/493 (57%), Gaps = 18/493 (3%)
Query: 22 SVISLVNRLQDIFSRVGNQS------TIID-LPQVAVVGSQSSGKSSVLESLVGRDFLPR 74
++ISLVN++Q + +G+ T+ D LP +AVVG QSSGKSSVLES+VG+DFLPR
Sbjct: 3 NLISLVNKIQRACTALGDHGDSSALPTLWDSLPAIAVVGGQSSGKSSVLESIVGKDFLPR 62
Query: 75 GTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVS 134
G+ I TRRPLVLQL E+ EFLHLP +KF DF +R EIQ ETDRE G +K +S
Sbjct: 63 GSGIVTRRPLVLQLQKIDDGTREYAEFLHLPRKKFTDFAAVRKEIQDETDRETGRSKAIS 122
Query: 135 EKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTP 194
I L I+SPNV+++TL+DLPG+TKV V Q I I M+ SYI+ P C+ILA++P
Sbjct: 123 SVPIHLSIYSPNVVNLTLIDLPGLTKVAVDGQSDSIVKDIENMVRSYIEKPNCIILAISP 182
Query: 195 ANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVN 254
AN DLA SDA+++++ DP G+RT GV+TK+D+MD+GTDA +L G+ L+ +VGVVN
Sbjct: 183 ANQDLATSDAIKISREVDPSGDRTFGVLTKIDLMDKGTDAVEILEGRSFKLKYPWVGVVN 242
Query: 255 RSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGL 314
RSQ DI N + A E E+F + Y LA+ G LAK L+K L + IK+ +PG+
Sbjct: 243 RSQADINKNVDMIAARKREREYFSNTTEYRHLANKMGSEHLAKMLSKHLERVIKSRIPGI 302
Query: 315 KARISTSLVALAKEHASYGE-ITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCEL 373
++ I+ +++ L E + G+ I G+ ++ I + F L+G
Sbjct: 303 QSLINKTVLELETELSRLGKPIAADAGGKLYSIMEICRLFDQIFKEHLDGVR-------- 354
Query: 374 SGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQ 433
+GG +++ +F + +L+ + + L D+IR + A G + L PE + +
Sbjct: 355 AGGEKVYNVFDNQLPAALKRLQFDKQLAMDNIRKLVTEADGYQPHLIAPEQGYRRLIESS 414
Query: 434 ISRLLDPSLQCARFIYDELMKISHRCM--FTELQRFPFLRKHMDEVIGNFLRDGLEPSET 491
I + P+ ++ L + H+ + EL+++P LR + L E S+
Sbjct: 415 IVSIRGPAEASVDTVHAILKDLVHKSVNETVELKQYPALRVEVTNAAIESLDKMREGSKK 474
Query: 492 MITHIIEMEMDYI 504
+++ME Y+
Sbjct: 475 ATLQLVDMECSYL 487
>At3g61760 dynamin-like protein B
Length = 610
Score = 312 bits (799), Expect = 5e-85
Identities = 188/494 (38%), Positives = 289/494 (58%), Gaps = 20/494 (4%)
Query: 22 SVISLVNRLQDIFSRVGNQS------TIID-LPQVAVVGSQSSGKSSVLESLVGRDFLPR 74
S+I+LVN++Q + +G+ T+ D LP +AVVG QSSGKSSVLES+VG+DFLPR
Sbjct: 3 SLIALVNKIQRACTALGDHGEGSSLPTLWDSLPAIAVVGGQSSGKSSVLESVVGKDFLPR 62
Query: 75 GTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGEN-KGV 133
G I TRRPLVLQL H E+ EF+HLP +KF DF +R EI ETDRE G + K +
Sbjct: 63 GAGIVTRRPLVLQL-HRIDEGKEYAEFMHLPKKKFTDFAAVRQEISDETDRETGRSSKVI 121
Query: 134 SEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVT 193
S I L IFSPNV+++TLVDLPG+TKV V QP I I M+ S+I+ P C+ILA++
Sbjct: 122 STVPIHLSIFSPNVVNLTLVDLPGLTKVAVDGQPESIVQDIENMVRSFIEKPNCIILAIS 181
Query: 194 PANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVV 253
PAN DLA SDA+++++ DP G+RT GV+TK+D+MD+GT+A ++L G+ LR +VGVV
Sbjct: 182 PANQDLATSDAIKISREVDPKGDRTFGVLTKIDLMDQGTNAVDILEGRGYKLRYPWVGVV 241
Query: 254 NRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPG 313
NRSQ DI + + A E ++F++ P Y L + G L K L+K L IK+ +PG
Sbjct: 242 NRSQADINKSVDMIAARRRERDYFQTSPEYRHLTERMGSEYLGKMLSKHLEVVIKSRIPG 301
Query: 314 LKARISTSLVALAKEHASYGE-ITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCE 372
L++ I+ ++ L E + G+ + G+ +++ I + F L+G
Sbjct: 302 LQSLITKTISELETELSRLGKPVAADAGGKLYMIMEICRAFDQTFKEHLDGTR------- 354
Query: 373 LSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRR 432
SGG +I+ +F + + +++ + + L+ D++R + A G + L PE + +
Sbjct: 355 -SGGEKINSVFDNQFPAAIKRLQFDKHLSMDNVRKLITEADGYQPHLIAPEQGYRRLIES 413
Query: 433 QISRLLDPSLQCARFIYDELMKISHRCM--FTELQRFPFLRKHMDEVIGNFLRDGLEPSE 490
+ + P+ ++ L + H+ M +EL+++P LR + + L + S
Sbjct: 414 CLVSIRGPAEAAVDAVHSILKDLIHKSMGETSELKQYPTLRVEVSGAAVDSLDRMRDESR 473
Query: 491 TMITHIIEMEMDYI 504
+++ME Y+
Sbjct: 474 KATLLLVDMESGYL 487
>At3g60190 dynamin-like protein 4 (ADL4)
Length = 624
Score = 311 bits (798), Expect = 7e-85
Identities = 202/524 (38%), Positives = 288/524 (54%), Gaps = 23/524 (4%)
Query: 22 SVISLVNRLQDIFSRVGN----------QSTIIDLPQVAVVGSQSSGKSSVLESLVGRDF 71
S+I LVNR+Q + +G+ S LP VAVVG QSSGKSSVLES+VGRDF
Sbjct: 6 SLIGLVNRIQRACTVLGDYGGGTGSNAFNSLWEALPTVAVVGGQSSGKSSVLESIVGRDF 65
Query: 72 LPRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENK 131
LPRG+ I TRRPLVLQL T E+ EFLHLP ++F DF +R EIQ ETDR G+NK
Sbjct: 66 LPRGSGIVTRRPLVLQLHKTDDGTEEYAEFLHLPKKQFTDFALVRREIQDETDRITGKNK 125
Query: 132 GVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILA 191
+S I L I+SPNV+++TL+DLPG+TKV V QP I I +M+ +Y+ P C+ILA
Sbjct: 126 QISPVPIHLSIYSPNVVNLTLIDLPGLTKVAVEGQPETIAEDIESMVRTYVDKPNCIILA 185
Query: 192 VTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVG 251
++PAN D+A SDA+++A+ DP G RT GV+TKLD+MD+GT+A +L G+ L+ +VG
Sbjct: 186 ISPANQDIATSDAIKLAKDVDPTGERTFGVLTKLDLMDKGTNALEVLEGRSYRLQHPWVG 245
Query: 252 VVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVL 311
+VNRSQ DI N + A E E+F + P Y LA G LAK L+K L I+ +
Sbjct: 246 IVNRSQADINKNVDMMLARRKEREYFDTSPDYGHLASKMGSEYLAKLLSKHLESVIRTRI 305
Query: 312 PGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTC 371
P + + I+ S+ L +E G A GA + IL + C AF + KE
Sbjct: 306 PSILSLINKSIEELERELDRMGRPVAVDA--GAQLYTIL-EMCRAFDKIF----KEHLDG 358
Query: 372 ELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVR 431
GG RI+ +F + +L+ + L+ ++ + A G + L PE + +
Sbjct: 359 GRPGGDRIYGVFDNQLPAALKKLPFDRHLSLQSVKKIVSEADGYQPHLIAPEQGYRRLIE 418
Query: 432 RQISRLLDP---SLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEP 488
+ P S+ ++ EL++ S EL+RFP L+ + + L E
Sbjct: 419 GALGYFRGPAEASVDAVHYVLKELVRKS-ISETEELKRFPSLQVELAAAANSSLEKFREE 477
Query: 489 SETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSP 532
S+ + +++ME Y+ F + +E V +K SP
Sbjct: 478 SKKSVIRLVDMESAYLTAEF--FRKLPQEIERPVTNSKNQTASP 519
Score = 32.3 bits (72), Expect = 1.0
Identities = 17/78 (21%), Positives = 38/78 (47%), Gaps = 2/78 (2%)
Query: 659 SYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYRED--LFEEMLQEPAEVAAKR 716
+Y ++ + + +PKA ++ V K L N F ++ + + ++L E + +R
Sbjct: 541 AYVNMVSDTLRNTIPKACVYCQVRQAKLALLNYFYSQISKREGKQLGQLLDEDPALMDRR 600
Query: 717 KRCRELLRAYQQAFRDLD 734
C + L Y++A ++D
Sbjct: 601 LECAKRLELYKKARDEID 618
>At1g14830 dynamin-like protein
Length = 614
Score = 304 bits (779), Expect = 1e-82
Identities = 188/492 (38%), Positives = 273/492 (55%), Gaps = 18/492 (3%)
Query: 22 SVISLVNRLQDIFSRVGNQ-----STIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGT 76
S+I L+N++Q + +G+ S LP VAVVG QSSGKSSVLES+VGRDFLPRG+
Sbjct: 6 SLIGLINKIQRACTVLGDHGGEGMSLWEALPTVAVVGGQSSGKSSVLESVVGRDFLPRGS 65
Query: 77 DICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEK 136
I TRRPLVLQL T+ E+ EFLH P ++F DF +R EI+ ETDR G++K +S
Sbjct: 66 GIVTRRPLVLQLHKTEDGTTEYAEFLHAPKKRFADFAAVRKEIEDETDRITGKSKQISNI 125
Query: 137 QIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPAN 196
I+L I+SPNV+++TL+DLPG+TKV V QP I I M+ SY++ P C+ILA++PAN
Sbjct: 126 PIQLSIYSPNVVNLTLIDLPGLTKVAVDGQPESIVQDIENMVRSYVEKPNCIILAISPAN 185
Query: 197 SDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRS 256
D+A SDA+++A+ DP G RT GV TKLDIMD+GTD ++L G+ L+ +VG+VNRS
Sbjct: 186 QDIATSDAIKLAREVDPTGERTFGVATKLDIMDKGTDCLDVLEGRSYRLQHPWVGIVNRS 245
Query: 257 QEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKA 316
Q DI + A E+E+F + P Y LA G LAK L++ L I+ +P + A
Sbjct: 246 QADINKRVDMIAARRKEQEYFETSPEYGHLASRMGSEYLAKLLSQHLETVIRQKIPSIVA 305
Query: 317 RISTSLVALAKEHASYGE-ITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSG 375
I+ S+ + E G I Q +L + + F L+G G
Sbjct: 306 LINKSIDEINAELDRIGRPIAVDSGAQLYTILELCRAFDRVFKEHLDGGR--------PG 357
Query: 376 GARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQIS 435
G RI+ +F +L+ + L+ +++ + A G + L PE + + IS
Sbjct: 358 GDRIYGVFDHQLPAALKKLPFDRHLSTKNVQKVVSEADGYQPHLIAPEQGYRRLIDGSIS 417
Query: 436 RLLDP---SLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETM 492
P ++ F+ EL++ S EL+RFP L + L + S
Sbjct: 418 YFKGPAEATVDAVHFVLKELVRKS-ISETEELKRFPTLASDIAAAANEALERFRDESRKT 476
Query: 493 ITHIIEMEMDYI 504
+ +++ME Y+
Sbjct: 477 VLRLVDMESSYL 488
Score = 35.8 bits (81), Expect = 0.094
Identities = 21/78 (26%), Positives = 40/78 (50%), Gaps = 2/78 (2%)
Query: 659 SYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYR--EDLFEEMLQEPAEVAAKR 716
+Y ++ + + +PKA+++ V KR L N F ++ R ++ ML E ++ +R
Sbjct: 532 AYINMVCDTLRNSLPKAVVYCQVREAKRSLLNFFYAQVGRKEKEKLGAMLDEDPQLMERR 591
Query: 717 KRCRELLRAYQQAFRDLD 734
+ L Y+QA D+D
Sbjct: 592 GTLAKRLELYKQARDDID 609
>At2g44590 dynamin-like protein D
Length = 596
Score = 273 bits (697), Expect = 3e-73
Identities = 180/493 (36%), Positives = 266/493 (53%), Gaps = 34/493 (6%)
Query: 22 SVISLVNRLQDIFSRVGNQ--------STIIDLPQVAVVGSQSSGKSSVLESLVGRDFLP 73
S+I L+N +Q + VG+ S LP VAVVG QSSGKSSVLES+VGRDFLP
Sbjct: 3 SLIVLINTIQRACTVVGDHGGDSNALSSLWEALPSVAVVGGQSSGKSSVLESIVGRDFLP 62
Query: 74 RGTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGV 133
RG+ I TRRPLVLQL T+ + EFLHL +KF +F+ +R EI+ ETDR G+NK +
Sbjct: 63 RGSGIVTRRPLVLQLHKTENGTEDNAEFLHLTNKKFTNFSLVRKEIEDETDRITGKNKQI 122
Query: 134 SEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVT 193
S I L IFSPN QP I I +M+ SY++ P CLILA++
Sbjct: 123 SSIPIHLSIFSPNE-----------------GQPETIVEDIESMVRSYVEKPNCLILAIS 165
Query: 194 PANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVV 253
PAN D+A SDA+++A+ DP G+RT GV+TKLD+MD+GT+A +++ G+ L+ +VG+V
Sbjct: 166 PANQDIATSDAMKLAKEVDPIGDRTFGVLTKLDLMDKGTNALDVINGRSYKLKYPWVGIV 225
Query: 254 NRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPG 313
NRSQ DI N + A E E+F + P Y LA G LAK L+K+L I++ +P
Sbjct: 226 NRSQADINKNVDMMVARRKEREYFETSPDYGHLATRMGSEYLAKLLSKLLESVIRSRIPS 285
Query: 314 LKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCEL 373
+ + I+ ++ L +E G A GA + IL C AF + KE
Sbjct: 286 ILSLINNNIEELERELDQLGRPIAIDA--GAQLYTILG-MCRAFEKIF----KEHLDGGR 338
Query: 374 SGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQ 433
GGARI+ IF +++ + L+ ++ + + G + L PE+ + +
Sbjct: 339 PGGARIYGIFDYNLPTAIKKLPFDRHLSLQSVKRIVSESDGYQPHLIAPELGYRRLIEGS 398
Query: 434 ISRLLDPSLQCARFIYDELMKISHRCM--FTELQRFPFLRKHMDEVIGNFLRDGLEPSET 491
++ P+ I+ L ++ + + EL+RFP L+ + + L E S
Sbjct: 399 LNHFRGPAEASVNAIHLILKELVRKAIAETEELKRFPSLQIELVAAANSSLDKFREESMK 458
Query: 492 MITHIIEMEMDYI 504
+ +++ME Y+
Sbjct: 459 SVLRLVDMESSYL 471
Score = 34.7 bits (78), Expect = 0.21
Identities = 19/79 (24%), Positives = 39/79 (49%), Gaps = 3/79 (3%)
Query: 659 SYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVF---IEKLYREDLFEEMLQEPAEVAAK 715
+Y + + + + +PKA++H V K L N F I + + ++L E + +
Sbjct: 512 AYIKMVAETLVNTIPKAVVHCQVRQAKLSLLNYFYAQISQSQQGKRLGQLLDENPALMER 571
Query: 716 RKRCRELLRAYQQAFRDLD 734
R +C + L Y++A ++D
Sbjct: 572 RMQCAKRLELYKKARDEID 590
>At1g60540
Length = 648
Score = 179 bits (454), Expect = 5e-45
Identities = 151/495 (30%), Positives = 240/495 (47%), Gaps = 45/495 (9%)
Query: 18 PLGSSVISLVNRLQDIFSRVGNQSTI---IDLPQVAVVGSQSSGKSSVLESLVGRDFLPR 74
P+ SS + L D ++ N + + I LP + VVG QSSGKSSVLESL G LPR
Sbjct: 35 PIVSSYNDHIRPLLDTVDKLRNLNVMQEGIQLPTIVVVGDQSSGKSSVLESLAGIS-LPR 93
Query: 75 GTDICTRRPLVLQLVHTKPSHPE----FGEFLHLPGRKFHDFTQIRHEIQAETDREAGEN 130
G ICTR PLV++L ++ PE +G+ + +P + H I I A TD AG
Sbjct: 94 GQGICTRVPLVMRLQRSRSPEPEIWLEYGDKI-VPTDEEH----IAQTICAATDVIAGSG 148
Query: 131 KGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLIL 190
KGVS+ + L + V DIT+VDLPGIT+VPV QP +I +I M+M YI+ +IL
Sbjct: 149 KGVSDTPLTLHVKKAGVPDITMVDLPGITRVPVNGQPENIYEQISRMVMKYIEPQESIIL 208
Query: 191 AVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKV----IPLR 246
V A D ++++M++ D G RT+ V+TK D+ G LL KV + +
Sbjct: 209 NVLSATVDFTTCESIRMSKQVDKTGERTLAVVTKADMAPEG------LLQKVTADDVSIG 262
Query: 247 LGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADS-CGVPQLAKKLNKILAQ 305
LGYV V NR E+ + ++A + EE FR+ P+ S + D G+P LA+KL +I
Sbjct: 263 LGYVCVRNRIGEE-----TYEEARMQEELLFRTHPMLSMINDEIVGIPVLAQKLTQIQGM 317
Query: 306 HIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSML---- 361
I LP ++ +I+ + E + ++ A G +++++ A S+L
Sbjct: 318 MISRCLPEIERKINVKMEISVLE---FNKLPIVMASTGEALMSLMDIIGSAKESLLRILV 374
Query: 362 -----EGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPK 416
E N + C ++ ++ + +AV E+ D+I+ L+
Sbjct: 375 HGDFSEYPNDQKMHCTARLAEMLNRFSDNLQAQP-QAV--TEEFLMDEIK-VLEERKCIG 430
Query: 417 SALFIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDE 476
FIP F + + + + D ++ + I+D + + + T FP ++ +
Sbjct: 431 LPNFIPRSSFLAILSQHVDGIQDKPVEFIKEIWDYIEVVLSSVITTYSDNFPQIQSSIKR 490
Query: 477 VIGNFLRDGLEPSET 491
N + E S+T
Sbjct: 491 AGRNLMSRTKERSKT 505
>At1g60500
Length = 669
Score = 178 bits (452), Expect = 9e-45
Identities = 156/516 (30%), Positives = 236/516 (45%), Gaps = 46/516 (8%)
Query: 18 PLGSSVISLVNRLQDIFSRVGNQSTI---IDLPQVAVVGSQSSGKSSVLESLVGRDFLPR 74
P+ SS + L D R+ N + + I LP + VVG QSSGKSSVLESL G LPR
Sbjct: 34 PIISSYNDRIRPLLDTVDRLRNLNVMREGIHLPTIVVVGDQSSGKSSVLESLAGIS-LPR 92
Query: 75 GTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFH-DFTQIRHEIQAETDREAGENKGV 133
G ICTR PLV++L + PE +L + D I I+A TD AG KGV
Sbjct: 93 GQGICTRVPLVMRLQRSSSPEPEI--WLEYNDKVVPTDEEHIAEAIRAATDVIAGSGKGV 150
Query: 134 SEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVT 193
S+ + L + V D+T+VDLPGIT+VPV QP +I +I MIM YI+ +IL V
Sbjct: 151 SDAPLTLHVKKAGVPDLTMVDLPGITRVPVNGQPENIYEQISGMIMEYIEPQESIILNVL 210
Query: 194 PANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKV----IPLRLGY 249
A D ++++M++ D G RT+ V+TK D+ G LL KV + + LGY
Sbjct: 211 SATVDFTTCESIRMSRKVDKTGQRTLAVVTKADMAPEG------LLQKVTADDVSIVLGY 264
Query: 250 VGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGL-ADSCGVPQLAKKLNKILAQHIK 308
V V NR E+ + ++A + EE FR+ PV S + D G+P LA+KL I + I
Sbjct: 265 VCVRNRIGEE-----TYEEARMQEELLFRTHPVLSLIDEDIVGIPVLAQKLMLIQSSMIA 319
Query: 309 AVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSML------- 361
LP + ++I+ L E ++ A G ++ ++ A S+L
Sbjct: 320 RCLPKIVSKINQKLDTAVLE---LNKLPMVMASTGEALMALMDIIGSAKESLLRILVQGD 376
Query: 362 --EGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCED---LTDDDIRTALQNATGPK 416
E + + C + S+ + E + D + D+ L N
Sbjct: 377 FSEYPDDQNMHCTARLADMLSQFSDSLQAKPKEVAEFLMDEIKILDECKCVGLPN----- 431
Query: 417 SALFIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDE 476
FIP F + + + + D ++ I+D + + FP ++ +
Sbjct: 432 ---FIPRSAFLAILSQHVDGIQDKPVEFINKIWDYIEDVLSSVTAKRSDNFPQIQSSIKR 488
Query: 477 VIGNFLRDGLEPSETMITHIIEMEMDYINTSHPNFI 512
N + E S + I+EME T +P ++
Sbjct: 489 AGRNLISKIKEQSVNRVMEIVEMEKLTDYTCNPEYM 524
>At1g10290 putative phragmoplastin
Length = 914
Score = 99.0 bits (245), Expect = 9e-21
Identities = 130/587 (22%), Positives = 245/587 (41%), Gaps = 56/587 (9%)
Query: 7 EAVTAASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESL 66
EA+ S + + + L + D S +T ++ V +G+ +GKS+VL SL
Sbjct: 2 EAIDELSQLSDSMKQAASLLADEDPDETSSSKRPATFLN---VVALGNVGAGKSAVLNSL 58
Query: 67 VGRDFLPRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFH-DFTQIRHEIQAETDR 125
+G LP G + TR P++++L + S L + + + +RH +Q DR
Sbjct: 59 IGHPVLPTGENGATRAPIIIEL-SRESSLSSKAIILQIDNKSQQVSASALRHSLQ---DR 114
Query: 126 EAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIP 185
+ G + +I LK+ + + LVDLPG+ + V + +MI Y +
Sbjct: 115 LSKGASGKNRDEINLKLRTSTAPPLKLVDLPGLDQRIVDE----------SMIAEYAQHN 164
Query: 186 TCLILAVTPAN--SDLANSDALQMAQVADPDGNRTIGVITKLDIM---DRGTDARNLLLG 240
++L + PA+ S++++S AL++A+ DP+ RTIG+I K+D + A LL
Sbjct: 165 DAILLVIVPASQASEISSSRALKIAKEYDPESTRTIGIIGKIDQAAENSKALAAVQALLS 224
Query: 241 KVIPLR---LGYVGVVNR-----SQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADS-CG 291
P + + +V V+ + S + S++ A AE E +S + +G S G
Sbjct: 225 NQGPPKTTDIPWVAVIGQSVSIASAQSGSGENSLETAWRAESESLKS--ILTGAPQSKLG 282
Query: 292 VPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYGE-ITESKAGQGALVLNIL 350
L L + +K LP + + + + E A GE + S G A+ L +
Sbjct: 283 RIALVDTLASQIRSRMKLRLPSVLSGLQGKSQIVQDELARLGEQLVNSAEGTRAIALELC 342
Query: 351 SKYCDAFSSMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQ 410
++ D F L G G ++ F+ + ++ + ++++ +
Sbjct: 343 REFEDKFLLHLAGGE--------GSGWKVVASFEGNFPNRIKQLPLDRHFDLNNVKRVVL 394
Query: 411 NATGPKSALFIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKI--SHRCMFTELQRFP 468
A G + L PE ++ + DP+ C ++ L+ I + L R+P
Sbjct: 395 EADGYQPYLISPEKGLRSLIKIVLELAKDPARLCVDEVHRVLVDIVSASANATPGLGRYP 454
Query: 469 FLRKHMDEVIGNFLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPS 528
++ + + L ++ M+ +++ME ++ H FI + Q +
Sbjct: 455 PFKREVVAIASAALDGFKNEAKKMVVALVDMERAFVPPQH--FI--------RLVQRRME 504
Query: 529 RVSPPVSLDALESDKGLASERNGKSRGIIARQANGVVADHGVRGASD 575
R L S KG +E++ SR + Q +G A ++ D
Sbjct: 505 RQRREEELKGRSSKKGQDAEQSLLSRA-TSPQPDGPTAGGSLKSMKD 550
>At1g59610 dynamin-like protein CF1
Length = 920
Score = 93.6 bits (231), Expect = 4e-19
Identities = 106/478 (22%), Positives = 204/478 (42%), Gaps = 42/478 (8%)
Query: 49 VAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRK 108
V +G+ +GKS+VL SL+G LP G + TR P+++ L + S L + +
Sbjct: 41 VVALGNVGAGKSAVLNSLIGHPVLPTGENGATRAPIIIDL-SREESLSSKAIILQIDNKN 99
Query: 109 FH-DFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQP 167
+ +RH +Q DR + G +I LK+ + + L+DLPG+ + V D
Sbjct: 100 QQVSASALRHSLQ---DRLSKGASGRGRDEIYLKLRTSTAPPLKLIDLPGLDQRIVDD-- 154
Query: 168 SDIEARIRTMIMSYIKIPTCLILAVTPAN--SDLANSDALQMAQVADPDGNRTIGVITKL 225
+MI + + ++L V PA+ S++++S AL++A+ DP+ RT+G+I+K+
Sbjct: 155 --------SMIGEHAQHNDAILLVVVPASQASEISSSRALKIAKEYDPESTRTVGIISKI 206
Query: 226 DIM---DRGTDARNLLLGKVIPLR---LGYVGVVNR-----SQEDIQMNRSIKDALVAEE 274
D + A LL P + + +V ++ + S + S++ A AE
Sbjct: 207 DQAAENPKSLAAVQALLSNQGPPKTTDIPWVALIGQSVSIASAQSGGSENSLETAWRAES 266
Query: 275 EFFRSRPVYSGLADS-CGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYG 333
E +S + +G S G L L + +K LP + + + E A G
Sbjct: 267 ESLKS--ILTGAPQSKLGRIALVDTLASQIRSRMKLRLPNILTGLQGKSQIVQDELARLG 324
Query: 334 E-ITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLE 392
E + S G A+ L + ++ D F L G G ++ F+ + ++
Sbjct: 325 EQLVSSAEGTRAIALELCREFEDKFLLHLAGGE--------GSGWKVVASFEGNFPNRIK 376
Query: 393 AVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPSLQCARFIYDEL 452
+ ++++ + A G + L PE ++ + DP+ C ++ L
Sbjct: 377 KLPLDRHFDLNNVKRIVLEADGYQPYLISPEKGLRSLIKTVLELAKDPARLCVDEVHRVL 436
Query: 453 MKI--SHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEMDYINTSH 508
+ I + L R+P ++ + + L ++ M+ +++ME ++ H
Sbjct: 437 VDIVSASANATPGLGRYPPFKREVVAIASAALDGFKNEAKKMVVALVDMERAFVPPQH 494
>At1g53140 dynamin-like protein
Length = 260
Score = 74.7 bits (182), Expect = 2e-13
Identities = 58/204 (28%), Positives = 97/204 (47%), Gaps = 8/204 (3%)
Query: 28 NRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPLVLQ 87
NRLQ G + + +P++ +G QS GKSS+LE+L+G F R ++ TRRPL+LQ
Sbjct: 47 NRLQAAAVAFGEK---LPIPEIVAIGGQSDGKSSLLEALLGFRFNVREVEMGTRRPLILQ 103
Query: 88 LVH----TKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENK-GVSEKQIRLKI 142
+VH +P E G T + I++ T+ + K VS K I ++
Sbjct: 104 MVHDLSALEPRCRFQDEDSEEYGSPIVSATAVADVIRSRTEALLKKTKTAVSPKPIVMRA 163
Query: 143 FSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANS 202
+ ++T++D PG +P I +M+ S P ++L + ++ + +S
Sbjct: 164 EYAHCPNLTIIDTPGFVLKAKKGEPETTPDEILSMVKSLASPPHRILLFLQQSSVEWCSS 223
Query: 203 DALQMAQVADPDGNRTIGVITKLD 226
L + D RTI V++K D
Sbjct: 224 LWLDAVREIDSSFRRTIVVVSKFD 247
>At1g60530 unknown protein
Length = 175
Score = 66.6 bits (161), Expect = 5e-11
Identities = 42/99 (42%), Positives = 58/99 (58%), Gaps = 8/99 (8%)
Query: 6 DEAVTAASTAATPLGSSVISLVNR----LQDIFSRVGNQSTI---IDLPQVAVVGSQSSG 58
+ +++ S TP+ + ++S N L D R+ N + + I LP + VVG QSSG
Sbjct: 15 EAGMSSLSIVNTPIEAPIVSSYNDRIRPLLDTVDRLRNLNVMREGIQLPTIVVVGDQSSG 74
Query: 59 KSSVLESLVGRDFLPRGTDICTRRPLVLQLVHTKPSHPE 97
KSSVLESL G + LPRG ICTR PLV++L + PE
Sbjct: 75 KSSVLESLAGIN-LPRGQGICTRVPLVMRLQRSSSPEPE 112
>At3g19730 dynamin-like protein
Length = 157
Score = 40.8 bits (94), Expect = 0.003
Identities = 22/64 (34%), Positives = 34/64 (52%)
Query: 25 SLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPL 84
SL ++ + T + P V VVG Q+ GKS+++E+L+G F G TRRP+
Sbjct: 27 SLYEAYNELHALAQELETPFEAPAVLVVGQQTDGKSALVEALMGFQFNHVGGGTKTRRPI 86
Query: 85 VLQL 88
L +
Sbjct: 87 TLHM 90
>At3g19720 hypothetical protein
Length = 649
Score = 34.7 bits (78), Expect = 0.21
Identities = 54/251 (21%), Positives = 106/251 (41%), Gaps = 31/251 (12%)
Query: 134 SEKQIRLKIFSPNVLDITLVDLPGITKVPVG--DQPSDIEAR-IRTMIMSYIKIPTCLIL 190
S K+I +K+ ++T++D PG+ G ++ ++AR + ++ + ++ +IL
Sbjct: 12 SAKEIIVKVQYKYCPNLTIIDTPGLIAPAPGLKNRALQVQARAVEALVRAKMQHKEFIIL 71
Query: 191 AVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDI----MDRGTDARNLLLGKVIPLR 246
+ + SD + + ++ DP+ +RTI V TKLD +D L L
Sbjct: 72 CLEDS-SDWSIATTRRIVMQVDPELSRTIVVSTKLDTKIPQFSCSSDVEVFLSPPASALD 130
Query: 247 LGYVGVVNRSQEDIQMNRSIKDALV--AEEEFFRSRPVYS---GLADSCGVPQLAKKLNK 301
+G D S+ V ++ ++S + L + + L KKL +
Sbjct: 131 SSLLG-------DSPFFTSVPSGRVGYGQDSVYKSNDEFKQAVSLREMEDIASLEKKLGR 183
Query: 302 ILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSML 361
+L + K+RI S + L E + ES L++ +L K + L
Sbjct: 184 LLTKQ-------EKSRIGISKLRLFLEELLWKRYKESV----PLIIPLLGKEYRSTVRKL 232
Query: 362 EGKNKEMSTCE 372
+ +KE+S+ +
Sbjct: 233 DTVSKELSSLD 243
>At3g19210 DNA repair protein, putative
Length = 688
Score = 33.1 bits (74), Expect = 0.61
Identities = 40/182 (21%), Positives = 73/182 (39%), Gaps = 22/182 (12%)
Query: 391 LEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPSLQCARFIYD 450
++ + CE T DD+ + + + T P+SAL + + +E F R S S C I D
Sbjct: 162 IQLIALCES-TRDDVLSGIDSFTRPRSALQVLIISYETF--RMHSSKFCQSESCDLLICD 218
Query: 451 ELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEMDYINTSHPN 510
E ++ + T K + G +++ LE M +N ++P
Sbjct: 219 EAHRLKNDQTLTNRALASLTCKRRVLLSGTPMQNDLEEFFAM-----------VNFTNPG 267
Query: 511 FIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGLASER----NGKSRGIIARQANGVVA 566
+G + + P A E +K LA++R + K I R+ N +++
Sbjct: 268 SLGDAAHFRHYYEAPIICGREP----TATEEEKNLAADRSAELSSKVNQFILRRTNALLS 323
Query: 567 DH 568
+H
Sbjct: 324 NH 325
>At1g32260 hypothetical protein
Length = 202
Score = 32.7 bits (73), Expect = 0.79
Identities = 19/52 (36%), Positives = 23/52 (43%)
Query: 81 RRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKG 132
R L +Q P HP+ +FLH RK D H+ E D E G N G
Sbjct: 21 RLALKIQRRLVTPKHPKIRQFLHRRTRKIFDVAISVHKNIQERDIEVGRNLG 72
>At4g24660 unknown protein
Length = 220
Score = 31.2 bits (69), Expect = 2.3
Identities = 34/140 (24%), Positives = 51/140 (36%), Gaps = 28/140 (20%)
Query: 528 SRVSPPVSLDALESDKGLASERNG--KSRGIIAR------------QANGVVAD------ 567
S V+PP D+L + +S G +S+G+ A+ G D
Sbjct: 13 SGVNPPCGYDSLSGEGATSSGGGGVGRSKGVGAKIRYRECLKNHAVNIGGHAVDGCCEFM 72
Query: 568 -HGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDNRVYVKENTSTKPHAEPVHNVQSSST 626
G G D K A G + + GGR +RV N +PH P +
Sbjct: 73 PSGEDGTLDALKCAACGCHRNFHRKETESIGGRAHRVPTYYNRPPQPHQPPGY------- 125
Query: 627 IHLREPPPVLRPSESNSEME 646
+HL P RP ++ + E
Sbjct: 126 LHLTSPAAPYRPPAASGDEE 145
>At2g16070 unknown protein
Length = 307
Score = 30.8 bits (68), Expect = 3.0
Identities = 24/71 (33%), Positives = 36/71 (49%), Gaps = 8/71 (11%)
Query: 527 PSRVSPPVSLDALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAG 586
P V PP+SL L+++ G S K + ANG + H VR ++ E +P+G +G
Sbjct: 154 PYPVHPPLSL-GLDNNNGYLSHLPSKKKS----DANGFGSGH-VR--NEAEAKSPNGGSG 205
Query: 587 GSSWRISSIFG 597
GSS + G
Sbjct: 206 GSSHGVIRFLG 216
>At2g12050 hypothetical protein
Length = 139
Score = 30.8 bits (68), Expect = 3.0
Identities = 23/73 (31%), Positives = 34/73 (46%), Gaps = 7/73 (9%)
Query: 252 VVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVL 311
+ N +E +M K +A FRS SG + +P AK + K +H AVL
Sbjct: 50 ITNYWKELEEMKHMTKQEFIAS---FRS----SGFSTGAYIPMCAKIIKKDFGKHGLAVL 102
Query: 312 PGLKARISTSLVA 324
PG+K +LV+
Sbjct: 103 PGIKIITLETLVS 115
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.135 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,594,435
Number of Sequences: 26719
Number of extensions: 719654
Number of successful extensions: 2206
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 2141
Number of HSP's gapped (non-prelim): 44
length of query: 773
length of database: 11,318,596
effective HSP length: 107
effective length of query: 666
effective length of database: 8,459,663
effective search space: 5634135558
effective search space used: 5634135558
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0100c.1


