

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100b.1
(354 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
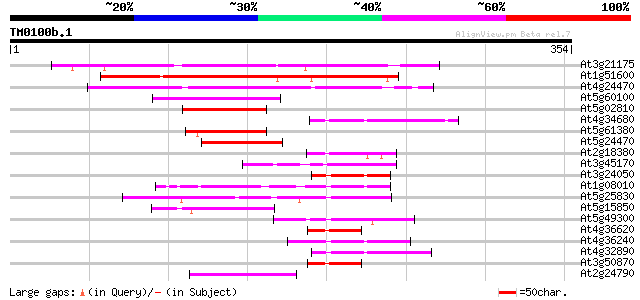
At3g21175 unknown protein 194 6e-50
At1g51600 putative zinc finger protein (At1g51600) 191 5e-49
At4g24470 unknown protein 160 1e-39
At5g60100 pseudo-response regulator 3 (APRR3) 61 8e-10
At5g02810 unknown protein 57 2e-08
At4g34680 GATA transcription factor 3 55 8e-08
At5g61380 pseudo-response regulator 1 54 1e-07
At5g24470 pseudo-response regulator 5 (APRR5) 52 4e-07
At2g18380 putative GATA-type zinc finger transcription factor 52 6e-07
At3g45170 putative protein 51 8e-07
At3g24050 GATA transcription factor 1 (AtGATA-1) 51 1e-06
At1g08010 GATA transcription factor 3, putative 51 1e-06
At5g25830 GATA transcription factor - like 50 1e-06
At5g15850 CONSTANS-like 1 50 2e-06
At5g49300 putative protein 48 9e-06
At4g36620 transcription factor like protein 47 1e-05
At4g36240 unknown protein 47 1e-05
At4g32890 unknown protein 47 1e-05
At3g50870 transcription factor-like protein 47 1e-05
At2g24790 CONSTANS-like B-box zinc finger protein 47 1e-05
>At3g21175 unknown protein
Length = 295
Score = 194 bits (493), Expect = 6e-50
Identities = 123/259 (47%), Positives = 150/259 (57%), Gaps = 26/259 (10%)
Query: 27 HFANGADEGQHA--PATVAASASNASAPHPRAGE----LTISFEGEVYVFPAVTPEKVQA 80
H G DEG P+ SA N R E LT+SF+G+VYVF V+PEKVQA
Sbjct: 42 HADGGMDEGVETDIPSHPGNSADNRGEVVDRGIENGDQLTLSFQGQVYVFDRVSPEKVQA 101
Query: 81 VLLLLGQKDIPNSTPNS-GLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCF 139
VLLLLG +++P++ P + G Q N +G++ Q ++ +R ASL+RFREKRK R F
Sbjct: 102 VLLLLGGREVPHTLPTTLGSPHQNN----RGLSGTPQRLSVPQRLASLLRFREKRKGRNF 157
Query: 140 EKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLDSSNGTPPC-------PESIERR 192
+K IRYT RKEVA RM RK GQF S K +G+ S G+ + E
Sbjct: 158 DKTIRYTVRKEVALRMQRKKGQFTSAKSS-NDDSGSTGSDWGSNQSWAVEGTETQKPEVL 216
Query: 193 CQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNERDTSADM 252
C+HCGTSEKSTP MRRGP GPR+LCNACGLMWANKGTLRDLSK T +
Sbjct: 217 CRHCGTSEKSTPMMRRGPDGPRTLCNACGLMWANKGTLRDLSKVPPP-------QTPQHL 269
Query: 253 KPSTTEPENSCADQAEEGT 271
+ E N ADQ E T
Sbjct: 270 SLNKNEDANLEADQMMEVT 288
>At1g51600 putative zinc finger protein (At1g51600)
Length = 302
Score = 191 bits (485), Expect = 5e-49
Identities = 108/199 (54%), Positives = 130/199 (65%), Gaps = 12/199 (6%)
Query: 58 ELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIPNSTPNSGLLQQKNFPQIKGINDPSQS 117
+LT+SF+G+VYVF +V PEKVQAVLLLLG +++P + P GL ++ + Q
Sbjct: 83 QLTLSFQGQVYVFDSVLPEKVQAVLLLLGGRELPQAAP-PGLGSPHQNNRVSSLPGTPQR 141
Query: 118 SNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKE--DYKSPAGN 175
++ +R ASLVRFREKRK R F+KKIRYT RKEVA RM R GQF S K D + AG+
Sbjct: 142 FSIPQRLASLVRFREKRKGRNFDKKIRYTVRKEVALRMQRNKGQFTSAKSNNDEAASAGS 201
Query: 176 LDSSNGTPPCPESI----ERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLR 231
SN T S E C+HCG EKSTP MRRGPAGPR+LCNACGLMWANKG R
Sbjct: 202 SWGSNQTWAIESSEAQHQEISCRHCGIGEKSTPMMRRGPAGPRTLCNACGLMWANKGAFR 261
Query: 232 DLSKAA-----KTPYEQNE 245
DLSKA+ P +NE
Sbjct: 262 DLSKASPQTAQNLPLNKNE 280
>At4g24470 unknown protein
Length = 309
Score = 160 bits (405), Expect = 1e-39
Identities = 101/219 (46%), Positives = 122/219 (55%), Gaps = 16/219 (7%)
Query: 50 SAPHPRAGELTISFEGEVYVFPAVTPEKVQAVLLLLG-QKDIPNSTPNSGLLQQKNFPQI 108
S P A +LTISF G+VYVF AV +KV AVL LLG ++ L QQ+N +
Sbjct: 75 SRPPEGANQLTISFRGQVYVFDAVGADKVDAVLSLLGGSTELAPGPQVMELAQQQNHMPV 134
Query: 109 KGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKED 168
+ +L +R SL RFR+KR RCFEKK+RY R+EVA RM R GQF S K
Sbjct: 135 V---EYQSRCSLPQRAQSLDRFRKKRNARCFEKKVRYGVRQEVALRMARNKGQFTSSKMT 191
Query: 169 YKSPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKG 228
+ D + I C HCG S K TP MRRGP+GPR+LCNACGL WAN+G
Sbjct: 192 DGAYNSGTDQDSAQDDAHPEIS--CTHCGISSKCTPMMRRGPSGPRTLCNACGLFWANRG 249
Query: 229 TLRDLSKAAKTPYEQNERDTSADMKPSTTEPENSCADQA 267
TLRDLSK + E + A MKP + S AD A
Sbjct: 250 TLRDLSK-------KTEENQLALMKP---DDGGSVADAA 278
>At5g60100 pseudo-response regulator 3 (APRR3)
Length = 495
Score = 61.2 bits (147), Expect = 8e-10
Identities = 32/81 (39%), Positives = 49/81 (59%)
Query: 91 PNSTPNSGLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKE 150
P+ +P + LL + S S ++R A+L++FR KRKERCFEKK+RY RK+
Sbjct: 410 PHDSPIAKLLGSSSSSDNPLKQQSSGSDRWAQREAALMKFRLKRKERCFEKKVRYHSRKK 469
Query: 151 VAQRMHRKNGQFASLKEDYKS 171
+A++ GQF ++D+KS
Sbjct: 470 LAEQRPHVKGQFIRKRDDHKS 490
>At5g02810 unknown protein
Length = 727
Score = 57.0 bits (136), Expect = 2e-08
Identities = 25/53 (47%), Positives = 38/53 (71%)
Query: 110 GINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQF 162
G + + + +S+R A+L +FR+KRKERCF KK+RY RK++A++ R GQF
Sbjct: 656 GSGNLADENKISQREAALTKFRQKRKERCFRKKVRYQSRKKLAEQRPRVRGQF 708
>At4g34680 GATA transcription factor 3
Length = 269
Score = 54.7 bits (130), Expect = 8e-08
Identities = 31/94 (32%), Positives = 48/94 (50%), Gaps = 4/94 (4%)
Query: 190 ERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNERDTS 249
+RRC HCGT+ +TP R GP GP++LCNACG+ + G L + A +P NE ++
Sbjct: 179 QRRCSHCGTN--NTPQWRTGPVGPKTLCNACGVRF-KSGRLCPEYRPADSPTFSNEIHSN 235
Query: 250 ADMKPSTTEPENSCADQAEEGTPEENKPVPMDSR 283
K ++ E + + + PV S+
Sbjct: 236 LHRKVLELRKSKELGEETGEASTKSD-PVKFGSK 268
>At5g61380 pseudo-response regulator 1
Length = 618
Score = 53.9 bits (128), Expect = 1e-07
Identities = 27/54 (50%), Positives = 35/54 (64%), Gaps = 3/54 (5%)
Query: 112 NDPSQS---SNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQF 162
N PS + L RR +L++FR KR +RCF+KKIRY RK +A+R R GQF
Sbjct: 519 NPPSNEVRVNKLDRREEALLKFRRKRNQRCFDKKIRYVNRKRLAERRPRVKGQF 572
>At5g24470 pseudo-response regulator 5 (APRR5)
Length = 667
Score = 52.4 bits (124), Expect = 4e-07
Identities = 23/51 (45%), Positives = 36/51 (70%)
Query: 122 RRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSP 172
+R A+L +FR KRK+RC+EKK+RY RK++A++ R GQF + ++P
Sbjct: 617 QREAALTKFRMKRKDRCYEKKVRYESRKKLAEQRPRIKGQFVRQVQSTQAP 667
>At2g18380 putative GATA-type zinc finger transcription factor
Length = 207
Score = 51.6 bits (122), Expect = 6e-07
Identities = 29/66 (43%), Positives = 38/66 (56%), Gaps = 11/66 (16%)
Query: 188 SIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMW---ANKGTLRDL------SKAAK 238
S+ RRC C T+ STP R GP GP+SLCNACG+ + + T R+L S AA+
Sbjct: 88 SLPRRCASCDTT--STPLWRNGPKGPKSLCNACGIRFKKEERRATARNLTISGGGSSAAE 145
Query: 239 TPYEQN 244
P E +
Sbjct: 146 VPVENS 151
>At3g45170 putative protein
Length = 204
Score = 51.2 bits (121), Expect = 8e-07
Identities = 31/97 (31%), Positives = 45/97 (45%), Gaps = 10/97 (10%)
Query: 148 RKEVAQRMHRKNGQFASLKEDYKSPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMR 207
R +R K G+ L +KSP+ DS G ++ C HCGT + TP R
Sbjct: 80 RLHAPKRSGNKRGRQKRLS--FKSPSDLFDSKFGIT------DKSCSHCGT--RKTPLWR 129
Query: 208 RGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQN 244
GP G +LCNACG+ + L + A+ ++ N
Sbjct: 130 EGPRGAGTLCNACGMRYRTGRLLPEYRPASSPDFKPN 166
>At3g24050 GATA transcription factor 1 (AtGATA-1)
Length = 274
Score = 50.8 bits (120), Expect = 1e-06
Identities = 24/50 (48%), Positives = 32/50 (64%), Gaps = 3/50 (6%)
Query: 191 RRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTP 240
R+CQHCG + TP R GPAGP++LCNACG+ + G L + A +P
Sbjct: 194 RKCQHCGAEK--TPQWRAGPAGPKTLCNACGVRY-KSGRLVPEYRPANSP 240
>At1g08010 GATA transcription factor 3, putative
Length = 303
Score = 50.8 bits (120), Expect = 1e-06
Identities = 42/148 (28%), Positives = 70/148 (46%), Gaps = 15/148 (10%)
Query: 93 STPNSGLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVA 152
ST N+G Q+ FP +KG+ + + R + L ++ E+ K C
Sbjct: 134 STHNNGS-QRLAFP-VKGMRSKRKRPT-TLRLSYLFPSEPRKPEKSTPGKPESECYFSSE 190
Query: 153 QRMHRKNGQFASLKEDYKSPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAG 212
Q +K + ++ + L++SN + I R+C HC T++ TP R GP+G
Sbjct: 191 QHAKKKR----KIHLTTRTVSSTLEASNS-----DGIVRKCTHCETTK--TPQWREGPSG 239
Query: 213 PRSLCNACGLMWANKGTLRDLSKAAKTP 240
P++LCNACG+ + G L + A +P
Sbjct: 240 PKTLCNACGVRF-RSGRLVPEYRPASSP 266
>At5g25830 GATA transcription factor - like
Length = 331
Score = 50.4 bits (119), Expect = 1e-06
Identities = 46/183 (25%), Positives = 80/183 (43%), Gaps = 20/183 (10%)
Query: 72 AVTPEKVQAVLLLLGQKDIPNSTPNSGLLQQKNFPQ---IKGINDPSQS-SNLSRRFASL 127
+++PE V + L+ G K P+ ++G + N ++ P+++ S SR A
Sbjct: 92 SLSPEDVHKLELISGFKSRPDPKSDTGSPENPNSSSPIFTTDVSVPAKARSKRSRAAACN 151
Query: 128 VRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLDSSNG------ 181
R KE ++ +T ++ + H L +P G + +G
Sbjct: 152 WASRGLLKETFYDSP--FTGETILSSQQHLSPPTSPPL---LMAPLGKKQAVDGGHRRKK 206
Query: 182 ---TPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAK 238
+P + ERRC HC T + TP R GP GP++LCNACG+ + + + + AA
Sbjct: 207 DVSSPESGGAEERRCLHCATDK--TPQWRTGPMGPKTLCNACGVRYKSGRLVPEYRPAAS 264
Query: 239 TPY 241
+
Sbjct: 265 PTF 267
>At5g15850 CONSTANS-like 1
Length = 355
Score = 49.7 bits (117), Expect = 2e-06
Identities = 30/83 (36%), Positives = 44/83 (52%), Gaps = 7/83 (8%)
Query: 90 IPNSTPNSGLLQQKNFPQIKGIND-----PSQSSNLSRRFASLVRFREKRKERCFEKKIR 144
+P ST + + P K + D P+Q + R A ++R+REK+K R FEK IR
Sbjct: 250 VPESTTSDATVSNPRSP--KAVTDQPPYPPAQMLSPRDREARVLRYREKKKMRKFEKTIR 307
Query: 145 YTCRKEVAQRMHRKNGQFASLKE 167
Y RK A++ R G+FA K+
Sbjct: 308 YASRKAYAEKRPRIKGRFAKKKD 330
>At5g49300 putative protein
Length = 139
Score = 47.8 bits (112), Expect = 9e-06
Identities = 32/91 (35%), Positives = 43/91 (47%), Gaps = 6/91 (6%)
Query: 167 EDYKSPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWAN 226
E K+ A ++ N T + ++ C CGTS+ TP R GP GP+SLCNACG+
Sbjct: 14 ETMKTRAEDMIEQNNTSVNDK--KKTCADCGTSK--TPLWRGGPVGPKSLCNACGIRNRK 69
Query: 227 K--GTLRDLSKAAKTPYEQNERDTSADMKPS 255
K G D K K+ R +K S
Sbjct: 70 KRRGGTEDNKKLKKSSSGGGNRKFGESLKQS 100
>At4g36620 transcription factor like protein
Length = 211
Score = 47.4 bits (111), Expect = 1e-05
Identities = 20/34 (58%), Positives = 25/34 (72%), Gaps = 2/34 (5%)
Query: 189 IERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGL 222
+ RRC +C T+ STP R GP GP+SLCNACG+
Sbjct: 73 LARRCANCDTT--STPLWRNGPRGPKSLCNACGI 104
>At4g36240 unknown protein
Length = 238
Score = 47.4 bits (111), Expect = 1e-05
Identities = 27/78 (34%), Positives = 41/78 (51%), Gaps = 3/78 (3%)
Query: 176 LDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSK 235
+D+S G + + R C HCG + TP R GP G ++LCNACG+ + G L +
Sbjct: 149 VDASYGGVVQQQQLRRCCSHCGVQK--TPQWRMGPLGAKTLCNACGVRF-KSGRLLPEYR 205
Query: 236 AAKTPYEQNERDTSADMK 253
A +P NE +++ K
Sbjct: 206 PACSPTFTNEIHSNSHRK 223
>At4g32890 unknown protein
Length = 308
Score = 47.4 bits (111), Expect = 1e-05
Identities = 26/76 (34%), Positives = 38/76 (49%), Gaps = 3/76 (3%)
Query: 191 RRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNERDTSA 250
RRC HC T + TP R GP GP++LCNACG+ + G L + A +P R +++
Sbjct: 197 RRCLHCATEK--TPQWRTGPMGPKTLCNACGVRY-KSGRLVPEYRPASSPTFVMARHSNS 253
Query: 251 DMKPSTTEPENSCADQ 266
K + D+
Sbjct: 254 HRKVMELRRQKEMRDE 269
>At3g50870 transcription factor-like protein
Length = 295
Score = 47.4 bits (111), Expect = 1e-05
Identities = 20/34 (58%), Positives = 25/34 (72%), Gaps = 2/34 (5%)
Query: 189 IERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGL 222
+ RRC +C T+ STP R GP GP+SLCNACG+
Sbjct: 150 LARRCANCDTT--STPLWRNGPRGPKSLCNACGI 181
>At2g24790 CONSTANS-like B-box zinc finger protein
Length = 294
Score = 47.4 bits (111), Expect = 1e-05
Identities = 25/68 (36%), Positives = 38/68 (55%)
Query: 114 PSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPA 173
P+ + + R A ++R+REKRK R FEK IRY RK A+ R G+FA + ++
Sbjct: 220 PAVQLSPAEREARVLRYREKRKNRKFEKTIRYASRKAYAEMRPRIKGRFAKRTDSRENDG 279
Query: 174 GNLDSSNG 181
G++ G
Sbjct: 280 GDVGVYGG 287
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.128 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,366,867
Number of Sequences: 26719
Number of extensions: 375335
Number of successful extensions: 1241
Number of sequences better than 10.0: 139
Number of HSP's better than 10.0 without gapping: 90
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 1090
Number of HSP's gapped (non-prelim): 174
length of query: 354
length of database: 11,318,596
effective HSP length: 100
effective length of query: 254
effective length of database: 8,646,696
effective search space: 2196260784
effective search space used: 2196260784
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0100b.1


