

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
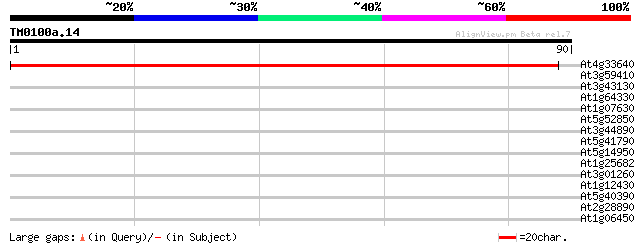
Query= TM0100a.14
(90 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g33640 unknown protein 146 2e-36
At3g59410 protein kinase like 32 0.042
At3g43130 putative protein 27 1.8
At1g64330 unknown protein 27 1.8
At1g07630 unknown protein 27 1.8
At5g52850 selenium-binding protein-like 26 3.0
At3g44890 RP19 gene for chloroplast ribosomal protein CL9 26 3.0
At5g41790 myosin heavy chain-like protein 25 5.1
At5g14950 alpha-mannosidase -like protein 25 5.1
At1g25682 unknown protein 25 5.1
At3g01260 putative aldose 1-epimerase 25 6.7
At1g12430 kinesin-like protein 25 6.7
At5g40390 raffinose synthase -like protein 25 8.8
At2g28890 unknown protein 25 8.8
At1g06450 CCR4-associated like factor 25 8.8
>At4g33640 unknown protein
Length = 95
Score = 146 bits (369), Expect = 2e-36
Identities = 72/88 (81%), Positives = 80/88 (90%)
Query: 1 MNVEEEVQRLKEEITRLGSVQTDGSYKVTFGTLFHDDRCANIFEALVGTLRAAKKRKIVA 60
MNV+EE+Q+L+EEI RLGS QTDGSYKVTFG LF+DDRCANIFEALVGTLRAAKKRKIVA
Sbjct: 1 MNVDEEIQKLEEEIHRLGSRQTDGSYKVTFGVLFNDDRCANIFEALVGTLRAAKKRKIVA 60
Query: 61 YDGELLLQGVHDNVEITLNPSPAAPAAA 88
++GELLLQGVHD VEITL P+P P AA
Sbjct: 61 FEGELLLQGVHDKVEITLRPTPPPPQAA 88
>At3g59410 protein kinase like
Length = 1271
Score = 32.3 bits (72), Expect = 0.042
Identities = 13/36 (36%), Positives = 22/36 (61%)
Query: 2 NVEEEVQRLKEEITRLGSVQTDGSYKVTFGTLFHDD 37
N +E+ + L EEIT LG ++ ++KV +F +D
Sbjct: 27 NADEDNELLSEEITALGMIEVAMTFKVVLSAIFQED 62
>At3g43130 putative protein
Length = 340
Score = 26.9 bits (58), Expect = 1.8
Identities = 17/44 (38%), Positives = 21/44 (47%)
Query: 43 FEALVGTLRAAKKRKIVAYDGELLLQGVHDNVEITLNPSPAAPA 86
FE + A+K K + G+ L Q VE NPSPA PA
Sbjct: 279 FEKYSAPRQKAQKDKEIGCWGKPLDQQPEQRVEHPTNPSPAQPA 322
>At1g64330 unknown protein
Length = 555
Score = 26.9 bits (58), Expect = 1.8
Identities = 15/54 (27%), Positives = 27/54 (49%)
Query: 4 EEEVQRLKEEITRLGSVQTDGSYKVTFGTLFHDDRCANIFEALVGTLRAAKKRK 57
E+E ++LKE + LG + + ++ H DRC + E L + A +R+
Sbjct: 498 EKEKEKLKETLLGLGEEKREAIRQLCIWIEHHRDRCEYLEEVLSKMVVARGQRR 551
>At1g07630 unknown protein
Length = 662
Score = 26.9 bits (58), Expect = 1.8
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 6/39 (15%)
Query: 2 NVEEEVQRLKEE----ITRLGSVQTDGSYKVT--FGTLF 34
N+EEEV+R++ E +T + + + GS KVT FG F
Sbjct: 495 NIEEEVERIRNEHPDDVTAVTNERVKGSLKVTRAFGAGF 533
>At5g52850 selenium-binding protein-like
Length = 893
Score = 26.2 bits (56), Expect = 3.0
Identities = 18/71 (25%), Positives = 33/71 (46%), Gaps = 9/71 (12%)
Query: 6 EVQRLKEEITRLGSVQTDGSYKVTFGTLFHDDRCANIFEALVGTLRAAKKRKIVAYDGEL 65
E++ +KEEI R GS Y+ FH + A ++ G + A+ + + ++
Sbjct: 797 EIESIKEEIKRFGS-----PYRGNENASFHSAKQAVVY----GFIYASPEAPVHVVKNKI 847
Query: 66 LLQGVHDNVEI 76
L + H+ V I
Sbjct: 848 LCKDCHEFVSI 858
>At3g44890 RP19 gene for chloroplast ribosomal protein CL9
Length = 197
Score = 26.2 bits (56), Expect = 3.0
Identities = 14/27 (51%), Positives = 19/27 (69%), Gaps = 1/27 (3%)
Query: 3 VEEEVQRLKEEITRLGSV-QTDGSYKV 28
+E E QR+KEE +L V QT G++KV
Sbjct: 103 IEAEKQRVKEEAQQLAMVFQTVGAFKV 129
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 25.4 bits (54), Expect = 5.1
Identities = 14/55 (25%), Positives = 27/55 (48%)
Query: 4 EEEVQRLKEEITRLGSVQTDGSYKVTFGTLFHDDRCANIFEALVGTLRAAKKRKI 58
E+E + +KE + LG + + ++ H RC + E L T+ A +R++
Sbjct: 1247 EKEKEMMKETLMGLGEEKREAIRQLCVWIDHHRSRCEYLEEVLSKTVVARGQRRV 1301
>At5g14950 alpha-mannosidase -like protein
Length = 1173
Score = 25.4 bits (54), Expect = 5.1
Identities = 20/64 (31%), Positives = 27/64 (41%), Gaps = 8/64 (12%)
Query: 35 HDDRCANIFEALVGTLRAAKKRKIVAYDGELLLQGVHDNVEITL--------NPSPAAPA 86
H + + G L R++V DG L QGV DN +T+ N S A PA
Sbjct: 939 HSRQSLGVASLKEGWLEIMLDRRLVRDDGRGLGQGVMDNRAMTVVFHLLAESNISQADPA 998
Query: 87 AAVN 90
+ N
Sbjct: 999 SNTN 1002
>At1g25682 unknown protein
Length = 310
Score = 25.4 bits (54), Expect = 5.1
Identities = 13/33 (39%), Positives = 18/33 (54%)
Query: 53 AKKRKIVAYDGELLLQGVHDNVEITLNPSPAAP 85
AK+RKI A LL+G ++ NPS + P
Sbjct: 268 AKRRKISAASASSLLRGGFKASSLSTNPSASKP 300
>At3g01260 putative aldose 1-epimerase
Length = 378
Score = 25.0 bits (53), Expect = 6.7
Identities = 13/38 (34%), Positives = 20/38 (52%)
Query: 32 TLFHDDRCANIFEALVGTLRAAKKRKIVAYDGELLLQG 69
T+ +R A+I L + A+KK K A DG+ + G
Sbjct: 112 TVKFTNRGASIMSLLFPNINASKKEKAAANDGKNTIHG 149
>At1g12430 kinesin-like protein
Length = 919
Score = 25.0 bits (53), Expect = 6.7
Identities = 10/28 (35%), Positives = 16/28 (56%)
Query: 1 MNVEEEVQRLKEEITRLGSVQTDGSYKV 28
M EEEV RLK ++ V+ G+ ++
Sbjct: 549 MAAEEEVNRLKHQLNEFKKVEASGNSEI 576
>At5g40390 raffinose synthase -like protein
Length = 783
Score = 24.6 bits (52), Expect = 8.8
Identities = 9/17 (52%), Positives = 15/17 (87%)
Query: 64 ELLLQGVHDNVEITLNP 80
+LLL G++D++E+TL P
Sbjct: 665 KLLLSGLNDDLELTLEP 681
>At2g28890 unknown protein
Length = 654
Score = 24.6 bits (52), Expect = 8.8
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 6/39 (15%)
Query: 2 NVEEEVQRLKEE----ITRLGSVQTDGSYKVT--FGTLF 34
NVEEEV R+++E + + + + GS KVT FG F
Sbjct: 487 NVEEEVNRIRKEHPDDASAVSNERVKGSLKVTRAFGAGF 525
>At1g06450 CCR4-associated like factor
Length = 360
Score = 24.6 bits (52), Expect = 8.8
Identities = 13/53 (24%), Positives = 22/53 (40%)
Query: 2 NVEEEVQRLKEEITRLGSVQTDGSYKVTFGTLFHDDRCANIFEALVGTLRAAK 54
NV+EE+ R+ E + R + D Y + D + A+ G + K
Sbjct: 16 NVDEEMARMAECLKRFPLIAFDTEYPGIIFRTYFDSSSDECYRAMKGNVENTK 68
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,901,272
Number of Sequences: 26719
Number of extensions: 64270
Number of successful extensions: 167
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 157
Number of HSP's gapped (non-prelim): 15
length of query: 90
length of database: 11,318,596
effective HSP length: 66
effective length of query: 24
effective length of database: 9,555,142
effective search space: 229323408
effective search space used: 229323408
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0100a.14


