

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098e.6
(402 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
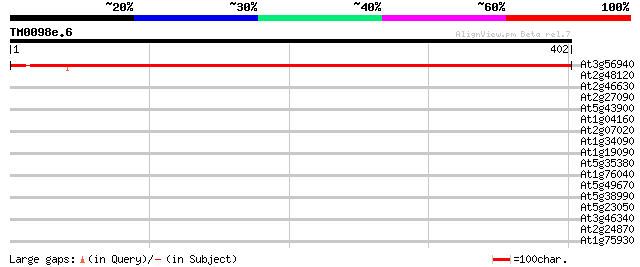
Score E
Sequences producing significant alignments: (bits) Value
At3g56940 putative dicarboxylate diiron protein (Crd1) 708 0.0
At2g48120 pale cress protein 33 0.36
At2g46630 putative extensin 30 2.3
At2g27090 unknown protein 30 2.3
At5g43900 myosin heavy chain MYA2 (pir||S51824) 30 3.1
At1g04160 myosin heavy chain MYA2 30 3.1
At2g07020 putative protein kinase 29 4.0
At1g34090 putative protein 29 4.0
At1g19090 receptor-like serine/threonine kinase (RKF2) 29 4.0
At5g35380 putative protein 29 5.2
At1g76040 calcium-dependent protein kinase (CPK29) 29 5.2
At5g49670 unknown protein 28 6.8
At5g38990 protein kinase - like protein 28 8.9
At5g23050 acetyl-CoA synthetase-like protein 28 8.9
At3g46340 receptor-like protein kinase homolog 28 8.9
At2g24870 unknown protein 28 8.9
At1g75930 family II lipase EXL6 (EXL6) 28 8.9
>At3g56940 putative dicarboxylate diiron protein (Crd1)
Length = 409
Score = 708 bits (1828), Expect = 0.0
Identities = 351/407 (86%), Positives = 376/407 (92%), Gaps = 7/407 (1%)
Query: 1 MALVKPISKFTNAAPKFSNP-RTISHSKFSTV-RMSATTTPP---SSTKPSKKANKTSIK 55
MALVKPISKF++ PK SNP + +S +FSTV RMSA+++PP ++T SKK K I+
Sbjct: 5 MALVKPISKFSS--PKLSNPSKFLSGRRFSTVIRMSASSSPPPPTTATSKSKKGTKKEIQ 62
Query: 56 ETLLTPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAAD 115
E+LLTPRFYTTDF+EME LFNTEINKNLNE EFEALLQEFKTDYNQTHFVRNKEFKEAAD
Sbjct: 63 ESLLTPRFYTTDFEEMEQLFNTEINKNLNEAEFEALLQEFKTDYNQTHFVRNKEFKEAAD 122
Query: 116 KLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFL 175
KL GPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFL
Sbjct: 123 KLQGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFL 182
Query: 176 NKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQC 235
NKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLK NPE+QC
Sbjct: 183 NKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKENPEFQC 242
Query: 236 YPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRT 295
YPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDW+AKLWSRFFCLSVYVTMYLNDCQRT
Sbjct: 243 YPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWQAKLWSRFFCLSVYVTMYLNDCQRT 302
Query: 296 DFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGES 355
+FYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR+LDRMV EK+LA+GE+
Sbjct: 303 NFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRKLDRMVVSYEKLLAIGET 362
Query: 356 DDIPFVKNLKRIPLIAALASELLATYLMPPIESGSVDLAEFEPQLVY 402
DD F+K LKRIPL+ +LASE+LA YLMPP+ESGSVD AEFEP LVY
Sbjct: 363 DDASFIKTLKRIPLVTSLASEILAAYLMPPVESGSVDFAEFEPNLVY 409
>At2g48120 pale cress protein
Length = 313
Score = 32.7 bits (73), Expect = 0.36
Identities = 27/102 (26%), Positives = 45/102 (43%), Gaps = 6/102 (5%)
Query: 11 TNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDE 70
T A+P FS+PR IS +K T +S +T + + N + + P +Y ++
Sbjct: 9 TCASPLFSSPRVISATKKLTTELSIST---AKFRRRCSGNNDEVLLEGMPPEYYDDEWQA 65
Query: 71 METLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKE 112
+ E+ + E E E +E K + + R KEF E
Sbjct: 66 RQREKTKELRRMQREEEEE---EERKIEEYREIGTRLKEFPE 104
>At2g46630 putative extensin
Length = 394
Score = 30.0 bits (66), Expect = 2.3
Identities = 23/88 (26%), Positives = 31/88 (35%)
Query: 16 KFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDEMETLF 75
K ++P + +HS ST S T PS ++ +KA + P E
Sbjct: 138 KPNSPPSPAHSSRSTTSESVKTRSPSESENHRKAPSPRVLSPYSLPASLLHSERETTQKN 197
Query: 76 NTEINKNLNEHEFEALLQEFKTDYNQTH 103
K HE Q DYNQ H
Sbjct: 198 ILTAEKTSQTHETNHHNQNHNHDYNQNH 225
>At2g27090 unknown protein
Length = 743
Score = 30.0 bits (66), Expect = 2.3
Identities = 17/49 (34%), Positives = 23/49 (46%)
Query: 27 KFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDEMETLF 75
K T +S TP +T +K NK E L PR + + E+E LF
Sbjct: 272 KSKTPELSPPVTPLVATPVNKTPNKGDHTENKLPPRDFLSSMKEIELLF 320
>At5g43900 myosin heavy chain MYA2 (pir||S51824)
Length = 1505
Score = 29.6 bits (65), Expect = 3.1
Identities = 24/105 (22%), Positives = 48/105 (44%), Gaps = 13/105 (12%)
Query: 63 FYTTDFDEM-ETLFNTEINKNLNEHEFEALLQEFKTD---YNQTHFVRNKEFKEAADKLD 118
F T F++ L N ++ ++ N+H F+ +E+K + ++ FV N++ + +K
Sbjct: 441 FKTNSFEQFCINLTNEKLQQHFNQHVFKMEQEEYKKEEINWSYIEFVDNQDILDLIEKKP 500
Query: 119 GPLRQIFVEFLERSC-----TAEFSGFLLYKELGRRLKKTNPVVA 158
G + + L+ +C T E LY+ + T P +A
Sbjct: 501 GGI----IALLDEACMFPRSTHETFAQKLYQTFKTHKRFTKPKLA 541
>At1g04160 myosin heavy chain MYA2
Length = 1519
Score = 29.6 bits (65), Expect = 3.1
Identities = 24/105 (22%), Positives = 48/105 (44%), Gaps = 13/105 (12%)
Query: 63 FYTTDFDEM-ETLFNTEINKNLNEHEFEALLQEFKTD---YNQTHFVRNKEFKEAADKLD 118
F T F++ L N ++ ++ N+H F+ +E+K + ++ FV N++ + +K
Sbjct: 442 FKTNSFEQFCINLTNEKLQQHFNQHVFKMEQEEYKKEEINWSYIEFVDNQDILDLIEKKP 501
Query: 119 GPLRQIFVEFLERSC-----TAEFSGFLLYKELGRRLKKTNPVVA 158
G + + L+ +C T E LY+ + T P +A
Sbjct: 502 GGI----IALLDEACMFPRSTHETFAQKLYQTYKNHKRFTKPKLA 542
>At2g07020 putative protein kinase
Length = 620
Score = 29.3 bits (64), Expect = 4.0
Identities = 16/50 (32%), Positives = 24/50 (48%), Gaps = 1/50 (2%)
Query: 14 APKFSNPRTISHSKFSTVRMSATTTPPSST-KPSKKANKTSIKETLLTPR 62
AP F IS K S+VR + ++ PP T +P A ++ +PR
Sbjct: 146 APNFCTVYAISKGKISSVRSATSSPPPLCTIRPQLPARSSNANNNNFSPR 195
>At1g34090 putative protein
Length = 761
Score = 29.3 bits (64), Expect = 4.0
Identities = 17/52 (32%), Positives = 26/52 (49%)
Query: 19 NPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDE 70
+P +I ++ S+ + T T SST K TSI T TP+ +T+ E
Sbjct: 68 DPASIGNNSTSSPPGTPTATQASSTSSEKSKTPTSIPSTPTTPQRSSTESTE 119
>At1g19090 receptor-like serine/threonine kinase (RKF2)
Length = 600
Score = 29.3 bits (64), Expect = 4.0
Identities = 23/107 (21%), Positives = 45/107 (41%), Gaps = 4/107 (3%)
Query: 50 NKTSIKETLLTPRFYTTDFDEMET-LFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNK 108
++ + +E L+ R D E F T + H+F E K D +Q +F+R+
Sbjct: 198 DENTCRECLVNARSSLRACDGHEARAFFTGCYLKYSTHKFFDDAAEHKPDADQRNFIRSS 257
Query: 109 EFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNP 155
F +D+ + ++ + + S F+ Y+ + R+ K P
Sbjct: 258 FFPHLSDR---DVTRLAIAAISLSILTSLGAFISYRRVSRKRKAQVP 301
>At5g35380 putative protein
Length = 731
Score = 28.9 bits (63), Expect = 5.2
Identities = 16/35 (45%), Positives = 20/35 (56%), Gaps = 1/35 (2%)
Query: 14 APKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKK 48
AP F IS K S +R SAT++PP S PS +
Sbjct: 136 APSFCTVYVISKGKISFLR-SATSSPPHSNMPSMR 169
>At1g76040 calcium-dependent protein kinase (CPK29)
Length = 561
Score = 28.9 bits (63), Expect = 5.2
Identities = 26/106 (24%), Positives = 48/106 (44%), Gaps = 11/106 (10%)
Query: 79 INKNLNEHEFEALLQEFKT-DYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEF 137
I +NL+E E + L Q FK D +++ + E + +L L + ++ L + +
Sbjct: 405 IAENLSEEEIKGLKQTFKNMDTDESGTITFDELRNGLHRLGSKLTESEIKQLMEAADVDK 464
Query: 138 SGFLLYKEL------GRRLKKTNPVVAEIFSLMSRDEARHAGFLNK 177
SG + Y E RL+K ++ E F +D +GF+ +
Sbjct: 465 SGTIDYIEFVTATMHRHRLEKEENLI-EAFKYFDKD---RSGFITR 506
>At5g49670 unknown protein
Length = 1184
Score = 28.5 bits (62), Expect = 6.8
Identities = 25/98 (25%), Positives = 39/98 (39%), Gaps = 7/98 (7%)
Query: 264 PQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTA-R 322
P + KL+ + F + Y CQ +G G + F + T+RT+
Sbjct: 1043 PAIIQMMNEKLYKQSF------SSYYKSCQSLRLSDGSGACKEGFQAGFKMSTSRTSLLS 1096
Query: 323 IFPAVLDVENPEFKRRLDRMVEINEKILAVGESDDIPF 360
+ LD+ M+EI +K+ V E DIPF
Sbjct: 1097 VSVTDLDLSLTAIGGGEAGMIEIVKKLDPVAEEKDIPF 1134
>At5g38990 protein kinase - like protein
Length = 880
Score = 28.1 bits (61), Expect = 8.9
Identities = 15/51 (29%), Positives = 25/51 (48%), Gaps = 9/51 (17%)
Query: 61 PRFYTTDFDEMETLFNTEINKNLNEHEF---------EALLQEFKTDYNQT 102
P Y +DFD +++ F+ +N+ H F +L++EF NQT
Sbjct: 110 PTRYGSDFDAVKSFFSVNVNRFTLLHNFSVKASIPESSSLIKEFIVPVNQT 160
>At5g23050 acetyl-CoA synthetase-like protein
Length = 668
Score = 28.1 bits (61), Expect = 8.9
Identities = 17/56 (30%), Positives = 25/56 (44%), Gaps = 4/56 (7%)
Query: 29 STVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDEMETLFNTEINKNLN 84
S + +A PP S P + K +P F D + ++ FN+EI K LN
Sbjct: 577 SVLETAAIGVPPPSGGPEQLVIAVVFK----SPEFSNPDLNLLKKSFNSEIQKKLN 628
>At3g46340 receptor-like protein kinase homolog
Length = 889
Score = 28.1 bits (61), Expect = 8.9
Identities = 34/144 (23%), Positives = 54/144 (36%), Gaps = 33/144 (22%)
Query: 39 PPSSTKPSKKANKTSIKETLLTPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQEFKTD 98
PPSS P + TSI +T + + + E+ E+ KNL E F
Sbjct: 548 PPSSNTPRENITSTSISDTSIETKRKRFSYSEV-----MEMTKNLQRPLGEG---GFGVV 599
Query: 99 YNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPV-- 156
Y+ ++G +Q+ V+ L +S T + F EL R+ N V
Sbjct: 600 YH--------------GDINGSSQQVAVKLLSQSSTQGYKEFKAEVELLLRVHHINLVSL 645
Query: 157 ---------VAEIFSLMSRDEARH 171
+A I+ MS + +H
Sbjct: 646 VGYCDERDHLALIYEYMSNKDLKH 669
>At2g24870 unknown protein
Length = 136
Score = 28.1 bits (61), Expect = 8.9
Identities = 20/64 (31%), Positives = 29/64 (45%), Gaps = 5/64 (7%)
Query: 182 FNLALDLGFLTKARKYTFFKPKFIFYATY--LSEKIGYWRYITIYRHLKANPEYQCYPIF 239
FN L +G +K K F PK I + ++ I +WR HL+ P Y+ Y F
Sbjct: 18 FNKLLRIGCYSKDDK---FGPKTIPIGKHVEINFMINFWRTTRFMCHLEQGPNYKHYQKF 74
Query: 240 KYFE 243
F+
Sbjct: 75 TAFK 78
>At1g75930 family II lipase EXL6 (EXL6)
Length = 343
Score = 28.1 bits (61), Expect = 8.9
Identities = 13/36 (36%), Positives = 19/36 (52%)
Query: 175 LNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATY 210
LN+ DFN+ L G + A +Y F KF++ Y
Sbjct: 241 LNRITEDFNMKLQKGLTSYAVEYDFKDAKFVYVDIY 276
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,911,002
Number of Sequences: 26719
Number of extensions: 381703
Number of successful extensions: 1096
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1088
Number of HSP's gapped (non-prelim): 18
length of query: 402
length of database: 11,318,596
effective HSP length: 102
effective length of query: 300
effective length of database: 8,593,258
effective search space: 2577977400
effective search space used: 2577977400
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0098e.6


