

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098c.1
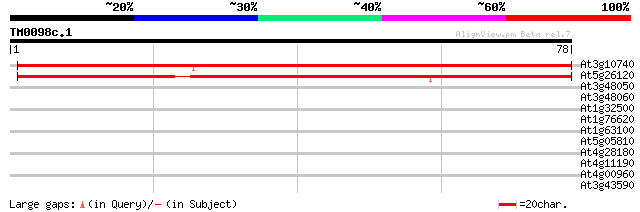
(78 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g10740 putative alpha-L-arabinofuranosidase 91 1e-19
At5g26120 arabinosidase - like protein 77 2e-15
At3g48050 putative protein 30 0.22
At3g48060 putative protein 27 1.8
At1g32500 unknown protein 27 1.8
At1g76620 unknown protein 26 3.1
At1g63100 transcription factor SCARECROW, putative 26 3.1
At5g05810 putative protein 25 5.3
At4g28180 putative protein 25 6.9
At4g11190 putative disease resistance response protein 25 9.1
At4g00960 25 9.1
At3g43590 putative protein 25 9.1
>At3g10740 putative alpha-L-arabinofuranosidase
Length = 678
Score = 90.9 bits (224), Expect = 1e-19
Identities = 45/78 (57%), Positives = 59/78 (74%), Gaps = 1/78 (1%)
Query: 2 VNFGSGSESFRISINGLKSKVKQ-SGSRKTVLTAANVMDENSFSEPNKIVPQQTPIEGAS 60
VNFG+ SE+ ++ + GL V + SGS+KTVLT+ NVMDENSFS+P K+VP ++ +E A
Sbjct: 579 VNFGANSENMQVLVTGLDPNVMRVSGSKKTVLTSTNVMDENSFSQPEKVVPHESLLELAE 638
Query: 61 TDMKVELPPHSVTSFDLL 78
DM V LPPHS +SFDLL
Sbjct: 639 EDMTVVLPPHSFSSFDLL 656
>At5g26120 arabinosidase - like protein
Length = 674
Score = 76.6 bits (187), Expect = 2e-15
Identities = 41/78 (52%), Positives = 55/78 (69%), Gaps = 3/78 (3%)
Query: 2 VNFGSGSESFRISINGLKSKVKQSGSRKTVLTAANVMDENSFSEPNKIVPQQTPIE-GAS 60
VNFG S + ++++ GL +K GS+K VLT+A+VMDENSFS PN IVPQ++ +E
Sbjct: 578 VNFGEQSVNLKVAVTGLMAKFY--GSKKKVLTSASVMDENSFSNPNMIVPQESLLEMTEQ 635
Query: 61 TDMKVELPPHSVTSFDLL 78
D+ LPPHS +SFDLL
Sbjct: 636 EDLMFVLPPHSFSSFDLL 653
>At3g48050 putative protein
Length = 1613
Score = 30.0 bits (66), Expect = 0.22
Identities = 18/65 (27%), Positives = 32/65 (48%), Gaps = 5/65 (7%)
Query: 15 INGLKSKVKQ-----SGSRKTVLTAANVMDENSFSEPNKIVPQQTPIEGASTDMKVELPP 69
+ +K+ VK S S K V ++ + + + + +QTP+EG D K E PP
Sbjct: 915 LKDIKTDVKSEADCTSDSTKRVASSMLTECRDVSKKVDSVAVEQTPLEGVDDDKKEEKPP 974
Query: 70 HSVTS 74
+++S
Sbjct: 975 TALSS 979
>At3g48060 putative protein
Length = 1611
Score = 26.9 bits (58), Expect = 1.8
Identities = 10/28 (35%), Positives = 17/28 (60%)
Query: 47 NKIVPQQTPIEGASTDMKVELPPHSVTS 74
+ + + TP+EG D K E PP +++S
Sbjct: 952 DSVAVEHTPLEGVDDDKKEEKPPTALSS 979
>At1g32500 unknown protein
Length = 475
Score = 26.9 bits (58), Expect = 1.8
Identities = 13/32 (40%), Positives = 19/32 (58%)
Query: 15 INGLKSKVKQSGSRKTVLTAANVMDENSFSEP 46
I L SK K +R+T T+ +V + SFS+P
Sbjct: 13 IPNLSSKPKLKSNRRTTSTSVSVRAQASFSDP 44
>At1g76620 unknown protein
Length = 527
Score = 26.2 bits (56), Expect = 3.1
Identities = 14/45 (31%), Positives = 25/45 (55%), Gaps = 5/45 (11%)
Query: 10 SFRISINGLKSK-----VKQSGSRKTVLTAANVMDENSFSEPNKI 49
SF S+ L S+ V + S +L +++++D N+F PNK+
Sbjct: 152 SFNTSLKALSSREGTRVVSGTHSLGELLGSSHIVDHNNFINPNKL 196
>At1g63100 transcription factor SCARECROW, putative
Length = 658
Score = 26.2 bits (56), Expect = 3.1
Identities = 11/34 (32%), Positives = 19/34 (55%)
Query: 40 ENSFSEPNKIVPQQTPIEGASTDMKVELPPHSVT 73
+NS + NK + ++T G S ++LPP + T
Sbjct: 66 DNSNNNNNKPIERKTKTSGCSLKQNIKLPPLATT 99
>At5g05810 putative protein
Length = 407
Score = 25.4 bits (54), Expect = 5.3
Identities = 15/56 (26%), Positives = 26/56 (45%), Gaps = 1/56 (1%)
Query: 3 NFGSGSESFRISINGLKSKVKQSGSRKTVLTAANVMDENSFSEPNKIVPQQTPIEG 58
++ S SFR S++ KV +G K+ A N +D + ++P + EG
Sbjct: 256 SYKSNPMSFRRSLDS-SLKVNDAGEEKSESVAVNCLDRLQRKDGLLLIPNRESFEG 310
>At4g28180 putative protein
Length = 256
Score = 25.0 bits (53), Expect = 6.9
Identities = 10/30 (33%), Positives = 20/30 (66%)
Query: 22 VKQSGSRKTVLTAANVMDENSFSEPNKIVP 51
V S S T TAA+++++++ S+P++ P
Sbjct: 91 VSHSYSTATTATAASILEDDALSKPHRPPP 120
>At4g11190 putative disease resistance response protein
Length = 184
Score = 24.6 bits (52), Expect = 9.1
Identities = 9/18 (50%), Positives = 13/18 (72%)
Query: 34 AANVMDENSFSEPNKIVP 51
A N+M EN+F EP + +P
Sbjct: 126 ALNIMGENAFMEPTRDLP 143
>At4g00960
Length = 372
Score = 24.6 bits (52), Expect = 9.1
Identities = 11/35 (31%), Positives = 19/35 (53%)
Query: 17 GLKSKVKQSGSRKTVLTAANVMDENSFSEPNKIVP 51
GL + GSR T+ + +++ NSF+ P + P
Sbjct: 302 GLLCVQENPGSRPTMASIVRMLNANSFTLPRPLQP 336
>At3g43590 putative protein
Length = 551
Score = 24.6 bits (52), Expect = 9.1
Identities = 10/17 (58%), Positives = 14/17 (81%)
Query: 13 ISINGLKSKVKQSGSRK 29
+ +NG+KSKVK+S S K
Sbjct: 76 VMVNGVKSKVKKSESSK 92
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.308 0.126 0.336
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,589,138
Number of Sequences: 26719
Number of extensions: 50196
Number of successful extensions: 130
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 119
Number of HSP's gapped (non-prelim): 12
length of query: 78
length of database: 11,318,596
effective HSP length: 54
effective length of query: 24
effective length of database: 9,875,770
effective search space: 237018480
effective search space used: 237018480
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0098c.1


