

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098b.1
(696 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
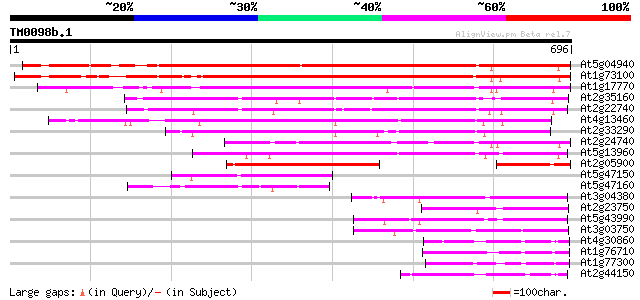
Score E
Sequences producing significant alignments: (bits) Value
At5g04940 SET-domain protein-like 615 e-176
At1g73100 putative protein 613 e-176
At1g17770 putative protein 416 e-116
At2g35160 similar to mammalian MHC III region protein G9a 372 e-103
At2g22740 similar to mammalian MHC III region protein G9a 357 1e-98
At4g13460 putative protein 351 7e-97
At2g33290 similar to mammalian MHC III region protein G9a 329 4e-90
At2g24740 pseudogene 329 4e-90
At5g13960 unknown protein (At5g13960) 308 7e-84
At2g05900 hypothetical protein 171 1e-42
At5g47150 putative protein 148 9e-36
At5g47160 putative protein 135 1e-31
At3g04380 Su(VAR)3-9-related protein 4 125 8e-29
At2g23750 putative SET-domain transcriptional regulator 125 8e-29
At5g43990 SET domain protein SUVR2 107 2e-23
At3g03750 unknown protein 98 2e-20
At4g30860 unknown protein 91 2e-18
At1g76710 ASH1-like protein 1 (ASHH1) 88 1e-17
At1g77300 hypothetical protein 85 1e-16
At2g44150 unknown protein 81 2e-15
>At5g04940 SET-domain protein-like
Length = 670
Score = 615 bits (1587), Expect = e-176
Identities = 325/696 (46%), Positives = 434/696 (61%), Gaps = 53/696 (7%)
Query: 17 DKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGPFPSGVAPFYPFFVSPE 76
DK+RVL++KPLRTL PVFPS + PFVC P GPFP G + FYPF S
Sbjct: 10 DKTRVLDIKPLRTLRPVFPSGNQ---------APPFVCAPPFGPFPPGFSSFYPFSSSQA 60
Query: 77 SQRLSEQNAQTPTAQRAAPISAAVPINSFRTPTGATNGDVGSSRRKSRGQLPEDDNFVDL 136
+Q + N Q P + P+ + P A+ + + R R D+
Sbjct: 61 NQHTPDLNQAQYPPQHQQPQNPP-PVYQQQPPQHASEPSLVTPLRSFRSP--------DV 111
Query: 137 SEVDGEGGTGDGKRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEILNQPDGSRDA 196
S + E KRR P+KR ++ G+ + ++ +G+R+
Sbjct: 112 SNGNAELEGSTVKRRIPKKR-------------PISRPENMNFESGI-NVADRENGNREL 157
Query: 197 VAYTLMIYEVMRRKLGQIDEKAKGSHSGAKRPDLKAGTLMNTKGIRANSRKRIGVVPGVE 256
V LM ++ +RR+ Q+++ + KRPDLK+G+ +G+R N++KR G+VPGVE
Sbjct: 158 VLSVLMRFDALRRRFAQLEDAKEAVSGIIKRPDLKSGSTCMGRGVRTNTKKRPGIVPGVE 217
Query: 257 VGDIFFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIY 316
+GD+FFFRFE+CLVGLH+PSMAGIDYL K EEEP+A SIVSSG Y+++ + DVLIY
Sbjct: 218 IGDVFFFRFEMCLVGLHSPSMAGIDYLVVKGETEEEPIATSIVSSGYYDNDEGNPDVLIY 277
Query: 317 SGQGGTS-REKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKI 375
+GQGG + ++K +SDQKLERGNLALE+SL R + VRVIRG+++ +H K+Y+YDGLY+I
Sbjct: 278 TGQGGNADKDKQSSDQKLERGNLALEKSLRRDSAVRVIRGLKEASH-NAKIYIYDGLYEI 336
Query: 376 QNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTSGAEKL 435
+ SWVEK KSG N FKYKLVR PGQP A+ W +I +W SR G+ILPD+TSG E +
Sbjct: 337 KESWVEKGKSGHNTFKYKLVRAPGQPPAFASWTAIQKWKTGVPSRQGLILPDMTSGVESI 396
Query: 436 PVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQKNGGY 495
PV LVN+VD + GPAYFTYS T+K ++ S GC C C+PG+ C C +KNGG
Sbjct: 397 PVSLVNEVDTDNGPAYFTYSTTVKYSESFKLMQPSFGCDCANLCKPGNLDCHCIRKNGGD 456
Query: 496 LPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPI 555
PY+ G+L K ++YEC PSC C +C+N+V+Q G+K+RLEVF+T +GWGLRSWD I
Sbjct: 457 FPYTGNGILVSRKPMIYECSPSCPC-STCKNKVTQMGVKVRLEVFKTANRGWGLRSWDAI 515
Query: 556 RAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQL-----------EVFSSDVE 604
RAG+FIC Y GE D ++V++ DDY FD+T +Y + E
Sbjct: 516 RAGSFICIYVGEAKDKSKVQQTMA--NDDYTFDTTNVYNPFKWNYEPGLADEDACEEMSE 573
Query: 605 APKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMME 664
+IP PL I+A+N GNVARFMNHSC+PNV W+PV EN ++ +HVAF+AI HIPPM E
Sbjct: 574 ESEIPLPLIISAKNVGNVARFMNHSCSPNVFWQPVSYENNSQLFVHVAFFAISHIPPMTE 633
Query: 665 LTYDYGIVLPLKVGQ-----KKKKCLCGSVKCRGYF 695
LTYDYG+ P K+KC CGS CRG F
Sbjct: 634 LTYDYGVSRPSGTQNGNPLYGKRKCFCGSAYCRGSF 669
>At1g73100 putative protein
Length = 669
Score = 613 bits (1582), Expect = e-176
Identities = 337/709 (47%), Positives = 437/709 (61%), Gaps = 65/709 (9%)
Query: 6 GQDPAPAAGSFDKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGPFPSGV 65
G + P +DKS VL++KPLR+L PVFP+ + G PFV P GP S
Sbjct: 6 GFNTVPNPNHYDKSIVLDIKPLRSLKPVFPNGNQ---------GPPFVGCPPFGPSSSEY 56
Query: 66 APFYPFFVSPESQRLSEQNAQTPTAQRAAPISAAVP-INSFRTPTGATNGDVGSSRRKSR 124
+ F+PF + + N T PI + VP + S+RTPT TNG SS K
Sbjct: 57 SSFFPFGAQQPTHDTPDLNQTQNT-----PIPSFVPPLRSYRTPT-KTNGPSSSSGTKR- 109
Query: 125 GQLPEDDNFVDLSEVDGEGGTGDGKRRKPQKRIREKRCSSDVDPDA-VANEILKTINPGV 183
G G K K+ +K +++ + D V + + G+
Sbjct: 110 -------------------GVGRPKGTTSVKKKEKKTVANEPNLDVQVVKKFSSDFDSGI 150
Query: 184 FEILNQPDGSRDAVAYTLMIYEVMRRKLGQIDEKAKGSHSGAKRPDLKAGTLMNTKGIRA 243
+ DG+ V+ LM ++ +RR+L Q+ E K + S A AGTLM+ G+R
Sbjct: 151 -SAAEREDGNAYLVSSVLMRFDAVRRRLSQV-EFTKSATSKA------AGTLMSN-GVRT 201
Query: 244 NSRKRIGVVPGVEVGDIFFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGG 303
N +KR+G VPG+EVGDIFF R E+CLVGLH +MAGIDY+ +K +EE LA SIVSSG
Sbjct: 202 NMKKRVGTVPGIEVGDIFFSRIEMCLVGLHMQTMAGIDYIISKAGSDEESLATSIVSSGR 261
Query: 304 YEDNVEDGDVLIYSGQGGTS-REKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHP 362
YE +D + LIYSGQGG + + + ASDQKLERGNLALE SL +GN VRV+RG D A
Sbjct: 262 YEGEAQDPESLIYSGQGGNADKNRQASDQKLERGNLALENSLRKGNGVRVVRGEEDAASK 321
Query: 363 TGKVYVYDGLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVG 422
TGK+Y+YDGLY I SWVEK KSG N FKYKLVR PGQP A+ WKS+ +W + +R G
Sbjct: 322 TGKIYIYDGLYSISESWVEKGKSGCNTFKYKLVRQPGQPPAFGFWKSVQKWKEGLTTRPG 381
Query: 423 VILPDLTSGAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPG 482
+ILPDLTSGAE PV LVNDVD +KGPAYFTY+ +LK + GC+C+G C PG
Sbjct: 382 LILPDLTSGAESKPVSLVNDVDEDKGPAYFTYTSSLKYSETFKLTQPVIGCSCSGSCSPG 441
Query: 483 SHKCGCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRT 542
+H C C +KN G LPY +L + V+YECGP+C C SC+NRV Q GLK RLEVF+T
Sbjct: 442 NHNCSCIRKNDGDLPYLNGVILVSRRPVIYECGPTCPCHASCKNRVIQTGLKSRLEVFKT 501
Query: 543 KGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQL------ 596
+ +GWGLRSWD +RAG+FICEYAGEV DN + + ED Y+FD++R++
Sbjct: 502 RNRGWGLRSWDSLRAGSFICEYAGEVKDNGNLR--GNQEEDAYVFDTSRVFNSFKWNYEP 559
Query: 597 EVFSSD--VEAPK---IPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHV 651
E+ D E P+ +PSPL I+A+ GNVARFMNHSC+PNV W+PV+RE E+ +H+
Sbjct: 560 ELVDEDPSTEVPEEFNLPSPLLISAKKFGNVARFMNHSCSPNVFWQPVIREGNGESVIHI 619
Query: 652 AFYAIRHIPPMMELTYDYGIVLPLKVGQK-----KKKCLCGSVKCRGYF 695
AF+A+RHIPPM ELTYDYGI + + ++ CLCGS +CRG F
Sbjct: 620 AFFAMRHIPPMAELTYDYGISPTSEARDESLLHGQRTCLCGSEQCRGSF 668
>At1g17770 putative protein
Length = 693
Score = 416 bits (1069), Expect = e-116
Identities = 264/699 (37%), Positives = 367/699 (51%), Gaps = 75/699 (10%)
Query: 35 PSPSNPSSSATPQGGAPFVCVSPSGPFPSGVAPFY-----PFFVSPESQRLSEQNAQTPT 89
P+ +P + P +P + V P +Y P P + S+ T
Sbjct: 31 PTMVSPVLTNMPSATSPLLMVPPLRTIWPSNKEWYDGDAGPSSTGPIKREASDNTNDTAH 90
Query: 90 AQRAAPISAAVPINSFRTPTGATNGDVGSSRRKSRGQLPEDDNFVDLSEVDGEGGTGDGK 149
A P +P+ + R ++N + S G + G G K
Sbjct: 91 NTFAPPPEMVIPLITIRPSDDSSNYSCDAGAGPSTGPVKR-----------GRGRPKGSK 139
Query: 150 RRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEI----LNQPDGSRDAVAYTLMIYE 205
P + + K DP++ LK + G F+ G+++ V +M ++
Sbjct: 140 NSTPTEPKKPKV----YDPNS-----LKVTSRGNFDSEITEAETETGNQEIVDSVMMRFD 190
Query: 206 VMRRKLGQID--EKAKGSHSGAKRPDLKAGTLMNTKGIRANSRKRIGVVPGVEVGDIFFF 263
+RR+L QI+ E + SG G++ N+R+RIG VPG+ VGDIF++
Sbjct: 191 AVRRRLCQINHPEDILTTASGN----------CTKMGVKTNTRRRIGAVPGIHVGDIFYY 240
Query: 264 RFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQGGTS 323
E+CLVGLH + GID+ S E A+ +V++G Y+ E D LIYSGQGGT
Sbjct: 241 WGEMCLVGLHKSNYGGIDFFTAAESAVEGHAAMCVVTAGQYDGETEGLDTLIYSGQGGTD 300
Query: 324 REKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKA 383
A DQ+++ GNLALE S+ +GNDVRV+RG+ K+Y+YDG+Y + W
Sbjct: 301 VYGNARDQEMKGGNLALEASVSKGNDVRVVRGVIHPHENNQKIYIYDGMYLVSKFWTVTG 360
Query: 384 KSGFNVFKYKLVRLPGQPQAYMIWKSI--LQWTDKSASRVGVILPDLTSGAEKLPVCLVN 441
KSGF F++KLVR P QP AY IWK++ L+ D SR G IL DL+ GAE L V LVN
Sbjct: 361 KSGFKEFRFKLVRKPNQPPAYAIWKTVENLRNHDLIDSRQGFILEDLSFGAELLRVPLVN 420
Query: 442 DVDNEKG--PAYFTYSPTLKNLNRLAPV----ESSEGCTCNGGCQPGSHK-CGCTQKNGG 494
+VD + P F Y P+ + + S GC N QP H+ C C Q+NG
Sbjct: 421 EVDEDDKTIPEDFDYIPSQCHSGMMTHEFHFDRQSLGCQ-NCRHQPCMHQNCTCVQRNGD 479
Query: 495 YLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDP 554
LPY +L K ++YECG SC CP C R+ Q GLKL LEVF+T+ GWGLRSWDP
Sbjct: 480 LLPYH-NNILVCRKPLIYECGGSCPCPDHCPTRLVQTGLKLHLEVFKTRNCGWGLRSWDP 538
Query: 555 IRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQL------EVFSSD-----V 603
IRAGTFICE+AG VEE +DDY+FD+++IYQ+ E+ D
Sbjct: 539 IRAGTFICEFAGLRKTKEEVEE-----DDDYLFDTSKIYQRFRWNYEPELLLEDSWEQVS 593
Query: 604 EAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMM 663
E +P+ + I+A+ +GNV RFMNHSC+PNV W+P+ EN+ + L + +A++HIPPM
Sbjct: 594 EFINLPTQVLISAKEKGNVGRFMNHSCSPNVFWQPIEYENRGDVYLLIGLFAMKHIPPMT 653
Query: 664 ELTYDYGIVL-------PLKVGQKKKKCLCGSVKCRGYF 695
ELTYDYG+ + + + KK CLCGSVKCRG F
Sbjct: 654 ELTYDYGVSCVERSEEDEVLLYKGKKTCLCGSVKCRGSF 692
>At2g35160 similar to mammalian MHC III region protein G9a
Length = 794
Score = 372 bits (954), Expect = e-103
Identities = 220/564 (39%), Positives = 327/564 (57%), Gaps = 49/564 (8%)
Query: 143 GGTGDGKRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEILNQPDGSRDAVAYTLM 202
G G+G RK +R+ +D +A++ + N G+ + D +R V T+
Sbjct: 264 GDYGEGSMRKNSERVA-------LDKKRLASKF-RLSNGGLPSCSSSGDSARYKVKETMR 315
Query: 203 IYEVMRRKLGQIDEKAKGSHSGAK-RPDLKAGTLMNTKGIRANSRKRI-GVVPGVEVGDI 260
++ +K+ Q +E G + +A ++ +KG S +I G VPGVEVGD
Sbjct: 316 LFHETCKKIMQEEEARPRKRDGGNFKVVCEASKILKSKGKNLYSGTQIIGTVPGVEVGDE 375
Query: 261 FFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQG 320
F +R EL L+G+H PS +GIDY+ E +A SIVSSGGY D +++ DVLIY+GQG
Sbjct: 376 FQYRMELNLLGIHRPSQSGIDYMK---DDGGELVATSIVSSGGYNDVLDNSDVLIYTGQG 432
Query: 321 GTSREKGAS----DQKLERGNLALERSLHRGNDVRVIRGMRD---EAHPTGKVYVYDGLY 373
G +K + DQ+L GNLAL+ S+++ N VRVIRG+++ ++ K YVYDGLY
Sbjct: 433 GNVGKKKNNEPPKDQQLVTGNLALKNSINKKNPVRVIRGIKNTTLQSSVVAKNYVYDGLY 492
Query: 374 KIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTSGAE 433
++ W E G VFK+KL R+PGQP+ + WK + + + KS R G+ D+T G E
Sbjct: 493 LVEEYWEETGSHGKLVFKFKLRRIPGQPE--LPWKEVAK-SKKSEFRDGLCNVDITEGKE 549
Query: 434 KLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQKNG 493
LP+C VN++D+EK P F Y+ + + P+ + C C GC S C C KNG
Sbjct: 550 TLPICAVNNLDDEKPPP-FIYTAKMIYPDWCRPIPP-KSCGCTNGCSK-SKNCACIVKNG 606
Query: 494 GYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWD 553
G +PY G + ++K +VYECGP C CPPSC RVSQ G+K++LE+F+T+ +GWG+RS +
Sbjct: 607 GKIPYYD-GAIVEIKPLVYECGPHCKCPPSCNMRVSQHGIKIKLEIFKTESRGWGVRSLE 665
Query: 554 PIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQLEVFSSDVEAPKIPSPLY 613
I G+FICEYAGE++++ + E L+G +D+Y+FD + D P
Sbjct: 666 SIPIGSFICEYAGELLEDKQAESLTG--KDEYLFD---------LGDED-------DPFT 707
Query: 614 ITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGI-- 671
I A +GN+ RF+NHSC+PN+ + V+ +++ H+ F+A+ +IPP+ EL+YDY
Sbjct: 708 INAAQKGNIGRFINHSCSPNLYAQDVLYDHEEIRIPHIMFFALDNIPPLQELSYDYNYKI 767
Query: 672 --VLPLKVGQKKKKCLCGSVKCRG 693
V KKK C CGS +C G
Sbjct: 768 DQVYDSNGNIKKKFCYCGSAECSG 791
>At2g22740 similar to mammalian MHC III region protein G9a
Length = 790
Score = 357 bits (916), Expect = 1e-98
Identities = 222/578 (38%), Positives = 315/578 (54%), Gaps = 48/578 (8%)
Query: 146 GDGKRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEILNQPDGSRDAVAYTLMIYE 205
G+G R+K K+ R +D + E L+ + G + D SR+ V TL ++
Sbjct: 226 GEGSRKKKSKKNLYWRDRESLD----SPEQLRILGVGTSSGSSSGDSSRNKVKETLRLFH 281
Query: 206 VMRRKLGQIDEKAKGSHSGAK----RPDLKAGTLMNTKGIRANSRKRI-GVVPGVEVGDI 260
+ RK+ Q DE AK K R D +A T++ G NS I G VPGVEVGD
Sbjct: 282 GVCRKILQEDE-AKPEDQRRKGKGLRIDFEASTILKRNGKFLNSGVHILGEVPGVEVGDE 340
Query: 261 FFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQG 320
F +R EL ++G+H PS AGIDY+ + +A SIV+SGGY+D++++ DVL Y+GQG
Sbjct: 341 FQYRMELNILGIHKPSQAGIDYM----KYGKAKVATSIVASGGYDDHLDNSDVLTYTGQG 396
Query: 321 GTSRE--------KGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKV--YVYD 370
G + K DQKL GNLAL S+ + VRVIRG H K YVYD
Sbjct: 397 GNVMQVKKKGEELKEPEDQKLITGNLALATSIEKQTPVRVIRGKHKSTHDKSKGGNYVYD 456
Query: 371 GLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTS 430
GLY ++ W + G NVFK++L R+PGQP+ + W + + KS R G+ D++
Sbjct: 457 GLYLVEKYWQQVGSHGMNVFKFQLRRIPGQPE--LSWVEVKK--SKSKYREGLCKLDISE 512
Query: 431 GAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHK-CGCT 489
G E+ P+ VN++D+EK P FTY+ L + PV + C C C + C C
Sbjct: 513 GKEQSPISAVNEIDDEK-PPLFTYTVKLIYPDWCRPVPP-KSCCCTTRCTEAEARVCACV 570
Query: 490 QKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGL 549
+KNGG +PY+ G + K +YECGP C CP SC RV+Q G+KL LE+F+TK +GWG+
Sbjct: 571 EKNGGEIPYNFDGAIVGAKPTIYECGPLCKCPSSCYLRVTQHGIKLPLEIFKTKSRGWGV 630
Query: 550 RSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIY-----QQLEVFSSDVE 604
R I G+FICEY GE+++++ E G D+Y+FD Y Q + +
Sbjct: 631 RCLKSIPIGSFICEYVGELLEDSEAERRIG--NDEYLFDIGNRYDNSLAQGMSELMLGTQ 688
Query: 605 APKI------PSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRH 658
A + S I A ++GNV RF+NHSC+PN+ + V+ ++++ HV F+A +
Sbjct: 689 AGRSMAEGDESSGFTIDAASKGNVGRFINHSCSPNLYAQNVLYDHEDSRIPHVMFFAQDN 748
Query: 659 IPPMMELTYDYGIVL----PLKVGQKKKKCLCGSVKCR 692
IPP+ EL YDY L K K+K C CG+ CR
Sbjct: 749 IPPLQELCYDYNYALDQVRDSKGNIKQKPCFCGAAVCR 786
>At4g13460 putative protein
Length = 650
Score = 351 bits (901), Expect = 7e-97
Identities = 242/666 (36%), Positives = 344/666 (51%), Gaps = 70/666 (10%)
Query: 49 GAPFVCVSPS-GPFPSGVAPFYPFFVSPESQRLSEQNAQTPTAQRAAPISAAVPINSFRT 107
G+ + + PS P PS + P V+ +Q L+ Q P A IS+AV + T
Sbjct: 2 GSSHIPLDPSLNPSPSLIPKLEP--VTESTQNLA---FQLPNTNPQALISSAVSDFNEAT 56
Query: 108 PTGATNGDVGSSRRKSRGQ-LPEDDNFVDLSEVDGE--------------GGTGDG---- 148
+ V S R + Q L D+ L + G T D
Sbjct: 57 DFSSDYNTVAESARSAFAQRLQRHDDVAVLDSLTGAIVPVEENPEPEPNPYSTSDSSPSV 116
Query: 149 --KRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEILNQPDGSRDAVAYTLMIYEV 206
+R +PQ R E +DV P++ R+ V T MIY+
Sbjct: 117 ATQRPRPQPRSSELVRITDVGPESERQ-------------------FREHVRKTRMIYDS 157
Query: 207 MRRKLGQIDEKAKGSHSGAKRPDLKAG---TLMNTKGIRANSRKRI-GVVPGVEVGDIFF 262
+R L + K G R D KAG ++M + N KRI G +PGV+VGDIFF
Sbjct: 158 LRMFLMMEEAKRNGVGGRRARADGKAGKAGSMMRDCMLWMNRDKRIVGSIPGVQVGDIFF 217
Query: 263 FRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQGGT 322
FRFELC++GLH +GID+L +S EP+A S++ SGGYED+ + GDV++Y+GQGG
Sbjct: 218 FRFELCVMGLHGHPQSGIDFLTGSLSSNGEPIATSVIVSGGYEDDDDQGDVIMYTGQGGQ 277
Query: 323 SR-EKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVE 381
R + A Q+LE GNLA+ERS++ G +VRVIRG++ E + +VYVYDGL++I +SW +
Sbjct: 278 DRLGRQAEHQRLEGGNLAMERSMYYGIEVRVIRGLKYENEVSSRVYVYDGLFRIVDSWFD 337
Query: 382 KAKSGFNVFKYKLVRLPGQPQ---AYMIWKSILQWTDKSASRVGVILPDLTSGAEKLPVC 438
KSGF VFKY+L R+ GQ + + + + L+ S G I D+++G E +PV
Sbjct: 338 VGKSGFGVFKYRLERIEGQAEMGSSVLKFARTLKTNPLSVRPRGYINFDISNGKENVPVY 397
Query: 439 LVNDVDNEKGPAYFTYSPTLKNLNRLAPVES--SEGCTCNGGCQPGSHKCGCTQKNGGYL 496
L ND+D+++ P Y+ Y L +S + GC C GC G C C KN G +
Sbjct: 398 LFNDIDSDQEPLYYEYLAQTSFPPGLFVQQSGNASGCDCVNGCGSG---CLCEAKNSGEI 454
Query: 497 PYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIR 556
Y G L K +++ECG +C CPPSCRNRV+Q GL+ RLEVFR+ GWG+RS D +
Sbjct: 455 AYDYNGTLIRQKPLIHECGSACQCPPSCRNRVTQKGLRNRLEVFRSLETGWGVRSLDVLH 514
Query: 557 AGTFICEYAGEVIDNARVEELSGENEDDYI----FDSTR--IYQQLEVFSSDVEAPKIPS 610
AG FICEYAG + + L+ N D + F S R + L +D E P P
Sbjct: 515 AGAFICEYAGVALTREQANILT-MNGDTLVYPARFSSARWEDWGDLSQVLADFERPSYPD 573
Query: 611 ----PLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELT 666
+ NVA +++HS PNV+ + V+ ++ + V +A +IPPM EL+
Sbjct: 574 IPPVDFAMDVSKMRNVACYISHSTDPNVIVQFVLHDHNSLMFPRVMLFAAENIPPMTELS 633
Query: 667 YDYGIV 672
DYG+V
Sbjct: 634 LDYGVV 639
>At2g33290 similar to mammalian MHC III region protein G9a
Length = 651
Score = 329 bits (843), Expect = 4e-90
Identities = 207/501 (41%), Positives = 281/501 (55%), Gaps = 33/501 (6%)
Query: 194 RDAVAYTLMIYEVMRRKLGQIDEKAKGSHSGA---KRPDLKAGTLMNTKGIRANSRKRI- 249
R + T M YE +R L + E K G +R D+ A +M +G+ N K I
Sbjct: 149 RQVMKRTRMTYESLRIHL--MAESMKNHVLGQGRRRRSDMAAAYIMRDRGLWLNYDKHIV 206
Query: 250 GVVPGVEVGDIFFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVE 309
G V GVEVGDIFF+R ELC++GLH + AGID L + S EP+A SIV SGGYED+ +
Sbjct: 207 GPVTGVEVGDIFFYRMELCVLGLHGQTQAGIDCLTAERSATGEPIATSIVVSGGYEDDED 266
Query: 310 DGDVLIYSGQGGTSRE-KGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYV 368
GDVL+Y+G GG + K +Q+L GNL +ERS+H G +VRVIRG++ E + KVYV
Sbjct: 267 TGDVLVYTGHGGQDHQHKQCDNQRLVGGNLGMERSMHYGIEVRVIRGIKYENSISSKVYV 326
Query: 369 YDGLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQ---AYMIWKSILQWTDKSASRVGVIL 425
YDGLYKI + W KSGF VFK++LVR+ GQP A M + L+ G +
Sbjct: 327 YDGLYKIVDWWFAVGKSGFGVFKFRLVRIEGQPMMGSAVMRFAQTLRNKPSMVRPTGYVS 386
Query: 426 PDLTSGAEKLPVCLVNDVDNEKGPAYFTY------SPTLKNLNRLAPVESSEGCTCNGGC 479
DL++ E +PV L NDVD ++ P ++ Y P + + S GC C C
Sbjct: 387 FDLSNKKENVPVFLYNDVDGDQEPRHYEYIAKAVFPPGIFGQGGI----SRTGCECKLSC 442
Query: 480 QPGSHKCGCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEV 539
+ C C +KNGG Y G L K VV+ECG C C PSC++RV+Q GL+ RLEV
Sbjct: 443 ---TDDCLCARKNGGEFAYDDNGHLLKGKHVVFECGEFCTCGPSCKSRVTQKGLRNRLEV 499
Query: 540 FRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIF-----DSTRIYQ 594
FR+K GWG+R+ D I AG FICEYAG V+ + E LS N D ++ D R +
Sbjct: 500 FRSKETGWGVRTLDLIEAGAFICEYAGVVVTRLQAEILS-MNGDVMVYPGRFTDQWRNWG 558
Query: 595 QLEVFSSDVEAPKIPS--PL--YITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLH 650
L D P PS PL + NVA +++HS PNV+ + V+ ++ +
Sbjct: 559 DLSQVYPDFVRPNYPSLPPLDFSMDVSRMRNVACYISHSKEPNVMVQFVLHDHNHLMFPR 618
Query: 651 VAFYAIRHIPPMMELTYDYGI 671
V +A+ +I P+ EL+ DYG+
Sbjct: 619 VMLFALENISPLAELSLDYGL 639
>At2g24740 pseudogene
Length = 429
Score = 329 bits (843), Expect = 4e-90
Identities = 202/454 (44%), Positives = 257/454 (56%), Gaps = 51/454 (11%)
Query: 267 LCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQGGTSREK 326
+CLVGLH + GID L K S + P A S+V+SG Y++ ED + LIYSG GG
Sbjct: 1 MCLVGLHRNTAGGIDSLLAKESGVDGPAATSVVTSGKYDNETEDLETLIYSGHGGKP--- 57
Query: 327 GASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKAKSG 386
DQ L+RGN ALE S+ R N+VRVIRG E + KVY+YDGLY + + W KSG
Sbjct: 58 --CDQVLQRGNRALEASVRRRNEVRVIRG---ELYNNEKVYIYDGLYLVSDCWQVTGKSG 112
Query: 387 FNVFKYKLVRLPGQPQAYMIWKSI--LQWTDKSASRVGVILPDLTSGAEKLPVCLVNDVD 444
F +++KL+R PGQP Y IWK + L+ + R G IL DL+ G E L V LVN+VD
Sbjct: 113 FKEYRFKLLRKPGQPPGYAIWKLVENLRNHELIDPRQGFILGDLSFGEEGLRVPLVNEVD 172
Query: 445 NEKG--PAYFTY--SPTLKNLNRLAPVESSEGCTCNGGCQPGSHK-CGCTQKNGGYLPYS 499
E P F Y S + V+S Q H+ C C KN G LPY
Sbjct: 173 EEDKTIPDDFDYIRSQCYSGMTNDVNVDSQSLV------QSYIHQNCTCILKNCGQLPYH 226
Query: 500 AAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGT 559
+L K ++YECG SC R+ + GLKL LEVF+T GWGLRSWDPIRAGT
Sbjct: 227 D-NILVCRKPLIYECGGSCP------TRMVETGLKLHLEVFKTSNCGWGLRSWDPIRAGT 279
Query: 560 FICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQL------EVFSSDV-----EAPKI 608
FICE+ G VEE +DDY+FD++RIY E+ D E +
Sbjct: 280 FICEFTGVSKTKEEVEE-----DDDYLFDTSRIYHSFRWNYEPELLCEDACEQVSEDANL 334
Query: 609 PSPLYITARNEGNVARFMNHSCTPNVLWRPV-VRENKNEADLHVAFYAIRHIPPMMELTY 667
P+ + I+A+ +GNV RFMNH+C PNV W+P+ +N + + +A++HIPPM ELTY
Sbjct: 335 PTQVLISAKEKGNVGRFMNHNCWPNVFWQPIEYDDNNGHIYVRIGLFAMKHIPPMTELTY 394
Query: 668 DYGIVLPLKVGQK------KKKCLCGSVKCRGYF 695
DYGI K G+ KK CLCGSVKCRG F
Sbjct: 395 DYGISCVEKTGEDEVIYKGKKICLCGSVKCRGSF 428
>At5g13960 unknown protein (At5g13960)
Length = 624
Score = 308 bits (789), Expect = 7e-84
Identities = 194/503 (38%), Positives = 266/503 (52%), Gaps = 45/503 (8%)
Query: 227 RPDLKAGTLMNTKGIRANSRKRIGVVPGVEVGDIFFFRFELCLVGLHAPSMAGIDYLGTK 286
RPDLK T M RK IG +PG++VG FF R E+C VG H + GIDY+ +
Sbjct: 126 RPDLKGVTEMIKAKAILYPRKIIGDLPGIDVGHRFFSRAEMCAVGFHNHWLNGIDYMSME 185
Query: 287 VSQEEE----PLAVSIVSSGGYEDNVEDGDVLIYSGQGG---TSREKGASDQKLERGNLA 339
+E PLAVSIV SG YED++++ D + Y+GQGG T ++ DQ LERGNLA
Sbjct: 186 YEKEYSNYKLPLAVSIVMSGQYEDDLDNADTVTYTGQGGHNLTGNKRQIKDQLLERGNLA 245
Query: 340 LERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKAKSGFNVFKYKLVRLPG 399
L+ VRV RG ++ T +VY YDGLYK++ W +K SGF V+KY+L RL G
Sbjct: 246 LKHCCEYNVPVRVTRGHNCKSSYTKRVYTYDGLYKVEKFWAQKGVSGFTVYKYRLKRLEG 305
Query: 400 QPQAYMIWKSILQWT-DKSASRV-GVILPDLTSGAEKLPVCLVNDVDNE--KGPAYFTYS 455
QP+ + + S S + G++ D++ G E + N VD+ + FTY
Sbjct: 306 QPELTTDQVNFVAGRIPTSTSEIEGLVCEDISGGLEFKGIPATNRVDDSPVSPTSGFTYI 365
Query: 456 PTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQKNGGYLPYSAA--GLLADLKSVVYE 513
+L + +SS GC C G C S KC C + NGG PY G L + + VV+E
Sbjct: 366 KSLIIEPNVIIPKSSTGCNCRGSCTD-SKKCACAKLNGGNFPYVDLNDGRLIESRDVVFE 424
Query: 514 CGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNAR 573
CGP C C P C NR SQ L+ LEVFR+ KGW +RSW+ I AG+ +CEY G V A
Sbjct: 425 CGPHCGCGPKCVNRTSQKRLRFNLEVFRSAKKGWAVRSWEYIPAGSPVCEYIGVVRRTAD 484
Query: 574 VEELSGENEDDYIFD--------------------STRIYQQLEVFSSDVEAPKIPSPLY 613
V+ +S +++YIF+ + + + S D AP+
Sbjct: 485 VDTIS---DNEYIFEIDCQQTMQGLGGRQRRLRDVAVPMNNGVSQSSEDENAPE----FC 537
Query: 614 ITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVL 673
I A + GN ARF+NHSC PN+ + V+ +++ V +A +I PM ELTYDYG L
Sbjct: 538 IDAGSTGNFARFINHSCEPNLFVQCVLSSHQDIRLARVVLFAADNISPMQELTYDYGYAL 597
Query: 674 PLKVGQ----KKKKCLCGSVKCR 692
G K+ C CG++ CR
Sbjct: 598 DSVHGPDGKVKQLACYCGALNCR 620
>At2g05900 hypothetical protein
Length = 312
Score = 171 bits (434), Expect = 1e-42
Identities = 92/193 (47%), Positives = 127/193 (65%), Gaps = 4/193 (2%)
Query: 269 LVGLHAPSMAGIDYLGTKVSQEEE--PLAVSIVSSGGYEDNVEDGDVLIYSGQGGTSREK 326
LVGLH+ ++ ++++G + +EE +AVS++SSG D ED D LI++G GGT
Sbjct: 3 LVGLHSGTI-DMEFIGVEDHGDEEGKQIAVSVISSGKNADKTEDPDSLIFTGFGGTDMYH 61
Query: 327 GAS-DQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKAKS 385
G +QKLER N+ LE + + + VRV+R M+DE G +Y+YDG Y I N W E+ ++
Sbjct: 62 GQPCNQKLERLNIPLEAAFRKKSIVRVVRCMKDEKRTNGNIYIYDGTYMITNRWEEEGQN 121
Query: 386 GFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTSGAEKLPVCLVNDVDN 445
GF VFK+KLVR P Q A+ IWKSI W + + R G+IL DL++GAE L VCLVN+VD
Sbjct: 122 GFIVFKFKLVREPDQKPAFGIWKSIQNWRNGLSIRPGLILEDLSNGAENLKVCLVNEVDK 181
Query: 446 EKGPAYFTYSPTL 458
E GPA F Y +L
Sbjct: 182 ENGPALFRYVTSL 194
Score = 94.4 bits (233), Expect = 2e-19
Identities = 45/92 (48%), Positives = 58/92 (62%), Gaps = 4/92 (4%)
Query: 604 EAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMM 663
E + S L I+A+ GNVARFMNHSC+PNV W+ + RE L++ F+A++HIPP+
Sbjct: 224 EKLSVSSSLVISAKKSGNVARFMNHSCSPNVFWQSIAREQNGLWCLYIGFFAMKHIPPLT 283
Query: 664 ELTYDYGIVLPLKVGQKKKKCLCGSVKCRGYF 695
EL YDYG G KK CLC + KC G F
Sbjct: 284 ELRYDYG----KSRGGGKKMCLCRTKKCCGSF 311
>At5g47150 putative protein
Length = 328
Score = 148 bits (374), Expect = 9e-36
Identities = 83/208 (39%), Positives = 124/208 (58%), Gaps = 13/208 (6%)
Query: 201 LMIYEVMRRKLGQIDEKAKGSHSG-----AKRPDLKAGTLMNTKGIRANSRKRIGVVPGV 255
L + + ++ Q+D K G R DLK T++ G + N+ KRIG VPG+
Sbjct: 122 LEVLSLFKQVYNQLDRDKKARRGGDFLDATSRIDLKTLTVLEKMGKQVNTEKRIGSVPGI 181
Query: 256 EVGDIFFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDN-VEDGDVL 314
+GD+F ++ EL +VGLH+ M GIDY + ++ + SIV+S GY N + V+
Sbjct: 182 NIGDVFQYKTELRVVGLHSKPMCGIDY----IKLGDDRITTSIVASEGYGYNDTYNSGVM 237
Query: 315 IYSGQGGT--SREKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGL 372
+Y+G+GG +++K DQKL +GNLAL S+ + + VRVIRG + GK YVYDGL
Sbjct: 238 VYTGEGGNVINKQKKTEDQKLVKGNLALATSMRQKSQVRVIRG-EERLDRKGKRYVYDGL 296
Query: 373 YKIQNSWVEKAKSGFNVFKYKLVRLPGQ 400
Y ++ WVE+ G +V+K+KL R+PGQ
Sbjct: 297 YMVEEYWVERDVRGKSVYKFKLCRIPGQ 324
>At5g47160 putative protein
Length = 415
Score = 135 bits (339), Expect = 1e-31
Identities = 95/255 (37%), Positives = 130/255 (50%), Gaps = 35/255 (13%)
Query: 147 DGKRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEILNQPDGSRDAVAYTLMIYEV 206
D +R I+ R S D+ P E+L+ +TL+ E+
Sbjct: 183 DERRSGVLSVIQRNRLSKDLTPRQKVQEVLRI--------------------FTLVFDEL 222
Query: 207 MRRKLGQIDEKAKGSHSGAKRPDLKAGTLMNTKGIRANSRKRIGVVPGVEVGDIFFFRFE 266
R K + GS + R D + T++ G++ NS+KRIG VPG++VGD F+
Sbjct: 223 DRNKAA----RRGGSETAKSRIDYQTWTILREMGMQVNSQKRIGSVPGIKVGDKIQFKAA 278
Query: 267 LCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGG--YEDNVEDGDVLIYSGQGGTSR 324
L ++GLH M+GIDY+ + + +A SIVSS G Y D + DV+IY GQGG R
Sbjct: 279 LSVIGLHFGIMSGIDYM----YKGNKEVATSIVSSEGNDYGDRFIN-DVMIYCGQGGNMR 333
Query: 325 ---EKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVE 381
K DQKL GNLAL S+ VRVIRG R GK YVYDGLY+++ W E
Sbjct: 334 SKDHKAIKDQKLVGGNLALANSIKEKTPVRVIRGER-RLDNRGKDYVYDGLYRVEKYWEE 392
Query: 382 KAKSGFNVFKYKLVR 396
+ G +FK+KL R
Sbjct: 393 RGPQGNILFKFKLRR 407
>At3g04380 Su(VAR)3-9-related protein 4
Length = 492
Score = 125 bits (314), Expect = 8e-29
Identities = 94/323 (29%), Positives = 142/323 (43%), Gaps = 63/323 (19%)
Query: 425 LPDLTSGAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLN--------RLAPVESSEGCT-C 475
+ D+T G+E + + LV+DV +E P FTY P N+ LA + + C C
Sbjct: 144 IADITKGSESVKIPLVDDVGSEAVPK-FTYIP--HNIVYQSAYLHVSLARISDEDCCANC 200
Query: 476 NGGCQPGSHKCGCTQKNGGYLPYSAAGLLAD----------------------------- 506
G C C C ++ G Y+ GLL +
Sbjct: 201 KGNCLSADFPCTCARETSGEYAYTKEGLLKEKFLDTCLKMKKEPDSFPKVYCKDCPLERD 260
Query: 507 -------------LKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTK-GKGWGLRSW 552
++ + EC C C C NRV Q G++ +L+V+ T+ GKGWGLR+
Sbjct: 261 HDKGTYGKCDGHLIRKFIKECWRKCGCDMQCGNRVVQRGIRCQLQVYFTQEGKGWGLRTL 320
Query: 553 DPIRAGTFICEYAGEVIDNARVEELSGENEDD-YIFDSTRIYQQLEVFSSDVEAPKIPSP 611
+ GTFICEY GE++ N + + + + + + + T L+ + K
Sbjct: 321 QDLPKGTFICEYIGEILTNTELYDRNVRSSSERHTYPVT-----LDADWGSEKDLKDEEA 375
Query: 612 LYITARNEGNVARFMNHSC-TPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYG 670
L + A GNVARF+NH C N++ P+ E + H+AF+ +R + M ELT+DY
Sbjct: 376 LCLDATICGNVARFINHRCEDANMIDIPIEIETPDRHYYHIAFFTLRDVKAMDELTWDYM 435
Query: 671 IVLPLKVGQKKK-KCLCGSVKCR 692
I K K +C CGS CR
Sbjct: 436 IDFNDKSHPVKAFRCCCGSESCR 458
>At2g23750 putative SET-domain transcriptional regulator
Length = 203
Score = 125 bits (314), Expect = 8e-29
Identities = 68/187 (36%), Positives = 97/187 (51%), Gaps = 11/187 (5%)
Query: 511 VYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVID 570
VYEC C C +C+NRV Q G++ +LEVFRT+ KGWGLR+ + I GTF+CEY GEV+D
Sbjct: 21 VYECNKFCGCSRTCQNRVLQNGIRAKLEVFRTESKGWGLRACEHILRGTFVCEYIGEVLD 80
Query: 571 NARVEELS---GENEDDYIFD-STRIYQQLEVFSSDVEAPKIPSPLYITARNEGNVARFM 626
+ G + YI D I + +++ I A GN++RF+
Sbjct: 81 QQEANKRRNQYGNGDCSYILDIDANINDIGRLMEEELD-------YAIDATTHGNISRFI 133
Query: 627 NHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQKKKKCLC 686
NHSC+PN++ V+ E+ H+ YA I E+T DYG + + C C
Sbjct: 134 NHSCSPNLVNHQVIVESMESPLAHIGLYASMDIAAGEEITRDYGRRPVPSEQENEHPCHC 193
Query: 687 GSVKCRG 693
+ CRG
Sbjct: 194 KATNCRG 200
>At5g43990 SET domain protein SUVR2
Length = 717
Score = 107 bits (267), Expect = 2e-23
Identities = 87/318 (27%), Positives = 135/318 (42%), Gaps = 62/318 (19%)
Query: 427 DLTSGAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNR-----LAPVESSEGCT-CNGGCQ 480
D++ G E + + VN+V+++ P + + +L + L + + C+ C G C
Sbjct: 395 DISLGKETVEIPWVNEVNDKVPPVFHYIAQSLVYQDAAVKFSLGNIRDDQCCSSCCGDCL 454
Query: 481 PGSHKCGCTQKNGGYLPYSAAGLLAD---------------------------------- 506
S C C G+ Y+ GLL +
Sbjct: 455 APSMACRCATAFNGFA-YTVDGLLQEDFLEQCISEARDPRKQMLLYCKECPLEKAKKEVI 513
Query: 507 --------LKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRT-KGKGWGLRSWDPIRA 557
+ + EC C C +C NRV Q G+ +L+VF T G+GWGLR+ + +
Sbjct: 514 LEPCKGHLKRKAIKECWSKCGCMKNCGNRVVQQGIHNKLQVFFTPNGRGWGLRTLEKLPK 573
Query: 558 GTFICEYAGEVIDNARV-EELSGENEDDYIFDSTRIYQQLEVFSSDVEAPKIPSPLYITA 616
G F+CE AGE++ + + +S I D+ Y E S D +A L +
Sbjct: 574 GAFVCELAGEILTIPELFQRISDRPTSPVILDA---YWGSEDISGDDKA------LSLEG 624
Query: 617 RNEGNVARFMNHSC-TPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPL 675
+ GN++RF+NH C N++ PV E + H+AF+ R I M ELT+DYG+
Sbjct: 625 THYGNISRFINHRCLDANLIEIPVHAETTDSHYYHLAFFTTREIDAMEELTWDYGVPFNQ 684
Query: 676 KVGQKKK-KCLCGSVKCR 692
V C CGS CR
Sbjct: 685 DVFPTSPFHCQCGSDFCR 702
>At3g03750 unknown protein
Length = 354
Score = 97.8 bits (242), Expect = 2e-20
Identities = 82/289 (28%), Positives = 120/289 (41%), Gaps = 31/289 (10%)
Query: 427 DLTSGAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCN---------- 476
D E + + N +D+++ AYF Y+P + P G N
Sbjct: 68 DAARSLENISIPFHNSIDSQRY-AYFIYTPFQIPASSPPPPRQWWGAAANECGSESRPCF 126
Query: 477 ---------GGCQPGSHKCGCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNR 527
G C C + GY A A ++ + ECG C C C NR
Sbjct: 127 DSVSESGRFGVSLVDESGCECERCEEGYCKCLA---FAGMEEIANECGSGCGCGSDCSNR 183
Query: 528 VSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVI--DNARVEELSGENEDDY 585
V+Q G+ + L++ R + KGW L + I+ G FICEYAGE++ D AR +N D
Sbjct: 184 VTQKGVSVSLKIVRDEKKGWCLYADQLIKQGQFICEYAGELLTTDEAR----RRQNIYDK 239
Query: 586 IFDSTRIYQQLEVFSSDVEAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKN 645
+ + L V + + + + I A GNVARF+NHSC L V+ +
Sbjct: 240 LRSTQSFASALLVVREHLPSGQACLRINIDATRIGNVARFINHSCDGGNL-STVLLRSSG 298
Query: 646 EADLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQKKK-KCLCGSVKCRG 693
+ F+A + I EL++ YG V + K C CGS C G
Sbjct: 299 ALLPRLCFFAAKDIIAEEELSFSYGDVSVAGENRDDKLNCSCGSSCCLG 347
>At4g30860 unknown protein
Length = 497
Score = 91.3 bits (225), Expect = 2e-18
Identities = 60/181 (33%), Positives = 86/181 (47%), Gaps = 24/181 (13%)
Query: 514 CGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNAR 573
C C CP SC NR + K++ + +T+ GWG+ + + I FI EY GEVI +A+
Sbjct: 307 CSKGCSCPESCGNRPFRKEKKIK--IVKTEHCGWGVEAAESINKEDFIVEYIGEVISDAQ 364
Query: 574 VEELSGENEDDYIFDSTRIYQQLEVFSSDVEAPKIPSPLYITARNEGNVARFMNHSCTPN 633
E+ R++ D +I I A +GN +RF+NHSC PN
Sbjct: 365 CEQ--------------RLWDMKHKGMKDFYMCEIQKDFTIDATFKGNASRFLNHSCNPN 410
Query: 634 VLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQKKKKCLCGSVKCRG 693
V+ + + E + V +A R I LTYDY V + G + KC CGS C+G
Sbjct: 411 C----VLEKWQVEGETRVGVFAARQIEAGEPLTYDYRFV---QFG-PEVKCNCGSENCQG 462
Query: 694 Y 694
Y
Sbjct: 463 Y 463
>At1g76710 ASH1-like protein 1 (ASHH1)
Length = 492
Score = 88.2 bits (217), Expect = 1e-17
Identities = 58/183 (31%), Positives = 87/183 (46%), Gaps = 22/183 (12%)
Query: 513 ECGPS-CHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDN 571
EC P C C C+N+ Q + ++ + +G+GWGL + + I+AG FI EY GEVI
Sbjct: 66 ECTPGYCPCGVYCKNQKFQKCEYAKTKLIKCEGRGWGLVALEEIKAGQFIMEYCGEVISW 125
Query: 572 ARVEELSGENEDDYIFDSTRIYQQLEVFSSDVEAPKIPSPLYITARNEGNVARFMNHSCT 631
++ + E + D+ I + + I A +G++ARF+NHSC
Sbjct: 126 KEAKKRAQTYETHGVKDAYII--------------SLNASEAIDATKKGSLARFINHSCR 171
Query: 632 PNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQKKKKCLCGSVKC 691
PN R+ ++ V +A I P EL YDY G K +CLCG+V C
Sbjct: 172 PNC----ETRKWNVLGEVRVGIFAKESISPRTELAYDYNFEW---YGGAKVRCLCGAVAC 224
Query: 692 RGY 694
G+
Sbjct: 225 SGF 227
>At1g77300 hypothetical protein
Length = 408
Score = 85.1 bits (209), Expect = 1e-16
Identities = 54/178 (30%), Positives = 85/178 (47%), Gaps = 20/178 (11%)
Query: 517 SCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEE 576
+C C N+ Q ++ E F++ KG+GLR + +R G F+ EY GEV+D E
Sbjct: 32 TCPAGDLCSNQQFQKRKYVKFERFQSGKKGYGLRLLEDVREGQFLIEYVGEVLDMQSYE- 90
Query: 577 LSGENEDDYIFDSTRIYQQLEVFSSDVEAPKIPSPLYITARNEGNVARFMNHSCTPNVLW 636
+ +Y F + + + + ++V I A +GN+ RF+NHSC PN
Sbjct: 91 ---TRQKEYAFKGQKHFYFMTLNGNEV----------IDAGAKGNLGRFINHSCEPNCR- 136
Query: 637 RPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQKKKKCLCGSVKCRGY 694
+ ++ V ++++ + ELT+DY V G KKC CGS CRGY
Sbjct: 137 ---TEKWMVNGEICVGIFSMQDLKKGQELTFDYNYVRVF--GAAAKKCYCGSSHCRGY 189
>At2g44150 unknown protein
Length = 363
Score = 80.9 bits (198), Expect = 2e-15
Identities = 57/207 (27%), Positives = 93/207 (44%), Gaps = 23/207 (11%)
Query: 486 CGCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGK 545
C C+ + G + G + C SC C C N+ Q ++++ +T+
Sbjct: 68 CSCSSSSPGSSS-TVCGSNCHCGMLFSSCSSSCKCGSECNNKPFQQRHVKKMKLIQTEKC 126
Query: 546 GWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQLEVFSSDVEA 605
G G+ + + I AG FI EY GEVID+ EE R+++ ++
Sbjct: 127 GSGIVAEEEIEAGEFIIEYVGEVIDDKTCEE--------------RLWKMKHRGETNFYL 172
Query: 606 PKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMEL 665
+I + I A ++GN +R++NHSC PN + + + + + +A R I L
Sbjct: 173 CEITRDMVIDATHKGNKSRYINHSCNPNTQMQKWI----IDGETRIGIFATRGIKKGEHL 228
Query: 666 TYDYGIVLPLKVGQKKKKCLCGSVKCR 692
TYDY V + G + C CG+V CR
Sbjct: 229 TYDYQFV---QFG-ADQDCHCGAVGCR 251
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,292,902
Number of Sequences: 26719
Number of extensions: 829062
Number of successful extensions: 2459
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 35
Number of HSP's successfully gapped in prelim test: 34
Number of HSP's that attempted gapping in prelim test: 2250
Number of HSP's gapped (non-prelim): 113
length of query: 696
length of database: 11,318,596
effective HSP length: 106
effective length of query: 590
effective length of database: 8,486,382
effective search space: 5006965380
effective search space used: 5006965380
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0098b.1


