

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098a.6
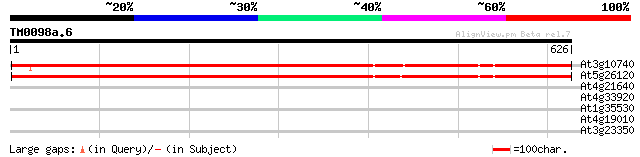
(626 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g10740 putative alpha-L-arabinofuranosidase 803 0.0
At5g26120 arabinosidase - like protein 768 0.0
At4g21640 subtilisin-like protease 33 0.48
At4g33920 unknown protein 31 2.4
At1g35530 ATP-dependent RNA helicase, putative 30 4.0
At4g19010 4-coumarate-CoA ligase - like 29 6.9
At3g23350 hypothetical protein 29 9.0
>At3g10740 putative alpha-L-arabinofuranosidase
Length = 678
Score = 803 bits (2073), Expect = 0.0
Identities = 391/632 (61%), Positives = 485/632 (75%), Gaps = 13/632 (2%)
Query: 3 VADAAGDQTSTLTVDLKSA---GRPIPETLFGVFYEEINHAGTGGLWAELVNNRGFEAGG 59
V DA D +T+ + ++ GRPIPETLFG+F+EEINHAG GGLWAELV+NRGFEAGG
Sbjct: 30 VVDAQEDPKPAVTLQVDASNGGGRPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGG 89
Query: 60 TQTPSNIAPWTIVGKESLVLLQTELSSCFERNKVALRMDVLCDQ--CPSDGVGVSNPGYW 117
TPSNI PW+IVG S + + T+ SSCFERNK+ALRMDVLCD CPS GVGV NPGYW
Sbjct: 90 QNTPSNIWPWSIVGDHSSIYVATDRSSCFERNKIALRMDVLCDSKGCPSGGVGVYNPGYW 149
Query: 118 GMNVVQKKEYKVVFFVKSTGSLDMTISFKKAEDGGILASQNVKASASEVANWKRMELKLV 177
GMN+ + K+YKV +V+STG +D+++S + LAS+ + ASAS+V+ W + E+ L
Sbjct: 150 GMNIEEGKKYKVALYVRSTGDIDLSVSLTSSNGSRTLASEKIIASASDVSKWIKKEVLLE 209
Query: 178 PTASNPKATLHLTTTQKGVIWLDQVSAMPVDTFKGHGFRTDLVNMVLELKPAFIRFPGGC 237
A++P A L LTTT+KG IW+DQVSAMPVDT KGHGFR DL M+ ++KP FIRFPGGC
Sbjct: 210 AKATDPSARLQLTTTKKGSIWIDQVSAMPVDTHKGHGFRNDLFQMMADIKPRFIRFPGGC 269
Query: 238 FIEGQTLRNAFRWKDSVGPWEERPGHFGDVWGSWTDEGLGYFEGLQLAEDIGAKPIWVFN 297
F+EG+ L NAFRWK++VGPWEERPGHFGDVW WTD+GLG+FE Q+AEDIGA PIWVFN
Sbjct: 270 FVEGEWLSNAFRWKETVGPWEERPGHFGDVWKYWTDDGLGHFEFFQMAEDIGAAPIWVFN 329
Query: 298 NGISHTDQIDTSVITPFVQEALDGIEFARGPATSKWGSLRKSMGHPEPFDLKYVAIGNED 357
NGISH D+++T+ I PFVQEALDGIEFARG A S WGS+R MG EPF+LKYVAIGNED
Sbjct: 330 NGISHNDEVETASIMPFVQEALDGIEFARGDANSTWGSVRAKMGRQEPFELKYVAIGNED 389
Query: 358 CGKKNYLGNYLAFYNAIRKAYPDIQMISNCDASSKPLDHPADLYDYHTYPNNPSNMFNNA 417
CGK Y GNY+ FY+AI+KAYPDI++ISNCD SS PLDHPAD YDYH Y + SN+F+
Sbjct: 390 CGKTYYRGNYIVFYDAIKKAYPDIKIISNCDGSSHPLDHPADYYDYHIY-TSASNLFSMY 448
Query: 418 HVFDKTPRKGPKAFVSEYALVGDQQAKLGTLIGGVSEAGFLIGLEKNSDHVAMAAYAPLF 477
H FD+T RKGPKAFVSEYA+ G + A G+L+ ++EA FLIGLEKNSD V MA+YAPLF
Sbjct: 449 HQFDRTSRKGPKAFVSEYAVTG-KDAGTGSLLASLAEAAFLIGLEKNSDIVEMASYAPLF 507
Query: 478 VNADDRKWNPDAIVFNSNQVYGTPSYWVTHMFKESNGATFLASTLQTPDPSSFDASTILW 537
VN +DR+WNPDAIVFNS+ +YGTPSYWV F ES+GAT L STL+ + +S AS I W
Sbjct: 508 VNTNDRRWNPDAIVFNSSHLYGTPSYWVQRFFAESSGATLLTSTLK-GNSTSLVASAISW 566
Query: 538 QNPQDKKTYLKIKVANLGNNQVKLGIVVHGLESS--KIIGT-KTVLTSKNALDENTFLEP 594
+N + K Y++IK N G N + ++V GL+ + ++ G+ KTVLTS N +DEN+F +P
Sbjct: 567 KN--NGKDYIRIKAVNFGANSENMQVLVTGLDPNVMRVSGSKKTVLTSTNVMDENSFSQP 624
Query: 595 RKIVPQQTPLEEASANMNVELPPLSVTSFDIL 626
K+VP ++ LE A +M V LPP S +SFD+L
Sbjct: 625 EKVVPHESLLELAEEDMTVVLPPHSFSSFDLL 656
>At5g26120 arabinosidase - like protein
Length = 674
Score = 768 bits (1983), Expect = 0.0
Identities = 378/628 (60%), Positives = 465/628 (73%), Gaps = 9/628 (1%)
Query: 3 VADAAGDQTSTLTVDLKSAGR-PIPETLFGVFYEEINHAGTGGLWAELVNNRGFEAGGTQ 61
+ DA D TL VD + R PIPETLFG+F+EEINHAG GGLWAELV+NRGFEAGG
Sbjct: 31 LVDAQEDAIVTLQVDASNVTRRPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGQI 90
Query: 62 TPSNIAPWTIVGKESLVLLQTELSSCFERNKVALRMDVLCDQ--CPSDGVGVSNPGYWGM 119
PSNI PW+I+G ES + + T+ SSCFERNK+ALRM+VLCD CP GVGV NPGYWGM
Sbjct: 91 IPSNIWPWSIIGDESSIYVVTDRSSCFERNKIALRMEVLCDSNSCPLGGVGVYNPGYWGM 150
Query: 120 NVVQKKEYKVVFFVKSTGSLDMTISFKKAEDGGILASQNVKASASEVANWKRMELKLVPT 179
N+ + K+YKVV +V+STG +D+++SF + LAS+N+ A AS++ NW + E+ L
Sbjct: 151 NIEEGKKYKVVLYVRSTGDIDVSVSFTSSNGSVTLASENIIALASDLLNWTKKEMLLEAN 210
Query: 180 ASNPKATLHLTTTQKGVIWLDQVSAMPVDTFKGHGFRTDLVNMVLELKPAFIRFPGGCFI 239
++ A L TTT+KG IW DQVSAMP+DT+KGHGFR DL M+++LKP FIRFPGGCF+
Sbjct: 211 GTDNGARLQFTTTKKGSIWFDQVSAMPMDTYKGHGFRNDLFQMMVDLKPRFIRFPGGCFV 270
Query: 240 EGQTLRNAFRWKDSVGPWEERPGHFGDVWGSWTDEGLGYFEGLQLAEDIGAKPIWVFNNG 299
EG L NAFRWK++V WEERPGH+GDVW WTD+GLG+FE QLAED+GA PIWVFNNG
Sbjct: 271 EGDWLGNAFRWKETVRAWEERPGHYGDVWKYWTDDGLGHFEFFQLAEDLGASPIWVFNNG 330
Query: 300 ISHTDQIDTSVITPFVQEALDGIEFARGPATSKWGSLRKSMGHPEPFDLKYVAIGNEDCG 359
ISH DQ++T + PFVQEA+DGIEFARG + S WGS+R +MGHPEPF+LKYVA+GNEDC
Sbjct: 331 ISHNDQVETKNVMPFVQEAIDGIEFARGDSNSTWGSVRAAMGHPEPFELKYVAVGNEDCF 390
Query: 360 KKNYLGNYLAFYNAIRKAYPDIQMISNCDASSKPLDHPADLYDYHTYPNNPSNMFNNAHV 419
K Y GNYL FYNAI+KAYPDI++ISNCDAS+KPLDHPAD +DYH Y ++F+ +H
Sbjct: 391 KSYYRGNYLEFYNAIKKAYPDIKIISNCDASAKPLDHPADYFDYHIY-TLARDLFSKSHD 449
Query: 420 FDKTPRKGPKAFVSEYALVGDQQAKLGTLIGGVSEAGFLIGLEKNSDHVAMAAYAPLFVN 479
FD TPR GPKAFVSEYA V AK G L+ + EA FL+GLEKNSD V M +YAPLFVN
Sbjct: 450 FDNTPRNGPKAFVSEYA-VNKADAKNGNLLAALGEAAFLLGLEKNSDIVEMVSYAPLFVN 508
Query: 480 ADDRKWNPDAIVFNSNQVYGTPSYWVTHMFKESNGATFLASTLQTPDPSSFDASTILWQN 539
+DR+W PDAIVFNS+ +YGTPSYWV H F ES+GAT L STL+ SS +AS I +Q
Sbjct: 509 TNDRRWIPDAIVFNSSHLYGTPSYWVQHFFTESSGATLLNSTLK-GKTSSVEASAISFQT 567
Query: 540 PQDKKTYLKIKVANLGNNQVKLGIVVHGLESSKIIGTKTVLTSKNALDENTFLEPRKIVP 599
+ K Y++IK N G V L + V GL + K VLTS + +DEN+F P IVP
Sbjct: 568 --NGKDYIQIKAVNFGEQSVNLKVAVTGLMAKFYGSKKKVLTSASVMDENSFSNPNMIVP 625
Query: 600 QQTPLE-EASANMNVELPPLSVTSFDIL 626
Q++ LE ++ LPP S +SFD+L
Sbjct: 626 QESLLEMTEQEDLMFVLPPHSFSSFDLL 653
>At4g21640 subtilisin-like protease
Length = 769
Score = 33.1 bits (74), Expect = 0.48
Identities = 24/67 (35%), Positives = 31/67 (45%), Gaps = 5/67 (7%)
Query: 5 DAAGDQTSTLTVDLKSAGRPIPETLFGVFYEEINHAGTGGLWAELVNNRGFEAGGTQTPS 64
DA DQ L+V S G IPE V + HA G+ +V G + G QT
Sbjct: 278 DAIHDQVDVLSV---SIGASIPEDSERVDFIAAFHAVAKGI--TVVAAAGNDGSGAQTIC 332
Query: 65 NIAPWTI 71
N+APW +
Sbjct: 333 NVAPWLL 339
>At4g33920 unknown protein
Length = 380
Score = 30.8 bits (68), Expect = 2.4
Identities = 17/54 (31%), Positives = 29/54 (53%)
Query: 551 VANLGNNQVKLGIVVHGLESSKIIGTKTVLTSKNALDENTFLEPRKIVPQQTPL 604
VANLG+++ LG VV G++S+K + + T N E E + + P + +
Sbjct: 144 VANLGDSRAVLGSVVSGVDSNKGAVAERLSTDHNVAVEEVRKEVKALNPDDSQI 197
>At1g35530 ATP-dependent RNA helicase, putative
Length = 1587
Score = 30.0 bits (66), Expect = 4.0
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query: 55 FEAGGTQTPSNIAPWTIVGKESLVLLQTELSSCFERNKVALRM 97
F AGG N+ T +G+E L +++ +L CF+ N LRM
Sbjct: 678 FRAGGF----NVIVATSIGEEGLDIMEVDLVICFDANVSPLRM 716
>At4g19010 4-coumarate-CoA ligase - like
Length = 566
Score = 29.3 bits (64), Expect = 6.9
Identities = 22/100 (22%), Positives = 39/100 (39%), Gaps = 4/100 (4%)
Query: 261 PGHFGDVWGSWTDEGLGYFEGLQLAEDIGAKPIWVFNNGISHTDQIDTSVITPFVQEALD 320
PG+ G++W GY + + + W+ I++ D+ I ++E +
Sbjct: 404 PGNRGELWIQGPGVMKGYLNNPKATQMSIVEDSWLRTGDIAYFDEDGYLFIVDRIKEIIK 463
Query: 321 GIEFARGPATSKWGSLRKSMGHPEPFDLKYVAIGNEDCGK 360
F PA + + HP D A NE+CG+
Sbjct: 464 YKGFQIAPADLE----AVLVSHPLIIDAAVTAAPNEECGE 499
>At3g23350 hypothetical protein
Length = 225
Score = 28.9 bits (63), Expect = 9.0
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 9/60 (15%)
Query: 407 PNNPSNMFNNAHVFDKTPRKGPKAFVSEYALVGDQ-------QAKLGTLIGGVSE--AGF 457
PN+ SN F++ + TP K Y+L+ D+ QA TL+ G+ AGF
Sbjct: 128 PNSLSNSFSSLKTSNSTPTKRIDLISESYSLIDDENKLSEKDQASKETLVSGICTKLAGF 187
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.135 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,852,243
Number of Sequences: 26719
Number of extensions: 666554
Number of successful extensions: 1356
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1341
Number of HSP's gapped (non-prelim): 8
length of query: 626
length of database: 11,318,596
effective HSP length: 105
effective length of query: 521
effective length of database: 8,513,101
effective search space: 4435325621
effective search space used: 4435325621
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0098a.6


