

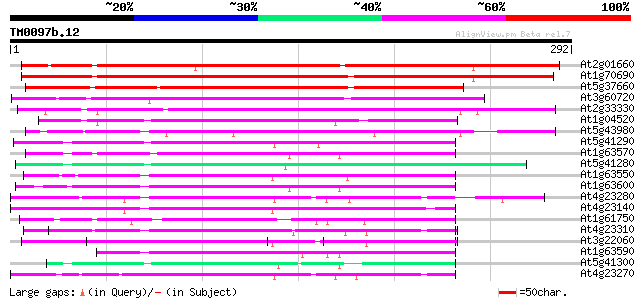
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0097b.12
(292 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g01660 unknown protein 336 1e-92
At1g70690 unknown protein 262 1e-70
At5g37660 putative protein 258 3e-69
At3g60720 secretory protein - like 175 2e-44
At2g33330 unknown protein 137 7e-33
At1g04520 unknown protein 119 1e-27
At5g43980 unknown protein 113 1e-25
At5g41290 predicted GPI-anchored protein 100 2e-21
At1g63570 unknown protein 97 1e-20
At5g41280 predicted GPI-anchored protein 95 5e-20
At1g63550 predicted GPI-anchored protein 91 6e-19
At1g63600 unknown protein 91 1e-18
At4g23280 serine /threonine kinase - like protein 87 8e-18
At4g23140 receptor-like protein kinase 5 (RLK5) 87 1e-17
At1g61750 hypothetical protein 83 2e-16
At4g23310 serine/threonine kinase - like protein 82 4e-16
At3g22060 unknown protein 80 1e-15
At1g63590 unknown protein 80 1e-15
At5g41300 predicted GPI-anchored protein 80 2e-15
At4g23270 serine/threonine kinase 79 2e-15
>At2g01660 unknown protein
Length = 288
Score = 336 bits (861), Expect = 1e-92
Identities = 168/285 (58%), Positives = 216/285 (74%), Gaps = 10/285 (3%)
Query: 7 LLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNY 66
+LFI +S + T S S+++DTFI+GGCSQ K P S YE+ VNSLLTS V+SAS Y
Sbjct: 7 VLFIAVVSLLGTFS-SAAVDTFIYGGCSQEKYFPGSP--YESNVNSLLTSFVSSASLYTY 63
Query: 67 NNFTVPATAGSGDTLYGLYQCRGDLNNDQ--CYRCVARAVSQLGTLCSAVRGGALQLEGC 124
NNFT +G ++YGLYQCRGDL++ C RCVARAVS+LG+LC+ GGALQLEGC
Sbjct: 64 NNFTTNGISGDSSSVYGLYQCRGDLSSGSGDCARCVARAVSRLGSLCAMASGGALQLEGC 123
Query: 125 FVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYGD 184
FVKYDN FLGVEDKT+VV++CGP +G SD +TRRD+V+ L S G ++R+ G+
Sbjct: 124 FVKYDNTTFLGVEDKTVVVRRCGPPVGYNSDEMTRRDSVVGSLAASSG--GSYRVGVSGE 181
Query: 185 FQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSR---H 241
QG AQCTGDLS +ECQDCL E+I RL+T+CG + WG++YLAKCYARYS G HSR +
Sbjct: 182 LQGVAQCTGDLSATECQDCLMEAIGRLRTDCGGAAWGDVYLAKCYARYSARGGHSRANGY 241
Query: 242 SDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKVCEKQRG 286
+ +N++DDEIEKTLAI++GLIAGV L++VFLSF++K CE+ +G
Sbjct: 242 GGNRNNNDDDEIEKTLAIIVGLIAGVTLLVVFLSFMAKSCERGKG 286
>At1g70690 unknown protein
Length = 299
Score = 262 bits (670), Expect = 1e-70
Identities = 136/287 (47%), Positives = 185/287 (64%), Gaps = 15/287 (5%)
Query: 7 LLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNY 66
L F+LT + PS+SS D +I+ CS K +P S YET +NSLL+S V S + T Y
Sbjct: 10 LCFLLTAVILMNPSSSSPTDNYIYAVCSPAKFSPSSG--YETNLNSLLSSFVTSTAQTRY 67
Query: 67 NNFTVPATAGSGD-TLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCF 125
NFTVP T+YG+YQCRGDL+ C CV+ AV+Q+G LCS G LQ+E C
Sbjct: 68 ANFTVPTGKPEPTVTVYGIYQCRGDLDPTACSTCVSSAVAQVGALCSNSYSGFLQMENCL 127
Query: 126 VKYDNVKFLGVEDKTMVVKKCGPSIGLT-SDALTRRDAVLAYLQTSDGVYRTWRISAYGD 184
++YDN FLGV+DKT+++ KCG + DALT+ V+ L T DG YRT G+
Sbjct: 128 IRYDNKSFLGVQDKTLILNKCGQPMEFNDQDALTKASDVIGSLGTGDGSYRT---GGNGN 184
Query: 185 FQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSR---- 240
QG AQC+GDLS S+CQDCLS++I RLK++CG + G +YL+KCYAR+S GG H+R
Sbjct: 185 VQGVAQCSGDLSTSQCQDCLSDAIGRLKSDCGMAQGGYVYLSKCYARFSVGGSHARQTPG 244
Query: 241 ----HSDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKVCEK 283
H + N +D+ + KTLAI+IG++ + L++VFL+F+ K C K
Sbjct: 245 PNFGHEGEKGNKDDNGVGKTLAIIIGIVTLIILLVVFLAFVGKCCRK 291
>At5g37660 putative protein
Length = 250
Score = 258 bits (658), Expect = 3e-69
Identities = 127/228 (55%), Positives = 163/228 (70%), Gaps = 6/228 (2%)
Query: 9 FILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNN 68
F++ +A + S +S+ DTF+FGGCSQ K +P S YE+ +NSLLTSLVNSA++++YNN
Sbjct: 16 FLIAATAPSLSSATSATDTFVFGGCSQQKFSPAS--AYESNLNSLLTSLVNSATYSSYNN 73
Query: 69 FTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKY 128
FT+ ++ S DT GL+QCRGDL+ C CVARAVSQ+G LC GGALQL GC++KY
Sbjct: 74 FTIMGSSSS-DTARGLFQCRGDLSMPDCATCVARAVSQVGPLCPFTCGGALQLAGCYIKY 132
Query: 129 DNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYGDFQGA 188
DN+ FLG EDKT+V+KKCG S G +D ++RRDAVL L G +R GD QG
Sbjct: 133 DNISFLGQEDKTVVLKKCGSSEGYNTDGISRRDAVLTELVNGGGYFRA---GGSGDVQGM 189
Query: 189 AQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGG 236
QC GDL+ SECQDCL +I RLK +CG + +G+M+LAKCYARYS G
Sbjct: 190 GQCVGDLTVSECQDCLGTAIGRLKNDCGTAVFGDMFLAKCYARYSTDG 237
>At3g60720 secretory protein - like
Length = 247
Score = 175 bits (444), Expect = 2e-44
Identities = 98/250 (39%), Positives = 150/250 (59%), Gaps = 10/250 (4%)
Query: 2 VLLLRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSA 61
+ L LLF+ S+ ++ S+S S FI+GGCS K TP + +E+ ++ L+S+V S+
Sbjct: 4 LFLFSLLFLFFYSSSSSRSSSES-HIFIYGGCSPEKYTP--NTPFESNRDTFLSSVVTSS 60
Query: 62 SFTNYNNFTV---PATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGA 118
S ++N+F V +++ S ++GLYQCR DL + C +C+ +V Q+ +C G +
Sbjct: 61 SDASFNSFAVGNDSSSSSSSSAVFGLYQCRDDLRSSDCSKCIQTSVDQITLICPYSYGAS 120
Query: 119 LQLEGCFVKYDNVKFLGVEDKTMVVKKC-GPSIGLTSDALTRRDAVLAYLQTSDGVYRTW 177
LQLEGCF++Y+ FLG D ++ KKC S+ D RRD VL+ L+++ Y
Sbjct: 121 LQLEGCFLRYETNDFLGKPDTSLRYKKCSSKSVENDYDFFKRRDDVLSDLESTQLGY--- 177
Query: 178 RISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGD 237
++S G +G AQC GDLSPS+C CL+ES+ +LK CG++ E+YLA+CYARY G
Sbjct: 178 KVSRSGLVEGYAQCVGDLSPSDCTACLAESVGKLKNLCGSAVAAEVYLAQCYARYWGSGY 237
Query: 238 HSRHSDDDSN 247
+ S N
Sbjct: 238 YDFSSGQSIN 247
>At2g33330 unknown protein
Length = 304
Score = 137 bits (345), Expect = 7e-33
Identities = 85/297 (28%), Positives = 144/297 (47%), Gaps = 22/297 (7%)
Query: 5 LRLLFILTLSAIT----TPSTSSSIDTFIFGGCSQPKNTPVSDP--VYETGVNSLLTSLV 58
L LL+I+ ++ + S+S I+ GC++ + +SDP +Y ++++ LV
Sbjct: 9 LLLLYIIIMALFSDLKLAKSSSPEYTNLIYKGCARQR---LSDPSGLYSQALSAMYGLLV 65
Query: 59 NSASFTNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGA 118
++ T + T T+ + T GL+QCRGDL+N+ CY CV+R G LC
Sbjct: 66 TQSTKTRFYKTTTGTTSQTSVT--GLFQCRGDLSNNDCYNCVSRLPVLSGKLCGKTIAAR 123
Query: 119 LQLEGCFVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWR 178
+QL GC++ Y+ F + ++ K CG + + RRD +Q +
Sbjct: 124 VQLSGCYLLYEISGFAQISGMELLFKTCGKNNVAGTGFEQRRDTAFGVMQNGVVQGHGFY 183
Query: 179 ISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS--EGG 236
+ Y QC GD+ S+C C+ ++QR + ECG+S G++YL KC+ YS G
Sbjct: 184 ATTYESVYVLGQCEGDIGDSDCSGCIKNALQRAQVECGSSISGQIYLHKCFVGYSFYPNG 243
Query: 237 DHSRHS---------DDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKVCEKQ 284
R S S+ + KT+AI++G AGV +++ L F+ + +K+
Sbjct: 244 VPKRSSPYPSSGSSGSSSSSSSSGTTGKTVAIIVGGTAGVGFLVICLLFVKNLMKKK 300
>At1g04520 unknown protein
Length = 270
Score = 119 bits (299), Expect = 1e-27
Identities = 69/224 (30%), Positives = 112/224 (49%), Gaps = 16/224 (7%)
Query: 16 ITTPSTSSSIDTFIFGGCSQPKNTPVSDP--VYETGVNSLLTSLVNSASFTNYNNFTVPA 73
+ S ++ T I+ GC++ + SDP +Y ++++ SLV+ ++ T + T
Sbjct: 23 VVVKSATTEYTTLIYKGCARQQ---FSDPSGLYSQALSAMFGSLVSQSTKTRFYKTT--- 76
Query: 74 TAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKF 133
T S T+ GL+QCRGDL+N CY CV+R LC +QL GC++ Y+ F
Sbjct: 77 TGTSTTTITGLFQCRGDLSNHDCYNCVSRLPVLSDKLCGKTIASRVQLSGCYLLYEVSGF 136
Query: 134 LGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQ----TSDGVYRTWRISAYGDFQGAA 189
+ M+ K CG + + RRD +Q + G Y T S Y
Sbjct: 137 SQISGMEMLFKTCGKNNIAGTGFEERRDTAFGVMQNGVVSGHGFYATTYESVY----VLG 192
Query: 190 QCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS 233
QC GD+ ++C C+ ++++ + ECG+S G++YL KC+ YS
Sbjct: 193 QCEGDVGDTDCSGCVKNALEKAQVECGSSISGQIYLHKCFIAYS 236
>At5g43980 unknown protein
Length = 303
Score = 113 bits (282), Expect = 1e-25
Identities = 79/304 (25%), Positives = 134/304 (43%), Gaps = 47/304 (15%)
Query: 9 FILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNN 68
F L AI S IF GC+ K+ P V+ + +L TSLV+ +S +++ +
Sbjct: 12 FFLPFFAI---SGDDDYKNLIFKGCANQKS-PDPTGVFSQNLKNLFTSLVSQSSQSSFAS 67
Query: 69 FTVPATAGSGDT--LYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVR------GGALQ 120
T+G+ +T + G++QCRGDL N QCY CV++ + LC R +
Sbjct: 68 ----VTSGTDNTTAVIGVFQCRGDLQNAQCYDCVSKIPKLVSKLCGGGRDDGNVVAARVH 123
Query: 121 LEGCFVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRIS 180
L GC+++Y++ F M+ + CG + +R+ + +
Sbjct: 124 LAGCYIRYESSGFRQTSGTEMLFRVCGKKDSNDPGFVGKRETAFGMAENGVKTGSSGGGG 183
Query: 181 AYGDFQGA--------AQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
G F QC G L S+C +C+ + ++ K+ECG S G++YL KC+ Y
Sbjct: 184 GGGGFYAGQYESVYVLGQCEGSLGNSDCGECVKDGFEKAKSECGESNSGQVYLQKCFVSY 243
Query: 233 S------------EGGDHSRHSDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKV 280
S GG+ +H+ E+T+A+ +G + + +IV L L
Sbjct: 244 SYYSHGVPNIEPLSGGEKRQHT-----------ERTIALAVGGVFVLGFVIVCLLVLRSA 292
Query: 281 CEKQ 284
+K+
Sbjct: 293 MKKK 296
>At5g41290 predicted GPI-anchored protein
Length = 287
Score = 99.8 bits (247), Expect = 2e-21
Identities = 65/242 (26%), Positives = 102/242 (41%), Gaps = 19/242 (7%)
Query: 3 LLLRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSAS 62
L L +T++ + S ++TF C + S+ Y +N+LL++L N +S
Sbjct: 7 LFALLCLFVTMNQAISVSDPDDMETF----CMKSSRNTTSNTTYNKNLNTLLSTLSNQSS 62
Query: 63 FTNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLE 122
F NY N T T + DT++G++ C GD+N C CV A ++ C+ R +
Sbjct: 63 FANYYNLT---TGLASDTVHGMFLCTGDVNRTTCNACVKNATIEIAKNCTNHREAIIYNV 119
Query: 123 GCFVKYDNVKFLGV---------EDKTMVVKKCGPSIGLTSDALTR---RDAVLAYLQTS 170
C V+Y + FL ++ K G SD + R ++L+ T
Sbjct: 120 DCMVRYSDKFFLTTLETNPSYWWSSNDLIPKSFGKFGQRLSDKMGEVIVRSSLLSSSFTP 179
Query: 171 DGVYRTWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYA 230
+ T R D + QCT DL P C CL ++Q L CG W +Y C
Sbjct: 180 YYLMDTTRFDNLYDLESIVQCTPDLDPRNCTTCLKLALQELTECCGNQVWAFIYTPNCMV 239
Query: 231 RY 232
+
Sbjct: 240 SF 241
Score = 30.8 bits (68), Expect = 0.90
Identities = 13/48 (27%), Positives = 21/48 (43%)
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSE 234
G CTGD++ + C C+ + + C +Y C RYS+
Sbjct: 80 GMFLCTGDVNRTTCNACVKNATIEIAKNCTNHREAIIYNVDCMVRYSD 127
Score = 30.4 bits (67), Expect = 1.2
Identities = 28/92 (30%), Positives = 39/92 (41%), Gaps = 11/92 (11%)
Query: 41 VSDPVYETGVNSLLTSLVNSASFTNYNNFTVPATAGSGDTLYGL---YQCRGDLNNDQCY 97
+SD + E V S L S +SFT Y + + T D LY L QC DL+ C
Sbjct: 159 LSDKMGEVIVRSSLLS----SSFTPY--YLMDTTRF--DNLYDLESIVQCTPDLDPRNCT 210
Query: 98 RCVARAVSQLGTLCSAVRGGALQLEGCFVKYD 129
C+ A+ +L C + C V +D
Sbjct: 211 TCLKLALQELTECCGNQVWAFIYTPNCMVSFD 242
>At1g63570 unknown protein
Length = 284
Score = 96.7 bits (239), Expect = 1e-20
Identities = 69/236 (29%), Positives = 108/236 (45%), Gaps = 21/236 (8%)
Query: 9 FILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNN 68
F +L + T S + TF CS T S YET N LLT+L ++S +Y N
Sbjct: 13 FFFSLLSHQTMSQPQHMHTF----CSVDSFTQTSS--YETNRNILLTTLSLTSSLVHYLN 66
Query: 69 FTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKY 128
T+ S DT+YG++ CRGD+N C CV A ++ T C+ + + + C V+Y
Sbjct: 67 ATIGL---SPDTVYGMFLCRGDINTTSCSDCVQTAAIEIATNCTLNKRAFIYYDECMVRY 123
Query: 129 DNVKFLG-VEDKTMVVK---KCGPSIGLTSDALTRR-DAVLAYLQTS-------DGVYRT 176
NV F E K ++V+ + P+ + L+ + D ++ + S + R
Sbjct: 124 SNVSFFSEFESKPVIVRYSLRSAPNSNRFNQTLSNKLDQLIPNVSPSTLIPYFVEDQERV 183
Query: 177 WRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
++ D QC+ DL PS C CL + + T CG + ++ KC RY
Sbjct: 184 TQLEGSYDLVSMIQCSPDLDPSNCTICLRFAYATVSTCCGVPSSALIFTPKCILRY 239
Score = 36.6 bits (83), Expect = 0.016
Identities = 34/145 (23%), Positives = 56/145 (38%), Gaps = 15/145 (10%)
Query: 153 TSDALTRRDAVLAYLQTSDGV--YRTWRISAYGD-FQGAAQCTGDLSPSECQDCLSESIQ 209
TS T R+ +L L + + Y I D G C GD++ + C DC+ +
Sbjct: 41 TSSYETNRNILLTTLSLTSSLVHYLNATIGLSPDTVYGMFLCRGDINTTSCSDCVQTAAI 100
Query: 210 RLKTECGASTWGEMYLAKCYARYSEGG------------DHSRHSDDDSNHNDDEIEKTL 257
+ T C + +Y +C RYS +S S +SN + + L
Sbjct: 101 EIATNCTLNKRAFIYYDECMVRYSNVSFFSEFESKPVIVRYSLRSAPNSNRFNQTLSNKL 160
Query: 258 AILIGLIAGVALIIVFLSFLSKVCE 282
LI ++ LI F+ +V +
Sbjct: 161 DQLIPNVSPSTLIPYFVEDQERVTQ 185
>At5g41280 predicted GPI-anchored protein
Length = 286
Score = 94.7 bits (234), Expect = 5e-20
Identities = 71/279 (25%), Positives = 111/279 (39%), Gaps = 20/279 (7%)
Query: 4 LLRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASF 63
+L LF AI+ + + TF C+ + Y T +N+LL++L N +SF
Sbjct: 9 VLLCLFFTMNQAISESDSDEHMATF----CNDSSGNFTRNTTYNTNLNTLLSTLSNQSSF 64
Query: 64 TNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEG 123
NY N T T DT++G++ C GD+N C CV A ++ C+ R +
Sbjct: 65 ANYYNLT---TGLGSDTVHGMFLCIGDVNRTTCNACVKNATIEIAKNCTNHREAIIYYFS 121
Query: 124 CFVKYDNVKFLG-VEDKTMV-----------VKKCGPSIGLTSDALTRRDAVLAYLQTSD 171
C V+Y + FL +E K K G + + R ++L+ T
Sbjct: 122 CMVRYSDKFFLSTLETKPNTYWSSDDPIPKSYDKFGQRLSDKMGEVIIRSSLLSSSFTPY 181
Query: 172 GVYRTWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYAR 231
+ T D + QC+ L P C CL ++Q L CG W ++ KC
Sbjct: 182 YLMDTTTFDNLYDLESVVQCSPHLDPKNCTTCLKLALQELTQCCGDQLWAFIFTPKCLVS 241
Query: 232 YSEGGDHSRHS-DDDSNHNDDEIEKTLAILIGLIAGVAL 269
+ S S I IL+G+I V++
Sbjct: 242 FDTSNSSSLPPLPPPSRSGSFSIRGNNKILVGMILAVSV 280
>At1g63550 predicted GPI-anchored protein
Length = 323
Score = 91.3 bits (225), Expect = 6e-19
Identities = 66/238 (27%), Positives = 108/238 (44%), Gaps = 19/238 (7%)
Query: 8 LFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYN 67
LF ++ + T S D IF C+ P N YET ++LL SL S+S +Y+
Sbjct: 12 LFYFFFFSLLSHQTMSQPD-HIFTVCN-PTNNFTQTSSYETNRDTLLASLRESSSLGHYS 69
Query: 68 NFTVPATAG-SGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFV 126
N AT G S DT++G++ CRGD+ C CV A +++ + C+ + + + C V
Sbjct: 70 N----ATEGLSPDTVHGMFLCRGDITTASCVDCVQTATTEIASNCTLNKRAVIYYDECMV 125
Query: 127 KYDNVKFLG----VEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVY-------- 174
+Y NV F V T+ + P+ + LT + + L + +S +
Sbjct: 126 RYSNVSFSSELEIVPSITIYSLRSAPNPTRFNQTLTEKFSELIFNVSSSSLVPYFVEDQE 185
Query: 175 RTWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
R + D QC+ DL C CL + R+ T CG ++ +++ KC R+
Sbjct: 186 RVTQSEGSYDLDTMVQCSPDLDIFNCTVCLRVAFFRISTCCGLPSYAKIFTPKCLLRF 243
Score = 39.7 bits (91), Expect = 0.002
Identities = 27/93 (29%), Positives = 40/93 (42%), Gaps = 5/93 (5%)
Query: 146 CGPSIGLT--SDALTRRDAVLAYLQTSD--GVYRTWRISAYGD-FQGAAQCTGDLSPSEC 200
C P+ T S T RD +LA L+ S G Y D G C GD++ + C
Sbjct: 36 CNPTNNFTQTSSYETNRDTLLASLRESSSLGHYSNATEGLSPDTVHGMFLCRGDITTASC 95
Query: 201 QDCLSESIQRLKTECGASTWGEMYLAKCYARYS 233
DC+ + + + C + +Y +C RYS
Sbjct: 96 VDCVQTATTEIASNCTLNKRAVIYYDECMVRYS 128
>At1g63600 unknown protein
Length = 302
Score = 90.5 bits (223), Expect = 1e-18
Identities = 69/245 (28%), Positives = 108/245 (43%), Gaps = 26/245 (10%)
Query: 4 LLRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASF 63
LL LLF L T S S I F C+Q + YET ++L+SL +S
Sbjct: 14 LLSLLFSTNL----TISESDHIHMSTF--CNQFSDNFTQTSTYETNRETVLSSLRLRSSL 67
Query: 64 TNYNNFTVPATAG-SGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLE 122
+Y+N ATAG S DT+ G++ CRGD++ C CV A ++ C+ + + E
Sbjct: 68 GSYSN----ATAGISPDTVRGMFLCRGDISETSCSDCVQTATLEISRNCTYQKEAFIFYE 123
Query: 123 GCFVKYDNVKFLG-VEDKTMVVK---KCGPSIGLTSDALTRRDAVLAYLQTS-------- 170
C V+Y + F V+++ +++ P++ L+ + L TS
Sbjct: 124 ECMVRYSDSSFFSLVDERPYIIRYSLSYAPNLDRFPQTLSDKMDELIINATSSPSLSSTP 183
Query: 171 ---DGVYRTWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAK 227
+ R + D AQC+ DL P C CL ++Q + C S W +++ K
Sbjct: 184 YFVEDQERVKQFEGSFDIDSMAQCSPDLDPRNCTTCLKLAVQEMLECCNQSRWAQIFTPK 243
Query: 228 CYARY 232
C RY
Sbjct: 244 CLLRY 248
Score = 37.7 bits (86), Expect = 0.007
Identities = 24/85 (28%), Positives = 38/85 (44%), Gaps = 3/85 (3%)
Query: 153 TSDALTRRDAVLAYLQ--TSDGVYRTWRISAYGD-FQGAAQCTGDLSPSECQDCLSESIQ 209
TS T R+ VL+ L+ +S G Y D +G C GD+S + C DC+ +
Sbjct: 47 TSTYETNRETVLSSLRLRSSLGSYSNATAGISPDTVRGMFLCRGDISETSCSDCVQTATL 106
Query: 210 RLKTECGASTWGEMYLAKCYARYSE 234
+ C ++ +C RYS+
Sbjct: 107 EISRNCTYQKEAFIFYEECMVRYSD 131
Score = 32.3 bits (72), Expect = 0.31
Identities = 21/89 (23%), Positives = 41/89 (45%), Gaps = 1/89 (1%)
Query: 41 VSDPVYETGVNSLLTSLVNSASFTNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCV 100
+SD + E +N+ + ++S + + V GS D + + QC DL+ C C+
Sbjct: 162 LSDKMDELIINATSSPSLSSTPYFVEDQERVKQFEGSFD-IDSMAQCSPDLDPRNCTTCL 220
Query: 101 ARAVSQLGTLCSAVRGGALQLEGCFVKYD 129
AV ++ C+ R + C ++Y+
Sbjct: 221 KLAVQEMLECCNQSRWAQIFTPKCLLRYE 249
>At4g23280 serine /threonine kinase - like protein
Length = 656
Score = 87.4 bits (215), Expect = 8e-18
Identities = 77/296 (26%), Positives = 138/296 (46%), Gaps = 38/296 (12%)
Query: 1 MVLLLRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLV-- 58
M L+ +F+ S IT+ + S+ +++ CS T S+ Y T + +LL+SL
Sbjct: 1 MSSLICFIFLFLFSFITSFTASAQNPFYLYHNCSIT-TTYSSNSTYSTNLKTLLSSLSSR 59
Query: 59 NSASFTNYNNFTVPATAGSG-DTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGG 117
N++ T + N ATAG D + GL+ CRG+++ + C C+A +V++ + C R
Sbjct: 60 NASYSTGFQN----ATAGQAPDMVTGLFLCRGNVSPEVCRSCIALSVNESLSRCPNEREA 115
Query: 118 ALQLEGCFVKYDNVKFLGV--EDKTMVVKKCGPSIGLTSDALTRRDAV-----LAYLQTS 170
E C ++Y N L D + ++ I + D RD V LA ++ +
Sbjct: 116 VFYYEQCMLRYSNRNILSTLNTDGGVFMQNARNPISVKQDRF--RDLVLNPMNLAAIEAA 173
Query: 171 DGVYR----TWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLA 226
+ R + ++A G QCT DL+ +C DCL +SI ++ + G +L
Sbjct: 174 RSIKRFAVTKFDLNALQSLYGMVQCTPDLTEQDCLDCLQQSINQVTYD---KIGGRTFLP 230
Query: 227 KCYARYSEGGDHSRHSDDDSNHNDDEIEK---TLAILIGLIAGV-ALIIVFLSFLS 278
C +RY D+ +N+ + K + I+I ++ + L ++F++F S
Sbjct: 231 SCTSRY----------DNYEFYNEFNVGKGGNSSVIVIAVVVPITVLFLLFVAFFS 276
>At4g23140 receptor-like protein kinase 5 (RLK5)
Length = 674
Score = 87.0 bits (214), Expect = 1e-17
Identities = 74/248 (29%), Positives = 112/248 (44%), Gaps = 24/248 (9%)
Query: 1 MVLLLRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLV-- 58
M L+ F+ S +T+ T+S+ D F T S+ Y T + +LL+SL
Sbjct: 1 MSSLISFNFLFLFSFLTSSFTASAQDPFYLNHYCPNTTTYSSNSTYSTNLRTLLSSLSSR 60
Query: 59 NSASFTNYNNFTVPATAGSG-DTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGG 117
N++ T + N ATAG D + GL+ CRGD++ + C CVA +V+Q LC VR
Sbjct: 61 NASYSTGFQN----ATAGKAPDRVTGLFLCRGDVSPEVCRNCVAFSVNQTLNLCPKVREA 116
Query: 118 ALQLEGCFVKYDNVKFLG---------VEDKTMVVKKCGPSI-GLTS-DALTRRDAVLAY 166
E C ++Y + L + T + I G TS + T +A
Sbjct: 117 VFYYEQCILRYSHKNILSTAITNEGEFILSNTNTISPNQKQIDGFTSFVSSTMSEAAGKA 176
Query: 167 LQTSDGVYR-TWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLK-TECGASTWGEMY 224
+S +Y ++AY + G QCT DL+ ++C CL SI + + GA +Y
Sbjct: 177 ANSSRKLYTVNTELTAYQNLYGLLQCTPDLTRADCLSCLQSSINGMALSRIGA----RLY 232
Query: 225 LAKCYARY 232
C ARY
Sbjct: 233 WPSCTARY 240
Score = 35.0 bits (79), Expect = 0.048
Identities = 15/47 (31%), Positives = 23/47 (48%)
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS 233
G C GD+SP C++C++ S+ + C Y +C RYS
Sbjct: 82 GLFLCRGDVSPEVCRNCVAFSVNQTLNLCPKVREAVFYYEQCILRYS 128
>At1g61750 hypothetical protein
Length = 352
Score = 83.2 bits (204), Expect = 2e-16
Identities = 66/248 (26%), Positives = 111/248 (44%), Gaps = 38/248 (15%)
Query: 6 RLLFILTLSAITTPSTSSSIDTFIFGGCSQPKN-TPVSDPVYETGVNSLLTSLVNSAS-- 62
+L+F+L+ I T + T++ C N TP S Y++ +++L++ L + +S
Sbjct: 10 KLIFLLSFLLIKTLNAQP---TYLLSYCYTSGNYTPNSS--YKSNLDTLISVLDSQSSNK 64
Query: 63 -FTNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQL 121
F +Y + + P T T+YG Y CRGD+++ C C++RA + C + +
Sbjct: 65 GFYSYASGSSPTT-----TVYGSYLCRGDISSSTCETCISRASKNVFIWCPVQKEAIIWY 119
Query: 122 EGCFVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALT------------RRDAVL--AYL 167
E CF++Y + K + D+ GP + TS T R D ++ AY
Sbjct: 120 EECFLRYSSRKIFSILDQ-------GPFVTWTSYDTTLYQFYFINTVEYRMDRLIQEAYS 172
Query: 168 QTSDGVYRTWRISAYG---DFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMY 224
+S T+ +S G D G QCT DL+ +C CL + K C + +Y
Sbjct: 173 SSSYFAEETYHVSYLGEVYDLNGLVQCTPDLNQYDCYRCLKSAYNETKDCCYGKRFALVY 232
Query: 225 LAKCYARY 232
+ C Y
Sbjct: 233 SSNCMLTY 240
Score = 35.0 bits (79), Expect = 0.048
Identities = 14/47 (29%), Positives = 25/47 (52%)
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS 233
G+ C GD+S S C+ C+S + + + C ++ +C+ RYS
Sbjct: 81 GSYLCRGDISSSTCETCISRASKNVFIWCPVQKEAIIWYEECFLRYS 127
>At4g23310 serine/threonine kinase - like protein
Length = 830
Score = 81.6 bits (200), Expect = 4e-16
Identities = 63/235 (26%), Positives = 102/235 (42%), Gaps = 11/235 (4%)
Query: 8 LFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVS-DPVYETGVNSLLTSLVNSASFTNY 66
+F+ S +T+ + T+ + C P T S D Y + + +LL+ L + + ++Y
Sbjct: 8 IFLFIFSFLTSFRVFAQDPTYRYHSC--PNTTIFSRDSAYFSNLQTLLSFLSSPDASSSY 65
Query: 67 NN-FTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCF 125
++ F A D + GL+ CRGDL + C+ CVA AV C R L + C
Sbjct: 66 SSGFRNDAVGTFPDRVTGLFDCRGDLPPEVCHNCVAFAVKDTLIRCPNERDVTLFYDECT 125
Query: 126 VKYDNVKFLGVEDKTMVVKKC--GPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYG 183
++Y N+ D T V C + +S +T +L+ L + Y T +A
Sbjct: 126 LRYSNLVVTSALDPTYVYHVCPSWATFPRSSTYMTNLITLLSTLSSPSASYSTGFQNATA 185
Query: 184 D-----FQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS 233
G C GD+SP C+ C+S ++ T C +Y +C RYS
Sbjct: 186 GKHPDRVTGLFNCRGDVSPEVCRRCVSFAVNETSTRCPIEKEVTLYYDQCTLRYS 240
Score = 71.6 bits (174), Expect = 5e-13
Identities = 63/225 (28%), Positives = 103/225 (45%), Gaps = 22/225 (9%)
Query: 21 TSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVN-SASF-TNYNNFTVPATAGSG 78
TS+ T+++ C P S Y T + +LL++L + SAS+ T + N ATAG
Sbjct: 134 TSALDPTYVYHVCPSWATFPRSS-TYMTNLITLLSTLSSPSASYSTGFQN----ATAGKH 188
Query: 79 -DTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKFLGVE 137
D + GL+ CRGD++ + C RCV+ AV++ T C + L + C ++Y N L
Sbjct: 189 PDRVTGLFNCRGDVSPEVCRRCVSFAVNETSTRCPIEKEVTLYYDQCTLRYSNRNILSTS 248
Query: 138 DKTMVVKKCGPSIGLTSDALTR-RDAVLAYLQTSDGV---------YRTWRISAYGDFQG 187
+ + S +TS+ R +D VL + + R+ +
Sbjct: 249 NTNGGIILAN-SQNMTSNEQARFKDLVLTTMNQATIAAANSSKRFDARSANFTTLHSLYT 307
Query: 188 AAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
QCT DL+ +C CL + I +L TE G+ + C +R+
Sbjct: 308 LVQCTHDLTRQDCLSCLQQIINQLPTE---KIGGQFIVPSCSSRF 349
>At3g22060 unknown protein
Length = 252
Score = 80.5 bits (197), Expect = 1e-15
Identities = 60/236 (25%), Positives = 99/236 (41%), Gaps = 15/236 (6%)
Query: 7 LLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNY 66
L + + +I T + S + F+F CS + + S +YE+ +N+L + L T
Sbjct: 13 LAIAIQILSIHTVLSQSQNNAFLFHKCSDIEGSFTSKSLYESNLNNLFSQLSYKVPSTG- 71
Query: 67 NNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFV 126
F +T + + + GL CRGD ++ C C+ A+ +L C + G + + C V
Sbjct: 72 --FAASSTGNTPNNVNGLALCRGDASSSDCRSCLETAIPELRQRCPNNKAGIVWYDNCLV 129
Query: 127 KYDNVKFLG---VEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYG 183
KY + F G E++ + S T ++ T+ A+L L ++ A G
Sbjct: 130 KYSSTNFFGKIDFENRFYLYNVKNVSDPSTFNSQTK--ALLTELTKKATTRDNQKLFATG 187
Query: 184 D-------FQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
+ G QCT DL C+ CL+ I L C G + C RY
Sbjct: 188 EKNIGKNKLYGLVQCTRDLKSITCKACLNGIIGELPNCCDGKEGGRVVGGSCNFRY 243
Score = 43.5 bits (101), Expect = 1e-04
Identities = 27/96 (28%), Positives = 41/96 (42%), Gaps = 2/96 (2%)
Query: 41 VSDP-VYETGVNSLLTSLVNSASFTNYNNFTVPATAGSG-DTLYGLYQCRGDLNNDQCYR 98
VSDP + + +LLT L A+ + G + LYGL QC DL + C
Sbjct: 154 VSDPSTFNSQTKALLTELTKKATTRDNQKLFATGEKNIGKNKLYGLVQCTRDLKSITCKA 213
Query: 99 CVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKFL 134
C+ + +L C GG + C +Y+ F+
Sbjct: 214 CLNGIIGELPNCCDGKEGGRVVGGSCNFRYEIYPFV 249
Score = 41.2 bits (95), Expect = 7e-04
Identities = 21/70 (30%), Positives = 33/70 (47%)
Query: 164 LAYLQTSDGVYRTWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEM 223
L+Y S G + + + G A C GD S S+C+ CL +I L+ C + G +
Sbjct: 63 LSYKVPSTGFAASSTGNTPNNVNGLALCRGDASSSDCRSCLETAIPELRQRCPNNKAGIV 122
Query: 224 YLAKCYARYS 233
+ C +YS
Sbjct: 123 WYDNCLVKYS 132
>At1g63590 unknown protein
Length = 268
Score = 80.1 bits (196), Expect = 1e-15
Identities = 59/203 (29%), Positives = 93/203 (45%), Gaps = 20/203 (9%)
Query: 46 YETGVNSLLTSLVNSASFTNYNNFTVPATAG-SGDTLYGLYQCRGDLNNDQCYRCVARAV 104
Y + +++L++L N +S +Y+N ATAG S +T+YG++ CRGDLN C CV
Sbjct: 45 YLSNRDTVLSTLRNRSSIGSYSN----ATAGLSPNTIYGMFLCRGDLNRTSCSDCVNATT 100
Query: 105 SQLGTLCSAVRGGALQLEGCFVKYDNVKFLG-VEDKTMVVK-KCGPSIG----LTSDALT 158
++ C + + C V+Y NV F VED + G S+ + L
Sbjct: 101 LEIYKSCFYRKSALVISNECIVRYSNVSFFTLVEDVPSTARFSTGNSLDSPQFFSQTLLE 160
Query: 159 RRDAVL--AYLQTS-------DGVYRTWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQ 209
+ DA++ A L +S D ++ D QC+ DL P C CL ++Q
Sbjct: 161 KLDALILRASLSSSLPVPYFVDDQQHVTQLEGSYDLHAMVQCSPDLDPRNCTVCLRLAVQ 220
Query: 210 RLKTECGASTWGEMYLAKCYARY 232
RL C + + ++ KC Y
Sbjct: 221 RLSGCCSHAQFARIFYTKCLITY 243
Score = 33.1 bits (74), Expect = 0.18
Identities = 22/67 (32%), Positives = 31/67 (45%), Gaps = 7/67 (10%)
Query: 76 GSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYD------ 129
GS D L+ + QC DL+ C C+ AV +L CS + + C + Y+
Sbjct: 192 GSYD-LHAMVQCSPDLDPRNCTVCLRLAVQRLSGCCSHAQFARIFYTKCLITYEISALQP 250
Query: 130 NVKFLGV 136
NV LGV
Sbjct: 251 NVTSLGV 257
>At5g41300 predicted GPI-anchored protein
Length = 287
Score = 79.7 bits (195), Expect = 2e-15
Identities = 58/235 (24%), Positives = 92/235 (38%), Gaps = 39/235 (16%)
Query: 20 STSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNNFTVPATAGSGD 79
S + +DTF C + Y +N++L++ N +S N N T T + D
Sbjct: 28 SETDHMDTF----CIDSSRNTTGNTTYNKNLNTMLSTFRNQSSIVNNYNLT---TGLASD 80
Query: 80 TLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKFLGVED- 138
T+YG++ C GD+N C CV A ++ C+ R + C V+Y + FL +
Sbjct: 81 TVYGMFLCTGDVNITTCNNCVKNATIEIVKNCTNHREAIIYYIDCMVRYSDKFFLSTFEK 140
Query: 139 -------------------KTMVVKKCGPSIGLTSDALTRRDAVLAYLQTS--DGVYRTW 177
K + KK G +I + S L+ YL + DG Y
Sbjct: 141 KPNSIWSGDDPIPKSLGPFKKRLYKKMGEAI-VRSSTLSSALTPYYYLDVTRFDGSY--- 196
Query: 178 RISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
D QC+ L+P C CL ++Q + C W ++ C+ Y
Sbjct: 197 ------DLDSLVQCSPHLNPENCTICLEYALQEIIDCCSDKFWAMIFTPNCFVNY 245
Score = 31.6 bits (70), Expect = 0.53
Identities = 13/48 (27%), Positives = 22/48 (45%)
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSE 234
G CTGD++ + C +C+ + + C +Y C RYS+
Sbjct: 84 GMFLCTGDVNITTCNNCVKNATIEIVKNCTNHREAIIYYIDCMVRYSD 131
>At4g23270 serine/threonine kinase
Length = 581
Score = 79.3 bits (194), Expect = 2e-15
Identities = 64/244 (26%), Positives = 112/244 (45%), Gaps = 22/244 (9%)
Query: 1 MVLLLRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNS 60
M L+ +F+ S+IT + ++ +++ CS T S+ Y T + +LL+SL +S
Sbjct: 1 MSSLISFIFLFLFSSITASAQNTF---YLYHNCSVT-TTFSSNSTYSTNLKTLLSSL-SS 55
Query: 61 ASFTNYNNFTVPATAGSG-DTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGAL 119
+ ++Y+ ATAG D + GL+ CR D++++ C CV AV++ T C + G
Sbjct: 56 LNASSYSTGFQTATAGQAPDRVTGLFLCRVDVSSEVCRSCVTFAVNETLTRCPKDKEGVF 115
Query: 120 QLEGCFVKYDNVKFLGV--EDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQ----TSDGV 173
E C ++Y N + D M ++ + + D RD VL + +
Sbjct: 116 YYEQCLLRYSNRNIVATLNTDGGMFMQSARNPLSVKQDQF--RDLVLTPMNLAAVEAARS 173
Query: 174 YRTWRI-----SAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKC 228
++ W + +A G +CT DL +C DCL I ++ + G + L C
Sbjct: 174 FKKWAVRKIDLNASQSLYGMVRCTPDLREQDCLDCLKIGINQVTYD---KIGGRILLPSC 230
Query: 229 YARY 232
+RY
Sbjct: 231 ASRY 234
Score = 35.0 bits (79), Expect = 0.048
Identities = 17/57 (29%), Positives = 27/57 (46%), Gaps = 3/57 (5%)
Query: 77 SGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKF 133
+ +LYG+ +C DL C C+ ++Q+ GG + L C +YDN F
Sbjct: 186 ASQSLYGMVRCTPDLREQDCLDCLKIGINQV---TYDKIGGRILLPSCASRYDNYAF 239
Score = 32.7 bits (73), Expect = 0.24
Identities = 14/47 (29%), Positives = 21/47 (43%)
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS 233
G C D+S C+ C++ ++ T C G Y +C RYS
Sbjct: 79 GLFLCRVDVSSEVCRSCVTFAVNETLTRCPKDKEGVFYYEQCLLRYS 125
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,629,122
Number of Sequences: 26719
Number of extensions: 276689
Number of successful extensions: 1178
Number of sequences better than 10.0: 109
Number of HSP's better than 10.0 without gapping: 100
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 815
Number of HSP's gapped (non-prelim): 247
length of query: 292
length of database: 11,318,596
effective HSP length: 98
effective length of query: 194
effective length of database: 8,700,134
effective search space: 1687825996
effective search space used: 1687825996
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0097b.12


