

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0096b.4
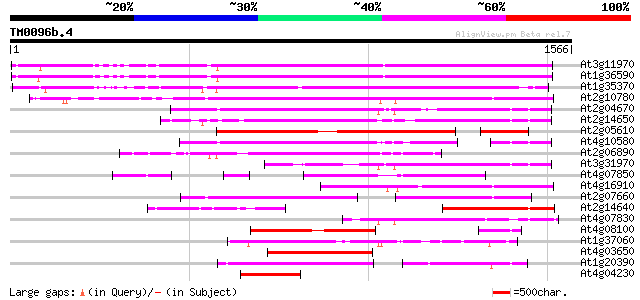
(1566 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g11970 hypothetical protein 1073 0.0
At1g36590 hypothetical protein 1068 0.0
At1g35370 hypothetical protein 969 0.0
At2g10780 pseudogene 711 0.0
At2g04670 putative retroelement pol polyprotein 650 0.0
At2g14650 putative retroelement pol polyprotein 634 0.0
At2g05610 putative retroelement pol polyprotein 586 e-167
At4g10580 putative reverse-transcriptase -like protein 493 e-139
At2g06890 putative retroelement integrase 481 e-135
At3g31970 hypothetical protein 473 e-133
At4g07850 putative polyprotein 395 e-109
At4g16910 retrotransposon like protein 387 e-107
At2g07660 putative retroelement pol polyprotein 381 e-105
At2g14640 putative retroelement pol polyprotein 319 9e-87
At4g07830 putative reverse transcriptase 296 8e-80
At4g08100 putative polyprotein 295 2e-79
At1g37060 Athila retroelment ORF 1, putative 292 9e-79
At4g03650 putative reverse transcriptase 274 3e-73
At1g20390 hypothetical protein 219 8e-57
At4g04230 putative transposon protein 191 2e-48
>At3g11970 hypothetical protein
Length = 1499
Score = 1073 bits (2774), Expect = 0.0
Identities = 615/1546 (39%), Positives = 893/1546 (56%), Gaps = 91/1546 (5%)
Query: 5 RQNKGASKEMEAKIATLEAELSGVKSTLTTMERNQ----ETLIALLEKSIGKTKVDDDST 60
++ G KE+E ++ L++ ++ +++T+ + L A+L +S VD+ ST
Sbjct: 7 QEGDGKGKEVELEVP-LDSRVTRLETTVAEQHKEMMKQFADLYAVLSRSTAGKMVDEQST 65
Query: 61 GDNVTPAKEADHGSGSGKGSMTR------LTGDVLSEFRQSAK--KVELPMFDGDDPAGW 112
D P + SG R + D + + + K++ P FDG W
Sbjct: 66 LDRSAPRSSQSMENRSGYPDPYRDARHQQVRSDHFNAYNNLTRLGKIDFPRFDGTRLKEW 125
Query: 113 ISRAEVYFRVQDTPPEVRASLAQLCMEG--PTIH--FFNSLLSEEENLTWERFKCALLER 168
+ + E +F V TP +++ +A + + T H F S + E W+ + L ER
Sbjct: 126 LFKVEEFFGVDSTPEDMKVKMAAIHFDSHASTWHQSFIQSGVGLEVLYDWKGYVKLLKER 185
Query: 169 YGGQGDGDVYEQLTELRQRGTVEEYITAFEYLTAQIPRLPEKQFLGYFLHGLKGEIRGRV 228
+ D D +L L++ + +Y FE + ++ L E+ + +L GL+ + + V
Sbjct: 186 FEDDCD-DPMAELKHLQETDGIIDYHQKFELIKTRV-NLSEEYLVSVYLAGLRTDTQMHV 243
Query: 229 RSMVTMADLSRMKILQIARAVERETMGDGGSGHARPTRSSLGGNRANRSGSNRSSDWVFV 288
R + L + + E+ + +P ++ NR+ +G +
Sbjct: 244 RMFQPQ---TVRHCLFLGKTYEK-------AHPKKPANTTWSTNRSAPTGG-------YN 286
Query: 289 KGSKETNSGSGYNNSRAGGNGPRNDRQAQPEKNRSTPRDRGFTHLSYNELMERRQKGLCF 348
K KE S + + G G QP+K +S E+ +RR KGLC+
Sbjct: 287 KYQKEGESKTDHY----GNKGNFKPVSQQPKK------------MSQQEMSDRRSKGLCY 330
Query: 349 KCGGAFHPMHQCPDKQLRVLIMEDEEEKEGGGNLLAVEVIEEEESSEGELSSMSLSQVEQ 408
C + P H K+ ++ M+ +EE E E++ +++ ++S ++S +
Sbjct: 331 FCDEKYTPEHYLVHKKTQLFRMDVDEEFEDARE----ELVNDDDEHMPQISVNAVSGIAG 386
Query: 409 VGKDKPQTIKLLGLIQGLPIVILIDSGATHNFVSTSLVHKLGKTVVDTPSLRITLGDGSQ 468
+T+++ G I ILIDSG+THNF+ + KLG V R+++ DG +
Sbjct: 387 Y-----KTMRVKGTYDKKIIFILIDSGSTHNFLDPNTAAKLGCKVDTAGLTRVSVADGRK 441
Query: 469 ARTKGKCKELMIIAGNHPLCVDAQLFELGNVDMVLGIEWLRTLGDMIVNWDKKTMSFWSG 528
R +GK + D L L +DMVLG++WL TLG + + K M F
Sbjct: 442 LRVEGKVTDFSWKLQTTTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFN 501
Query: 529 HKWVTLQGHEEQEGLLVALQTMISRAGFSGYLGKEKVQL-----EKDNKGVTGV------ 577
++ V L G V Q + L +++VQL ++ ++ G
Sbjct: 502 NQKVLLHGLTSGSVREVKAQKLQK-------LQEDQVQLAMLCVQEVSESTEGELCTINA 554
Query: 578 ------QQAELDMILERHSVVFQAPKGLPPKRNKQ-HAITLKEGEGPVNVRPYRYPHHQK 630
+++ ++ +L + +F P LPP R K H I L EG PVN RPYRY HQK
Sbjct: 555 LTSELGEESVVEEVLNEYPDIFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQK 614
Query: 631 NEIENQVKELLEGGVIRHSTSSFSSPVILVKKKDHSWRMCVDYRALNKATIPDKFPIPII 690
NEI+ V++LL G ++ S+S ++SPV+LVKKKD +WR+CVDYR LN T+ D FPIP+I
Sbjct: 615 NEIDKLVEDLLTNGTVQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLI 674
Query: 691 EELLDELHGARYFSKLDLKSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTF 750
E+L+DEL GA FSK+DL++GYHQVR+ +D+ KTAF+TH GH+E+LVMPFGL NAP+TF
Sbjct: 675 EDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATF 734
Query: 751 QSLMNDIFRHLLRKRVLVFFDDILVYSKDWPSHLEHLQEVLGILREQGLVANRKKCLFGR 810
Q LMN IF+ LRK VLVFFDDILVYS H +HL++V ++R L A KC F
Sbjct: 735 QGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAV 794
Query: 811 EKVEYLGHMISGQGVEVDPSKVESVTSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKP 870
KVEYLGH IS QG+E DP+K+++V WP P +K +RGFLGL GYYR+F+R +G IA P
Sbjct: 795 PKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGP 854
Query: 871 LTELTKKNGFEWSEKAQEAFETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQE 930
L LTK + FEW+ AQ+AFE LK L +PVL+LP F K+F +E DA G G+GA+LMQE
Sbjct: 855 LHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQE 914
Query: 931 KRPIAYFSKALGVRNLSKSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKELLQQK 990
P+AY S+ L + L S YEKEL+A+ A++ WR YLL HF + TDQRSLK LL+Q+
Sbjct: 915 GHPLAYISRQLKGKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQR 974
Query: 991 VVTMEQQNWAAKLLGFDFEISYKPGKLNKGADALSRV--NETLELRQMGSHVDWLGGKDL 1048
+ T QQ W KLL FD+EI Y+ GK N ADALSRV +E L + D L KD+
Sbjct: 975 LNTPIQQQWLPKLLEFDYEIQYRQGKENVVADALSRVEGSEVLHMAMTVVECDLL--KDI 1032
Query: 1049 KEEVSKDEELQRIIKSVHEKKDSSLGYTYENGILLYEGRLVLPRESPLIHTMLTEFHTTP 1108
+ + D +LQ II ++ DS +++ IL + ++V+P + +T+L H +
Sbjct: 1033 QAGYANDSQLQDIITALQRDPDSKKYFSWSQNILRRKSKIVVPANDNIKNTILLWLHGSG 1092
Query: 1109 QGGHSGFYRTYRRLAANVYWRGMKSAVQDFVKQCDVCQRQKYLASSPGGLLQPLPIPERI 1168
GGHSG T++R+ YW+GM +Q +++ C CQ+ K ++ GLLQPLPIP+ I
Sbjct: 1093 VGGHSGRDVTHQRVKGLFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTI 1152
Query: 1169 WEDLSMDFITGLPKSKGFEAILVVVDRLSKYAHFIPLKHPYTAKSVAEVFGKEIVRLHGV 1228
W ++SMDFI GLP S G I+VVVDRLSK AHFI L HPY+A +VA + + +LHG
Sbjct: 1153 WSEVSMDFIEGLPVSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNVFKLHGC 1212
Query: 1229 PSSIVSDRDPIFVSNFWRELFKLQGTKLKMSTAYHPESDGQSEVVNRCLETYLRCFIADQ 1288
P+SIVSDRD +F S FWRE F LQG LK+++AYHP+SDGQ+EVVNRCLETYLRC D+
Sbjct: 1213 PTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDR 1272
Query: 1289 PKTWVIWIPWAEYWYNTCFHASTGVTPFEVVYGRPPPTITRWIQGETRVEAVQKELLERD 1348
P+ W W+ AEYWYNT +H+S+ +TPFE+VYG+ PP ++ GE++V V + L ER+
Sbjct: 1273 PQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQERE 1332
Query: 1349 EALRQLRLQLARAQDRMKQFADRKRSDRSFSIGEWVFVKLRAHRQKSVVTRIYAKLAAKY 1408
+ L L+ L RAQ RMKQFAD+ R++R F IG++V+VKL+ +RQ+SVV R KL+ KY
Sbjct: 1333 DMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKY 1392
Query: 1409 YGPYPVVARVGAVAYQLKLPPGSKVHPVFHVSLLKKAVGTYHEGEELPDLEGDGGILIEP 1468
+GPY ++ R G VAY+L LP S+VHPVFHVS LK VG LP + D + P
Sbjct: 1393 FGPYKIIDRCGEVAYKLALPSYSQVHPVFHVSQLKVLVGNVSTTVHLPSVMQDVFEKV-P 1451
Query: 1469 TEVLATRTVQLQGQSIKQILIQWKGQQPEEATWEDVDMIKSQFPSF 1514
+V+ + V QG+++ ++L++W + EEATWE + ++ FP F
Sbjct: 1452 EKVVERKMVNRQGKAVTKVLVKWSNEPLEEATWEFLFDLQKTFPEF 1497
>At1g36590 hypothetical protein
Length = 1499
Score = 1068 bits (2763), Expect = 0.0
Identities = 613/1546 (39%), Positives = 893/1546 (57%), Gaps = 91/1546 (5%)
Query: 5 RQNKGASKEMEAKIATLEAELSGVKSTLTTMERNQ----ETLIALLEKSIGKTKVDDDST 60
++ G KE+E ++ L++ ++ +++T+ + L A+L +S VD+ ST
Sbjct: 7 QEGDGKGKEVELEVP-LDSRVTRLETTVAEQHKEMMKQFADLYAVLSRSTAGKMVDEQST 65
Query: 61 GDNVTPAKEADHGSGSG------KGSMTRLTGDVLSEFRQSAK--KVELPMFDGDDPAGW 112
D P + SG ++ D + + + K++ P FDG W
Sbjct: 66 LDRSAPRSSQSMENRSGYLDPYRDARHQQVRSDHFNAYNNLTRLGKIDFPRFDGTRLKEW 125
Query: 113 ISRAEVYFRVQDTPPEVRASLAQLCMEG--PTIH--FFNSLLSEEENLTWERFKCALLER 168
+ + E +F V TP +++ +A + + T H F S + E W+ + L ER
Sbjct: 126 LFKVEEFFGVDSTPEDMKVKMAAIHFDSHASTWHQSFIQSGVGLEVLYDWKGYVKLLKER 185
Query: 169 YGGQGDGDVYEQLTELRQRGTVEEYITAFEYLTAQIPRLPEKQFLGYFLHGLKGEIRGRV 228
+ D D +L L++ + +Y FE + ++ L E+ + +L GL+ + + V
Sbjct: 186 FEDDCD-DPMAELKHLQETDGIIDYHQKFELIKTRV-NLSEEYLVSVYLAGLRTDTQMHV 243
Query: 229 RSMVTMADLSRMKILQIARAVERETMGDGGSGHARPTRSSLGGNRANRSGSNRSSDWVFV 288
R + L + + E+ + +P ++ NR+ +G +
Sbjct: 244 RMFQPQ---TVRHCLFLGKTYEKAHL-------KKPANTTWSTNRSAPTGG-------YN 286
Query: 289 KGSKETNSGSGYNNSRAGGNGPRNDRQAQPEKNRSTPRDRGFTHLSYNELMERRQKGLCF 348
K KE S + + G G QP+K +S E+ +RR KGLC+
Sbjct: 287 KYQKEGESKTDHY----GNKGNFKPVSQQPKK------------MSQQEMSDRRSKGLCY 330
Query: 349 KCGGAFHPMHQCPDKQLRVLIMEDEEEKEGGGNLLAVEVIEEEESSEGELSSMSLSQVEQ 408
C + P H K+ ++ M+ +EE E E++ +++ ++S ++S +
Sbjct: 331 FCDEKYTPEHYLVHKKTQLFRMDVDEEFEDARE----ELVNDDDEHMPQISVNAVSGIAG 386
Query: 409 VGKDKPQTIKLLGLIQGLPIVILIDSGATHNFVSTSLVHKLGKTVVDTPSLRITLGDGSQ 468
+T+++ G I ILIDSG+THNF+ + KLG V R+++ DG +
Sbjct: 387 Y-----KTMRVKGTYDKKIIFILIDSGSTHNFLDPNTAAKLGCKVDTAGLTRVSVADGRK 441
Query: 469 ARTKGKCKELMIIAGNHPLCVDAQLFELGNVDMVLGIEWLRTLGDMIVNWDKKTMSFWSG 528
R +GK + D L L +DMVLG++WL TLG + + K M F
Sbjct: 442 LRVEGKVTDFSWKLQTTTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFN 501
Query: 529 HKWVTLQGHEEQEGLLVALQTMISRAGFSGYLGKEKVQL-----EKDNKGVTGV------ 577
++ V L G V Q + L +++VQL ++ ++ G
Sbjct: 502 NQKVLLHGLTSGSVREVKAQKLQK-------LQEDQVQLAMLCVQEVSESTEGELCTINA 554
Query: 578 ------QQAELDMILERHSVVFQAPKGLPPKRNKQ-HAITLKEGEGPVNVRPYRYPHHQK 630
+++ ++ +L + +F P LPP R K H I L EG PVN RPYRY HQK
Sbjct: 555 LTSELGEESVVEEVLNEYPDIFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQK 614
Query: 631 NEIENQVKELLEGGVIRHSTSSFSSPVILVKKKDHSWRMCVDYRALNKATIPDKFPIPII 690
NEI+ V++LL G ++ S+S ++SPV+LVKKKD +WR+CVDYR LN T+ D FPIP+I
Sbjct: 615 NEIDKLVEDLLTNGTVQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLI 674
Query: 691 EELLDELHGARYFSKLDLKSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTF 750
E+L+DEL GA FSK+DL++GYHQVR+ +D+ KTAF+TH GH+E+LVMPFGL NAP+TF
Sbjct: 675 EDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATF 734
Query: 751 QSLMNDIFRHLLRKRVLVFFDDILVYSKDWPSHLEHLQEVLGILREQGLVANRKKCLFGR 810
Q LMN IF+ LRK VLVFFDDILVYS H +HL++V ++R L A KC F
Sbjct: 735 QGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAV 794
Query: 811 EKVEYLGHMISGQGVEVDPSKVESVTSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKP 870
KVEYLGH IS QG+E DP+K+++V WP P +K +RGFLGL GYYR+F+R +G IA P
Sbjct: 795 PKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGP 854
Query: 871 LTELTKKNGFEWSEKAQEAFETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQE 930
L LTK + FEW+ AQ+AFE LK L +PVL+LP F K+F +E DA G G+GA+LMQE
Sbjct: 855 LHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQE 914
Query: 931 KRPIAYFSKALGVRNLSKSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKELLQQK 990
P+AY S+ L + L S YEKEL+A+ A++ WR YLL HF + TDQRSLK LL+Q+
Sbjct: 915 GHPLAYISRQLKGKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQR 974
Query: 991 VVTMEQQNWAAKLLGFDFEISYKPGKLNKGADALSRV--NETLELRQMGSHVDWLGGKDL 1048
+ T QQ W KLL FD+EI Y+ GK N ADALSRV +E L + D L KD+
Sbjct: 975 LNTPIQQQWLPKLLEFDYEIQYRQGKENVVADALSRVEGSEVLHMAMTVVECDLL--KDI 1032
Query: 1049 KEEVSKDEELQRIIKSVHEKKDSSLGYTYENGILLYEGRLVLPRESPLIHTMLTEFHTTP 1108
+ + D +LQ II ++ DS +++ IL + ++V+P + +T+L H +
Sbjct: 1033 QAGYANDSQLQDIITALQRDPDSKKYFSWSQNILRRKSKIVVPANDNIKNTILLWLHGSG 1092
Query: 1109 QGGHSGFYRTYRRLAANVYWRGMKSAVQDFVKQCDVCQRQKYLASSPGGLLQPLPIPERI 1168
GGHSG T++R+ Y +GM +Q +++ C CQ+ K ++ GLLQPLPIP+ I
Sbjct: 1093 VGGHSGRDVTHQRVKGLFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTI 1152
Query: 1169 WEDLSMDFITGLPKSKGFEAILVVVDRLSKYAHFIPLKHPYTAKSVAEVFGKEIVRLHGV 1228
W ++SMDFI GLP S G I+VVVDRLSK AHFI L HPY+A +VA+ + + +LHG
Sbjct: 1153 WSEVSMDFIEGLPVSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAQAYLDNVFKLHGC 1212
Query: 1229 PSSIVSDRDPIFVSNFWRELFKLQGTKLKMSTAYHPESDGQSEVVNRCLETYLRCFIADQ 1288
P+SIVSDRD +F S FWRE F LQG LK+++AYHP+SDGQ+EVVNRCLETYLRC D+
Sbjct: 1213 PTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDR 1272
Query: 1289 PKTWVIWIPWAEYWYNTCFHASTGVTPFEVVYGRPPPTITRWIQGETRVEAVQKELLERD 1348
P+ W W+ AEYWYNT +H+S+ +TPFE+VYG+ PP ++ GE++V V + L ER+
Sbjct: 1273 PQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQERE 1332
Query: 1349 EALRQLRLQLARAQDRMKQFADRKRSDRSFSIGEWVFVKLRAHRQKSVVTRIYAKLAAKY 1408
+ L L+ L RAQ RMKQFAD+ R++R F IG++V+VKL+ +RQ+SVV R KL+ KY
Sbjct: 1333 DMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKY 1392
Query: 1409 YGPYPVVARVGAVAYQLKLPPGSKVHPVFHVSLLKKAVGTYHEGEELPDLEGDGGILIEP 1468
+GPY ++ R G VAY+L LP S+VHPVFHVS LK VG LP + D + P
Sbjct: 1393 FGPYKIIDRCGEVAYKLALPSYSQVHPVFHVSQLKVLVGNVSTTVHLPSVMQDVFEKV-P 1451
Query: 1469 TEVLATRTVQLQGQSIKQILIQWKGQQPEEATWEDVDMIKSQFPSF 1514
+V+ + V QG+++ ++L++W + EEATWE + ++ FP F
Sbjct: 1452 EKVVERKMVNRQGKAVTKVLVKWSNEPLEEATWEFLFDLQKTFPEF 1497
>At1g35370 hypothetical protein
Length = 1447
Score = 969 bits (2504), Expect = 0.0
Identities = 583/1537 (37%), Positives = 845/1537 (54%), Gaps = 158/1537 (10%)
Query: 8 KGASKEMEAKIATLEAELSG-VKSTLTTMERNQETLIALLEKSIGKTKVDDDSTGDNVTP 66
KG E E IAT +L V +M ++ +++ + ++ K + + D P
Sbjct: 13 KGKGVEKEDLIATRMTKLEEMVAEQHKSMLKHMADMMSAMSRTTAKRVTEGEKVLDRSVP 72
Query: 67 AKEADHGSGSGKGSMTRLTGDVLSEFRQSAK---------------KVELPMFDGDDPAG 111
+ SG R D +FR K++ P FDG
Sbjct: 73 RSSTSSMARSGFVEHQR---DFQRDFRPEMVRQEVNNQYGNLTRLGKIDFPRFDGSRINE 129
Query: 112 WISRAEVYFRVQDTPPEVRASLAQLCMEGPTIHFFNSLLSEEENL----TWERFKCALLE 167
W+ + E +F V TP E++ + + + + +S + L W + L +
Sbjct: 130 WLFKVEEFFGVDFTPEEMKVKMVAIHFDSHAATWHHSFIQSGIGLDVFFNWPEYVKLLKD 189
Query: 168 RYGGQGDGDVYEQLTELRQRGTVEEYITAFEYLTAQIPRLPEKQFLGYFLHGLKGEIRGR 227
R+ D D +L +L++ + EY FE + ++ L E+ + +L GL+ + +
Sbjct: 190 RFEDACD-DPMAELKKLQETDGIVEYHQQFELIKVRL-NLSEEYLVSVYLAGLRTDTQMH 247
Query: 228 VRSMV--TMADLSRMKILQIARAVERETMGDGGSGHARPTRSSLGGNRANRSGSNRSSDW 285
VR T+ D L++ + ER A P ++ SS W
Sbjct: 248 VRMFEPKTVRDC-----LRLGKYYER----------AHPKKTV-------------SSTW 279
Query: 286 VFVKGSKETNSGSGYNNSRAGGNGPRNDRQAQPEKNRSTPRDRGFTHLSYNELMERRQKG 345
K T SG Y +P K D HL G
Sbjct: 280 ----SQKGTRSGGSY----------------RPVKEVEQKSD----HL-----------G 304
Query: 346 LCFKCGGAFHPMHQCPDKQLRVLIMEDEEEKEGGGNLLAVEVIEEEESSEGELSSMSLSQ 405
LC+ C F P H K+ ++ M+ +EE E AVEV+ +++ + + +S++
Sbjct: 305 LCYFCDEKFTPEHYLVHKKTQLFRMDVDEEFED-----AVEVLSDDDHEQKPMPQISVNA 359
Query: 406 VEQVGKDKPQTIKLLGLIQGLPIVILIDSGATHNFVSTSLVHKLGKTVVDTPSLRITLGD 465
V + K +K G + + ILIDSG+THNF+ +++ KLG V ++ + D
Sbjct: 360 VSGISGYKTMGVK--GTVDKRDLFILIDSGSTHNFIDSTVAAKLGCHVESAGLTKVAVAD 417
Query: 466 GSQARTKGKCKELMIIAGNHPLCVDAQLFELGNVDMVLGIEWLRTLGDMIVNWDKKTMSF 525
G + G+ K + D L L VDMVLG++WL TLG + + K M F
Sbjct: 418 GRKLNVDGQIKGFTWKLQSTTFQSDILLIPLQGVDMVLGVQWLETLGRISWEFKKLEMQF 477
Query: 526 WSGHKWVTLQG-----------HEEQEGLLVALQTMISRAGFSGYLGKEKVQLEKDNKGV 574
+ ++ V L G H+ Q+ T + + +E V E+ G
Sbjct: 478 FYKNQRVWLHGIITGSVRDIKAHKLQK-------TQADQIQLAMVCVREVVSDEEQEIGS 530
Query: 575 TG------VQQAELDMILERHSVVFQAPKGLPPKRNKQ-HAITLKEGEGPVNVRPYRYPH 627
V+++ + I+E VF P LPP R K H I L EG PVN RPYRY
Sbjct: 531 ISALTSDVVEESVVQNIVEEFPDVFAEPTDLPPFREKHDHKIKLLEGANPVNQRPYRYVV 590
Query: 628 HQKNEIENQVKELLEGGVIRHSTSSFSSPVILVKKKDHSWRMCVDYRALNKATIPDKFPI 687
HQK+EI+ V+++++ G I+ S+S F+SPV+LVKKKD +WR+CVDY LN T+ D+F I
Sbjct: 591 HQKDEIDKIVQDMIKSGTIQVSSSPFASPVVLVKKKDGTWRLCVDYTELNGMTVKDRFLI 650
Query: 688 PIIEELLDELHGARYFSKLDLKSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAP 747
P+IE+L+DEL G+ FSK+DL++GYHQVR+ +D+ KTAF+TH GH+E+LVM FGL NAP
Sbjct: 651 PLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHFEYLVMLFGLTNAP 710
Query: 748 STFQSLMNDIFRHLLRKRVLVFFDDILVYSKDWPSHLEHLQEVLGILREQGLVANRKKCL 807
+TFQSLMN +FR LRK VLVFFDDIL+YS H EHL+ V ++R L A K
Sbjct: 711 ATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVMRLHKLFAKGSK-- 768
Query: 808 FGREKVEYLGHMISGQGVEVDPSKVESVTSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKI 867
E+LGH IS + +E DP+K+++V WPTP VK VRGFLG GYYR+F+R++G I
Sbjct: 769 ------EHLGHFISAREIETDPAKIQAVKEWPTPTTVKQVRGFLGFAGYYRRFVRNFGVI 822
Query: 868 AKPLTELTKKNGFEWSEKAQEAFETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAIL 927
A PL LTK +GF WS +AQ AF+TLK L +PVLALP F K+F +E DA G G+ A+L
Sbjct: 823 AGPLHALTKTDGFCWSLEAQSAFDTLKAVLCNAPVLALPVFDKQFMVETDACGQGIRAVL 882
Query: 928 MQEKRPIAYFSKALGVRNLSKSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKELL 987
MQ+ P+AY S+ L + L S YEKEL+A A++ WR YLL HF + TDQRSLK LL
Sbjct: 883 MQKGHPLAYISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLL 942
Query: 988 QQKVVTMEQQNWAAKLLGFDFEISYKPGKLNKGADALSRVNETLELRQMGSHVDWLGGKD 1047
+Q++ T QQ W KLL FD+EI Y+ GK N ADALSRV + L S V+ K+
Sbjct: 943 EQRLNTPVQQQWLPKLLEFDYEIQYRQGKENLVADALSRVEGSEVLHMALSIVECDFLKE 1002
Query: 1048 LKEEVSKDEELQRIIKSVHEKKDSSLGYTYENGILLYEGRLVLPRESPLIHTMLTEFHTT 1107
++ D L+ II ++ + D+ Y++ IL + ++V+P + + + +L H +
Sbjct: 1003 IQVAYESDGVLKDIISALQQHPDAKKHYSWSQDILRRKSKIVVPNDVEITNKLLQWLHCS 1062
Query: 1108 PQGGHSGFYRTYRRLAANVYWRGMKSAVQDFVKQCDVCQRQKYLASSPGGLLQPLPIPER 1167
GG SG +++R+ + YW+GM +Q F++ C CQ+ K ++ GLLQPLPIP++
Sbjct: 1063 GMGGRSGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPLPIPDK 1122
Query: 1168 IWEDLSMDFITGLPKSKGFEAILVVVDRLSKYAHFIPLKHPYTAKSVAEVFGKEIVRLHG 1227
IW D+SMDFI GLP S G I+VVVDRLSK AHF+ L HPY+A +VA+ F + + HG
Sbjct: 1123 IWCDVSMDFIEGLPNSGGKSVIMVVVDRLSKAAHFVALAHPYSALTVAQAFLDNVYKHHG 1182
Query: 1228 VPSSIVSDRDPIFVSNFWRELFKLQGTKLKMSTAYHPESDGQSEVVNRCLETYLRCFIAD 1287
P+SIVSDRD +F S+FW+E FKLQG +L+MS+AYHP+SDGQ+EVVNRCLE YLRC
Sbjct: 1183 CPTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCLENYLRCMCHA 1242
Query: 1288 QPKTWVIWIPWAEYWYNTCFHASTGVTPFEVVYGRPPPTITRWIQGETRVEAVQKELLER 1347
+P W W+P AEYWYNT +H+S+ +TPFE+VYG+ PP ++ G+++V V + L ER
Sbjct: 1243 RPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAPPIHLPYLPGKSKVAVVARSLQER 1302
Query: 1348 DEALRQLRLQLARAQDRMKQFADRKRSDRSFSIGEWVFVKLRAHRQKSVVTRIYAKLAAK 1407
+ L L+ L RAQ RMKQFAD+ R++R+F IG++V+VKL+ +RQ+SVV R+ KL+ K
Sbjct: 1303 ENMLLFLKFHLMRAQHRMKQFADQHRTERTFDIGDFVYVKLQPYRQQSVVLRVNQKLSPK 1362
Query: 1408 YYGPYPVVARVGAVAYQLKLPPGSKVHPVFHVSLLKKAVGTYHEGEELPDLEGDGGILIE 1467
Y+GPY ++ + G V VG +LP + D + E
Sbjct: 1363 YFGPYKIIEKCGEV-----------------------MVGNVTTSTQLPSVLPD---IFE 1396
Query: 1468 --PTEVLATRTVQLQGQSIKQILIQWKGQQPEEATWE 1502
P +L + V+ QG++ +L++W G+ EEATW+
Sbjct: 1397 KAPEYILERKLVKRQGRAATMVLVKWIGEPVEEATWK 1433
>At2g10780 pseudogene
Length = 1611
Score = 711 bits (1835), Expect = 0.0
Identities = 500/1524 (32%), Positives = 740/1524 (47%), Gaps = 147/1524 (9%)
Query: 56 DDDSTGDNVTPAKEADHGSGSGKGSMTRLTGDVLSEFRQSAKKVELPMFDGDDPA---GW 112
DDDS V P + +HG R GD LS ++ DP W
Sbjct: 146 DDDSEVVEVNP--QPNHG---------RKRGDYLSLLEHVSRLGTRHFMGSTDPIVADEW 194
Query: 113 ISRAEVYFRVQDTPPEVRASLAQLCMEGPTIHFFNSL-------------LSEEENL--- 156
SR + F+ P + + +A +EG +++ ++ +E N
Sbjct: 195 RSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVEKRRGDEVRSFADFEDEFNKKYF 254
Query: 157 ---TWERFKCALLERYGGQGDGDVYEQLTELRQRGTVEEYITAFEYLTAQIPR-LPEKQF 212
W+R +CA L+ G TV EY F L + R L E+Q
Sbjct: 255 PPEAWDRLECAYLDLVQGNR---------------TVREYDEEFNRLRRYVGRELEEEQA 299
Query: 213 -LGYFLHGLKGEIRGR--VRSMVTMADLSRMKILQIARAVERETMGDGGSGHARPTRSSL 269
L F+ GL+ EIR VR+ ++++L VER M + G R
Sbjct: 300 QLRRFIRGLRIEIRNHCLVRTFNSVSEL-----------VERAAMIEEGIEEERYLNREK 348
Query: 270 GGNRANRSGSNRSSDWVFVKGSKETNSGSGYNNSRAGGNGPRNDRQAQPEKNRSTPRDRG 329
R N+S F K + G N +A R +D
Sbjct: 349 APIRNNQSTKPADKKRKFDKVDNTKSDAKTGECVTCGKNHSGTCWKAIGACGRCGSKDHA 408
Query: 330 FTHLSYNELMERRQKG----LCFKCGGAFHPMHQCPDKQLRVLIMEDEEEKEGGGNLLAV 385
E + + G CF CG H +CP +L + + GG L
Sbjct: 409 IQSCPKMEPGQSKVLGEETRTCFYCGKTGHLKRECP--KLTAEKQAGQRDNRGGNGLPPP 466
Query: 386 EVIEEEESSEGELSSMSLSQVEQVGKDKPQTIKLLGLIQGLPIVILIDSGATHNFVSTSL 445
+ S ELS + D + G + P
Sbjct: 467 PKRQAVASRVYELS--------EEANDAGNFRAITGGFRKEP------------------ 500
Query: 446 VHKLGKTVVDTPSLRITLGDGSQARTKGKCKELMIIAGNHPLCVDAQLFELGNVDMVLGI 505
+T + G G + + ++ + D + L D++LG+
Sbjct: 501 ---------NTDYGMVRAAGGQAMYPTGLVRGISVVVNGVNMPADLIIVPLKKHDVILGM 551
Query: 506 EWLRTLGDMIVNWDKKTMSFWSGHKWVTLQGHEEQEGLLVALQTMISRAGFSGYLGK--E 563
+WL I + + + F + QG G LV IS LGK E
Sbjct: 552 DWLGKYKAHI-DCHRGRIQFERDEGMLKFQGIRTTSGSLV-----ISAIQAERMLGKGCE 605
Query: 564 KVQLEKDNKGVTGVQQAELDMILERHSVVFQAPKGLPPKRNKQHAITLKEGEGPVNVRPY 623
K V + + I+ S VF A G+PP R+ I L+ G P++ PY
Sbjct: 606 AYLATITTKEVGASAELKDIPIVNEFSDVFAAVSGVPPDRSDPFTIELEPGTTPISKAPY 665
Query: 624 RYPHHQKNEIENQVKELLEGGVIRHSTSSFSSPVILVKKKDHSWRMCVDYRALNKATIPD 683
R + +++ Q++ELL+ G IR S+S + +PV+ VKKKD S+R+C+DYR LNK T+ +
Sbjct: 666 RMAPAEMAKLKKQLEELLDKGFIRPSSSPWGAPVLFVKKKDGSFRLCIDYRGLNKVTVKN 725
Query: 684 KFPIPIIEELLDELHGARYFSKLDLKSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGL 743
K+P+P I+EL+D+L GA++FSK+DL SGYHQ+ ++ DV KTAFRT H+EF+VMPFGL
Sbjct: 726 KYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPFGL 785
Query: 744 MNAPSTFQSLMNDIFRHLLRKRVLVFFDDILVYSKDWPSHLEHLQEVLGILREQGLVANR 803
NAP+ F +MN +FR L + V++F +DILVYSK W +H EHL+ VL LRE L A
Sbjct: 786 TNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHLRAVLERLREHELFAKL 845
Query: 804 KKCLFGREKVEYLGHMISGQGVEVDPSKVESVTSWPTPKNVKGVRGFLGLTGYYRKFIRD 863
KC F + V +LGH+IS QGV VDP K+ S+ WP P+N +R FLGL GYYR+F+
Sbjct: 846 SKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMS 905
Query: 864 YGKIAKPLTELT-KKNGFEWSEKAQEAFETLKKKLTTSPVLALPDFSKEFTIECDASGVG 922
+ +A+PLT LT K F WS++ +++F LK LT +PVL LP+ + +T+ DAS VG
Sbjct: 906 FASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVG 965
Query: 923 VGAILMQEKRPIAYFSKALGVRNLSKSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQRS 982
+G +LMQ+ IAY S+ L + ++ E+ A+ ++ WR YL G ++ TD +S
Sbjct: 966 LGCVLMQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFFLKIWRSYLYGAKVQIYTDHKS 1025
Query: 983 LKELLQQKVVTMEQQNWAAKLLGFDFEISYKPGKLNKGADALSRVNETLE--------LR 1034
LK + Q + + Q+ W + ++ +I+Y PGK N+ ADALSR +E +
Sbjct: 1026 LKYIFTQPELNLRQRRWMELVADYNLDIAYHPGKANQVADALSRRRSEVEAERSQVDLVN 1085
Query: 1035 QMGS-HVDWLGG--KDLKEEVSKDEELQRIIKSVHEKKDSSLGY---------TYENGIL 1082
MG+ HV+ L + L + +L I+ E+ + G+ T NG +
Sbjct: 1086 MMGTLHVNALSKEVEPLGLGAADQADLLSRIRLAQERDEEIKGWAQNNKTEYQTSNNGTI 1145
Query: 1083 LYEGRLVLPRESPLIHTMLTEFHTTPQGGHSGFYRTYRRLAANVYWRGMKSAVQDFVKQC 1142
+ GR+ +P + L +L E H + H G + YR L +W GMK V +V +C
Sbjct: 1146 VVNGRVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKC 1205
Query: 1143 DVCQRQKYLASSPGGLLQPLPIPERIWEDLSMDFITGLP---KSKGFEAILVVVDRLSKY 1199
CQ K P GLLQ LPIPE W+ ++MDF+TGLP KSK A+ VVVDRL+K
Sbjct: 1206 PTCQLVKAEHQVPSGLLQNLPIPEWKWDHITMDFVTGLPTGIKSK-HNAVWVVVDRLTKS 1264
Query: 1200 AHFIPLKHPYTAKSVAEVFGKEIVRLHGVPSSIVSDRDPIFVSNFWRELFKLQGTKLKMS 1259
AHF+ + A+ +AE + EIVRLHG+P SIVSDRD F S FW+ K GT++ +S
Sbjct: 1265 AHFMAISDKDGAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLS 1324
Query: 1260 TAYHPESDGQSEVVNRCLETYLRCFIADQPKTWVIWIPWAEYWYNTCFHASTGVTPFEVV 1319
TAYHP++D QSE + LE LR + D W ++ E+ YN F AS G++P+E +
Sbjct: 1325 TAYHPQTDEQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEAL 1384
Query: 1320 YGRPPPTITRWIQ-GETRV--EAVQKELLERDEALRQLRLQLARAQDRMKQFADRKRSDR 1376
YGR T W GE R+ + E ER ++ L+++L AQDR K +A+++R +
Sbjct: 1385 YGRACRTPLCWTPVGERRLFGPTIVDETTER---MKFLKIKLKEAQDRQKSYANKRRKEL 1441
Query: 1377 SFSIGEWVFVKLRAHRQKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLKLPPG-SKVHP 1435
F +G+ V++K ++ T KL+ +Y GPY V+ RVGAVAY+L LPP + H
Sbjct: 1442 EFQVGDLVYLKAMTYKGAGRFTS-RKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNAFHN 1500
Query: 1436 VFHVSLLKKAVGTYHEGEE--LPDLEGDGGILIEPTEVLATRTVQLQGQSIKQILIQWKG 1493
VFHVS L+K + E E P L+ + + P ++ T +G++ + + W
Sbjct: 1501 VFHVSQLRKCLSDQEESVEDIPPGLKENMTVEAWPVRIMDRMTKGTRGKARDLLKVLWNC 1560
Query: 1494 QQPEEATWEDVDMIKSQFPSFCLE 1517
+ EE TWE + +K+ FP + E
Sbjct: 1561 RGREEYTWETENKMKANFPEWFKE 1584
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 650 bits (1677), Expect = 0.0
Identities = 402/1103 (36%), Positives = 597/1103 (53%), Gaps = 91/1103 (8%)
Query: 449 LGKTVVDTPS---LRITLGDGSQARTKGKCKELMIIAGNHPLCVDAQLFELGNVDMVLGI 505
L TV D L + + G G+ K + I + D + + D++LG+
Sbjct: 350 LDHTVADCTERGHLHVKVAGGQFLAVLGRAKGVDIQIAGESMPADLIISPVELYDVILGM 409
Query: 506 EWLRTLGDMIVNWDKKTMSFWSGHKWVTLQGHEEQEGLLV--ALQT--MISRAGFSGYLG 561
+WL + ++ + +SF + QG G LV A+Q MI + G YL
Sbjct: 410 DWLDHYR-VHLDCHRGRVSFERPEGRLVYQGVRPTSGSLVISAVQAKKMIEK-GCEAYLV 467
Query: 562 KEKVQLEKDNKGVTGVQQAELDMILERHSVVFQAPKGLPPKRNKQHAITLKEGEGPVNVR 621
+ V+ ++ +++ VFQ+ +GLPP R+ I L+ G P++
Sbjct: 468 TISMPESLGQVAVSDIR------VIQEFEDVFQSLQGLPPSRSDPFTIELEPGTAPLSKA 521
Query: 622 PYRYPHHQKNEIENQVKELLEGGVIRHSTSSFSSPVILVKKKDHSWRMCVDYRALNKATI 681
PYR + E++ Q+++LL G IR STS + +PV+ VKKKD S+R+C+DYR LN T+
Sbjct: 522 PYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKKKDGSFRLCIDYRGLNWVTV 581
Query: 682 PDKFPIPIIEELLDELHGARYFSKLDLKSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPF 741
+K+P+P I+ELLD+L GA FSK+DL SGYHQ+ + E DV KTAFRT GH+EF+VMPF
Sbjct: 582 KNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPF 641
Query: 742 GLMNAPSTFQSLMNDIFRHLLRKRVLVFFDDILVYSKDWPSHLEHLQEVLGILREQGLVA 801
L NAP+ F LMN +F+ L + V++F DDILVYSK H HL+ V+ LREQ L A
Sbjct: 642 ALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLREQKLFA 701
Query: 802 NRKKCLFGREKVEYLGHMISGQGVEVDPSKVESVTSWPTPKNVKGVRGFLGLTGYYRKFI 861
KC F + ++ +LGH++S +GV VDP K+E++ WP P N +R FL LTGYYR+F+
Sbjct: 702 KLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLRLTGYYRRFV 761
Query: 862 RDYGKIAKPLTELTKKN-GFEWSEKAQEAFETLKKKLTTSPVLALPDFSKEFTIECDASG 920
+ + +A+P+T+LT K+ F WS + +E F +LK+ LT++PVLALP+ + + + DAS
Sbjct: 762 KGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASR 821
Query: 921 VGVGAILMQEKRPIAYFSKALGVRNLSKSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQ 980
VG+G +LMQ + IAY S+ L + ++ E+ + A++ WR YL G +V TD
Sbjct: 822 VGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYLYGGKVQVFTDH 881
Query: 981 RSLKELLQQKVVTMEQQNWAAKLLGFDFEISYKPGKLNKGADALSR-------------- 1026
+SLK + Q + + Q W + +D EI+Y PGK N ADALSR
Sbjct: 882 KSLKYIFNQPELNLRQMRWMELVADYDLEIAYHPGKANVVADALSRKRVGAAPGQSVEAL 941
Query: 1027 VNETLELRQMGSHVDWLGGKDLKEEVSKDEELQRIIKSVHEKKDSSL-------GYTYE- 1078
V+E LR + LG E V + + L R + ++KD L G Y+
Sbjct: 942 VSEIGALRLCVVAREPLG----LEAVDRADLLTR--ARLAQEKDEGLIAASKAEGSEYQF 995
Query: 1079 --NGILLYEGRLVLPRESPLIHTMLTEFHTTPQGGHSGFYRTYRRLAANVYWRGMKSAVQ 1136
NG + GR+ +P++ L +L+E H + H G + YR L W GMK V
Sbjct: 996 AANGTIFVYGRVCVPKDEELRREILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVA 1055
Query: 1137 DFVKQCDVCQRQKYLASSPGGLLQPLPIPERIWEDLSMDFITGLPKSKGFEAILVVVDRL 1196
++V +CDVCQ K +P+ IW V++DRL
Sbjct: 1056 NWVAECDVCQLVK----------AEHQVPDAIW---------------------VIMDRL 1084
Query: 1197 SKYAHFIPLKHPYTAKSVAEVFGKEIVRLHGVPSSIVSDRDPIFVSNFWRELFKLQGTKL 1256
+K AHF+ ++ A +A+ + EIV+LHGVP SIVSDRD F FWR GTK+
Sbjct: 1085 TKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTFAFWRAFQAKMGTKV 1144
Query: 1257 KMSTAYHPESDGQSEVVNRCLETYLRCFIADQPKTWVIWIPWAEYWYNTCFHASTGVTPF 1316
+MSTAYHP++DGQSE + LE LR + D W + E+ YN + AS G+ PF
Sbjct: 1145 QMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQASIGMAPF 1204
Query: 1317 EVVYGRPPPTITRWIQGETRVEAVQKELLERDEALRQLRLQLARAQDRMKQFADRKRSDR 1376
E +YGRP T RW Q E R + E E +R L+L + AQ R + +AD++R +
Sbjct: 1205 EALYGRPCWTPLRWTQVEERSIYGADYVQETTERIRVLKLNMKEAQARQRSYADKRRREL 1264
Query: 1377 SFSIGEWVFVKLRAHR--QKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLKLPPGSKV- 1433
F +G+ V++K+ R +S+ + KL+ +Y GP+ +V RVG VAY+L+LP +
Sbjct: 1265 EFEVGDRVYLKMAMLRGPNRSI---LETKLSPRYMGPFRIVERVGPVAYRLELPDVMRAF 1321
Query: 1434 HPVFHVSLLKKAVGTYHEGEEL-----PDLEGDGGILIEPTEVLATRTVQLQGQSIKQIL 1488
H VFHV +L+K + H+ +E+ DL+ + + P VL R +L+ + I I
Sbjct: 1322 HKVFHVLMLRKCL---HKDDEVLVKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIK 1378
Query: 1489 IQWKGQQPEEATWEDVDMIKSQF 1511
+ W E TWE +K++F
Sbjct: 1379 VLWDCDGVTEETWEPEARMKARF 1401
Score = 37.0 bits (84), Expect = 0.090
Identities = 43/187 (22%), Positives = 77/187 (40%), Gaps = 9/187 (4%)
Query: 107 DDPAGWISRAEVYFRVQDTPPEVRASLAQLCMEGPTIHFFNSLLS--EEENLTWERFKCA 164
++ W SR + F P E R LA +EG ++ S+ + + +++W F
Sbjct: 145 EEADSWRSRVQRNFGSSRCPAEYRVDLAVHFLEGDAHLWWRSVTARRRQADMSWADFVAE 204
Query: 165 LLERYGGQGDGDVYE-QLTELRQ-RGTVEEYITAFEYLTAQIPRLPE--KQFLGYFLHGL 220
+Y Q D E + EL Q +V EY F L A R E + + FL GL
Sbjct: 205 FNAKYFPQEALDRMEARFLELTQGEWSVREYDREFNRLLAYAGRGMEDDQAQMRRFLRGL 264
Query: 221 KGEIR--GRVRSMVTMADLSRMKILQIARAVERETMGDGGSGHARPTRSSLGGNRANRSG 278
+ ++R RV T A L ++ ++R+ +G + + T+ + ++ +
Sbjct: 265 RPDLRVQCRVSQYATKAALVE-TAAEVEEDLQRQVVGVSPAVQTKNTQQQVTPSKGGKPA 323
Query: 279 SNRSSDW 285
+ W
Sbjct: 324 QGQKRKW 330
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 634 bits (1636), Expect = 0.0
Identities = 403/1110 (36%), Positives = 592/1110 (53%), Gaps = 115/1110 (10%)
Query: 422 LIQGLPIVILIDSGATHNFVSTSLVHKLGKTVVDTPSLR---ITLGDGSQARTKGKCKEL 478
L+ G+ +L DSGA+H F++ + + P + + + G G+ K +
Sbjct: 307 LVGGVEAHVLFDSGASHCFITPESASR--GNIRGDPGEQLGAVKVAGGQFVAVLGRTKGV 364
Query: 479 MIIAGNHPLCVDAQLFELGNVDMVLGIEWLRTLGDMIVNWDKKTMSFWSGHKWVTLQGH- 537
I + D + + D++LG++WL H V L H
Sbjct: 365 DIQIAGESMPADLIISPVELYDVILGMDWL-------------------DHYRVHLDCHR 405
Query: 538 -----EEQEGLLVALQTMISRAGFSGYLGKEKVQLEKD-NKGVTGVQQAELDMILERHSV 591
E EG LV + SG L VQ EK KG + E
Sbjct: 406 GRVSFERPEGSLVYQGVRPT----SGSLVISAVQAEKMIEKGCEAYLEFE---------D 452
Query: 592 VFQAPKGLPPKRNKQHAITLKEGEGPVNVRPYRYPHHQKNEIENQVKELLEGGVIRHSTS 651
VFQ+ +GLPP R+ I L+ G P++ PYR + E++ Q+++LL
Sbjct: 453 VFQSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLEDLL---------- 502
Query: 652 SFSSPVILVKKKDHSWRMCVDYRALNKATIPDKFPIPIIEELLDELHGARYFSKLDLKSG 711
+PV+ VKKKD S+R+C+DYR LN T+ +K+P+P I+ELLD+L GA FSK+DL SG
Sbjct: 503 --GAPVLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSG 560
Query: 712 YHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMNDIFRHLLRKRVLVFFD 771
YH + + E DV KTAFRT GH+EF+VMPFGL NAP+ F LMN +F+ +L + V++F D
Sbjct: 561 YHLIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFID 620
Query: 772 DILVYSKDWPSHLEHLQEVLGILREQGLVANRKKCLFGREKVEYLGHMISGQGVEVDPSK 831
DILVYSK H HL+ V+ LREQ L A KC F + ++ +LGH++S +GV VDP K
Sbjct: 621 DILVYSKSLEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEK 680
Query: 832 VESVTSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKPLTELTKKN-GFEWSEKAQEAF 890
+E++ W TP N +R FLGL GYYR+F++ + +A+P+T+LT K+ F WS + +E F
Sbjct: 681 IEAIRDWHTPTNATEIRSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGF 740
Query: 891 ETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQEKRPIAYFSKALGVRNLSKSA 950
+LK+ LT++PVLALP+ + + + DASGVG+G +LMQ + IAY S+ L +
Sbjct: 741 VSLKEMLTSTPVLALPEHGEPYMVYTDASGVGLGCVLMQRGKVIAYASRQLRKHEGNYPT 800
Query: 951 YEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKELLQQKVVTMEQQNWAAKLLGFDFEI 1010
++ E+ A+ A++ WR YL G + +V TD +SLK + Q + + Q+ W + +D EI
Sbjct: 801 HDLEMAAVIFALKIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQRQWMELVADYDLEI 860
Query: 1011 SYKPGKLNKGADALSRVNETLELRQMGSHVDWLGGKDLKEEVSKDEELQRIIKSVHEKKD 1070
+Y PGK N ADALS V G+ ++ VS+ L R+ E
Sbjct: 861 AYHPGKANVVADALSH-----------KRVGAAPGQSVEALVSEIGAL-RLCAVARE--- 905
Query: 1071 SSLGY-TYENGILLYEGRLVLPRESPLIHTMLTEFHTTPQGGHSGFYRTYRRLAANVYWR 1129
LG + LL RL ++ LI E
Sbjct: 906 -PLGLEAVDRADLLTRVRLAQEKDEGLIAAYKAE-------------------------- 938
Query: 1130 GMKSAVQDFVKQCDVCQRQKYLASSPGGLLQPLPIPERIWEDLSMDFITGLPKSKGFEAI 1189
V ++V +CDVCQ K PGG+LQ LPIPE W+ +++DF+ GLP S+ +AI
Sbjct: 939 -GSEDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRTKDAI 997
Query: 1190 LVVVDRLSKYAHFIPLKHPYTAKSVAEVFGKEIVRLHGVPSSIVSDRDPIFVSNFWRELF 1249
V+VDRL+K AHF+ ++ A +A+ + EIV+LHGVP SIVSDRD F S FWR
Sbjct: 998 WVIVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQ 1057
Query: 1250 KLQGTKLKMSTAYHPESDGQSEVVNRCLETYLRCFIADQPKTWVIWIPWAEYWYNTCFHA 1309
GTK++MSTAYHP++ GQSE + LE LR + D W + E+ YN + A
Sbjct: 1058 AEMGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPA 1117
Query: 1310 STGVTPFEVVYGRPPPTITRWIQGETRVEAVQKELLERDEALRQLRLQLARAQDRMKQFA 1369
S G+ PFE +Y RP T Q R + E E +R L+L + AQDR + +A
Sbjct: 1118 SIGMAPFEALYERPCRTPLCLTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQRSYA 1177
Query: 1370 DRKRSDRSFSIGEWVFVKLRAHR--QKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLKL 1427
D++R + F +G+ V++K+ R +S+ KL+ +Y GP+ +V RVG VAY+L+L
Sbjct: 1178 DKRRRELEFEVGDRVYLKMAMLRGPNRSISE---TKLSPRYMGPFRIVERVGPVAYRLEL 1234
Query: 1428 PPGSKV-HPVFHVSLLKKAVGTYHEGEEL-----PDLEGDGGILIEPTEVLATRTVQLQG 1481
P + H VFHVS+L+K + H+ +E+ DL+ + + P VL R +L+
Sbjct: 1235 PDVMRAFHKVFHVSMLRKCL---HKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKELRR 1291
Query: 1482 QSIKQILIQWKGQQPEEATWEDVDMIKSQF 1511
+ I I + W + TWE +K++F
Sbjct: 1292 KKIPLIKVLWDCDGVTKETWEPEARMKARF 1321
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 586 bits (1510), Expect = e-167
Identities = 305/667 (45%), Positives = 412/667 (61%), Gaps = 52/667 (7%)
Query: 578 QQAELDMILERHSVVFQAPKGLPPKRNKQ-HAITLKEGEGPVNVRPYRYPHHQKNEIENQ 636
+++ ++ ++ +F P LPP R H I L E PVN RPYRY HQKNEI+
Sbjct: 3 EESFVEKVVTEFPDIFVEPTDLPPFRAPHDHKIELLEDSNPVNQRPYRYVVHQKNEIDKI 62
Query: 637 VKELLEGGVIRHSTSSFSSPVILVKKKDHSWRMCVDYRALNKATIPDKFPIPIIEELLDE 696
V ++L G I+ S+S ++SPV+LVKKKD +WR+CVDYR LN T+ D+FPIP+IE+L+DE
Sbjct: 63 VDDMLASGTIQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDRFPIPLIEDLMDE 122
Query: 697 LHGARYFSKLDLKSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMND 756
L G+ +SK+DL++GYHQVR+ D+HKTAF+TH GHYE+LVMPFGL NAP++FQSLMN
Sbjct: 123 LGGSNVYSKIDLRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMNS 182
Query: 757 IFRHLLRKRVLVFFDDILVYSKDWPSHLEHLQEVLGILREQGLVANRKKCLFGREKVEYL 816
F+ LRK VLVFFDDIL+YS H +HL+ V ++R L A KC F +VEYL
Sbjct: 183 FFKPFLRKFVLVFFDDILIYSTSMEEHKKHLEAVFEVMRVHHLFAKMSKCAFAVPRVEYL 242
Query: 817 GHMISGQGVEVDPSKVESVTSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKPLTELTK 876
GH ISG+G+ DP+K+++V WP P N+K + GFLGLTGYYR+F
Sbjct: 243 GHFISGEGIATDPAKIKAVQDWPVPVNLKQLCGFLGLTGYYRRF---------------- 286
Query: 877 KNGFEWSEKAQEAFETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQEKRPIAY 936
F +E DA G G+GA+LMQE P+A+
Sbjct: 287 -----------------------------------FVVEMDACGHGIGAVLMQEGHPLAF 311
Query: 937 FSKALGVRNLSKSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKELLQQKVVTMEQ 996
S+ L + L S YEKEL+A+ ++ WR YL+ HF + TDQRSLK LL+Q++ T Q
Sbjct: 312 ISRQLKGKQLHLSIYEKELLAVIFVVRKWRHYLISSHFIIKTDQRSLKYLLEQRLNTPIQ 371
Query: 997 QNWAAKLLGFDFEISYKPGKLNKGADALSRVNETLELRQMGSHVDWLGGKDLKEEVSKDE 1056
Q W KLL FD+EI YK GK N ADALSRV + L S V+ K+++ D
Sbjct: 372 QQWLPKLLEFDYEIQYKQGKENLVADALSRVEGSEVLHMALSVVECDLLKEIQAGYVTDG 431
Query: 1057 ELQRIIKSVHEKKDSSLGYTYENGILLYEGRLVLPRESPLIHTMLTEFHTTPQGGHSGFY 1116
++Q II + ++ DS YT+ G+L + ++V+P S + T+L H + GGHSG
Sbjct: 432 DIQGIITILQQQADSKKHYTWSQGVLRRKNKIVVPNNSGIKDTILRWLHCSGMGGHSGKE 491
Query: 1117 RTYRRLAANVYWRGMKSAVQDFVKQCDVCQRQKYLASSPGGLLQPLPIPERIWEDLSMDF 1176
T++R+ YW+ M +Q F++ C CQ+ K ++ GLLQPLPIP+RIW D+SMDF
Sbjct: 492 VTHQRVKGLFYWKSMVKDIQAFIRSCGTCQQCKSDNAASPGLLQPLPIPDRIWSDVSMDF 551
Query: 1177 ITGLPKSKGFEAILVVVDRLSKYAHFIPLKHPYTAKSVAEVFGKEIVRLHGVPSSIVSDR 1236
I GLP S G I+VVVDRLSK AHFI L HPY+A +VA+ + + +LHG PSSIV
Sbjct: 552 IDGLPLSNGKTVIMVVVDRLSKAAHFIALAHPYSAMTVAQAYLDNVFKLHGCPSSIVVSS 611
Query: 1237 DPIFVSN 1243
+ V +
Sbjct: 612 RRVLVQH 618
Score = 144 bits (363), Expect = 4e-34
Identities = 69/136 (50%), Positives = 97/136 (70%)
Query: 1313 VTPFEVVYGRPPPTITRWIQGETRVEAVQKELLERDEALRQLRLQLARAQDRMKQFADRK 1372
+TP+E VYG+PPP ++ GE++V V + + ER+ + L+ L RAQ RMKQ AD+
Sbjct: 625 MTPYEAVYGQPPPLHLPYLPGESKVAVVARSMQERESMILFLKFHLMRAQHRMKQLADQH 684
Query: 1373 RSDRSFSIGEWVFVKLRAHRQKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLKLPPGSK 1432
++R F +G++VFVKL+ +RQ+SVV R KL+ KY+GPY V+ R G VAY+L+LP S+
Sbjct: 685 ITEREFEVGDYVFVKLQPYRQQSVVMRSTQKLSPKYFGPYKVIDRCGEVAYKLQLPANSQ 744
Query: 1433 VHPVFHVSLLKKAVGT 1448
VHPVFHVS L+ VGT
Sbjct: 745 VHPVFHVSQLRVLVGT 760
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 493 bits (1270), Expect = e-139
Identities = 294/784 (37%), Positives = 443/784 (56%), Gaps = 49/784 (6%)
Query: 473 GKCKELMIIAGNHPLCVDAQLFELGNVDMVLGIEWLRTLGDMIVNWDKKTMSFWSGHKWV 532
G+ K + I + D + + D++LG++WL + ++W + + F +
Sbjct: 351 GRAKGVDIQIAGESMPADLIISPVELYDVILGMDWLDYYR-VHLDWHRGRVFFERPEGRL 409
Query: 533 TLQGHEEQEGLLVA----LQTMISRAGFSGYLGKEKVQLEKDNKGVTGVQQAELDMILER 588
QG G LV + MI + G YL + V+ ++ +++
Sbjct: 410 VYQGVRPISGSLVISAVQAEKMIEK-GCEAYLVTISMPESVGQVAVSDIR------VVQE 462
Query: 589 HSVVFQAPKGLPPKRNKQHAITLKEGEGPVNVRPYRYPHHQKNEIENQVKELLEGGVIRH 648
VFQ+ +GLPP ++ I L+ G P++ PYR + E++ Q+K+LL G IR
Sbjct: 463 FQDVFQSLQGLPPSQSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRP 522
Query: 649 STSSFSSPVILVKKKDHSWRMCVDYRALNKATIPDKFPIPIIEELLDELHGARYFSKLDL 708
STS + +PV+ VKKKD S+R+C+DYR LN+ T+ +++P+P I+ELLD+L GA FSK+DL
Sbjct: 523 STSPWGAPVLFVKKKDGSFRLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDL 582
Query: 709 KSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMNDIFRHLLRKRVLV 768
SGYHQ+ + E DV KTAFRT GH+EF+VMPFGL NAP+ F LMN +F+ L + V++
Sbjct: 583 TSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVII 642
Query: 769 FFDDILVYSKDWPSHLEHLQEVLGILREQGLVANRKKCLFGREKVEYLGHMISGQGVEVD 828
F DDILVYSK HL+ V+ LREQ L A KC F + ++ +LGH++S +GV VD
Sbjct: 643 FIDDILVYSKSPEEQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVD 702
Query: 829 PSKVESVTSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKPLTELTKKN-GFEWSEKAQ 887
P K+E++ WP P N +R FLG GYYR+F++ + +A+P+T+LT K+ F WS++ +
Sbjct: 703 PEKIEAIRDWPRPTNATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECE 762
Query: 888 EAFETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQEKRPIAYFSKALGVRNLS 947
E F +LK+ LT++PVLALP+ + + + DAS VG+G +LMQ + IAY S+ L +
Sbjct: 763 EGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLMQHGKVIAYASRQLMKHEGN 822
Query: 948 KSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKELLQQKVVTMEQQNWAAKLLGFD 1007
++ E+ A+ A++ WR YL G +V TD +SLK + Q + + Q+ W + +D
Sbjct: 823 YPTHDLEMAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYD 882
Query: 1008 FEISYKPGKLNKGADALSRVNETLELRQMGSHVDWLGGKDLKEEVSKDEELQRIIKSVHE 1067
EI+Y PGK N DALSR +++G+ + G+ ++ VS+ L R+ E
Sbjct: 883 LEIAYHPGKANVVVDALSR-------KRVGAAL----GQSVEVLVSEIGAL-RLCAVARE 930
Query: 1068 KKDSSLGY-TYENGILLYEGRLVLPRESPLIHTMLTEFHTTPQGGHSGFYRTYRRLAANV 1126
LG + LL RL ++ L T + YR L
Sbjct: 931 ----PLGLEAVDRADLLTRVRLAQKKDEGLRAT-----------------KMYRDLKRYY 969
Query: 1127 YWRGMKSAVQDFVKQCDVCQRQKYLASSPGGLLQPLPIPERIWEDLSMDFITGLPKSKGF 1186
W GMK V ++V +CDVCQ K GG+LQ LPIPE W+ ++MD + GL S+
Sbjct: 970 QWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWKWDFITMDLVVGLRVSRTK 1029
Query: 1187 EAILVVVDRLSKYAHFIPLKHPYTAKSVAEVFGKEIVRLHGVPSSI--VSDRDPIFVSNF 1244
+AI V+VDRL+K AHF+ ++ A +A+ F EIV+LHGVP ++ DR +
Sbjct: 1030 DAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHGVPLNMKEAQDRQRSYADKR 1089
Query: 1245 WREL 1248
REL
Sbjct: 1090 RREL 1093
Score = 86.7 bits (213), Expect = 1e-16
Identities = 56/178 (31%), Positives = 94/178 (52%), Gaps = 14/178 (7%)
Query: 1342 KELLERDEALRQLRLQLARAQDRMKQFADRKRSDRSFSIGEWVFVKLRAHR--QKSVVTR 1399
K+ + L + L + AQDR + +AD++R + F +G+ V++K+ R +S+
Sbjct: 1059 KKFVSEIVKLHGVPLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISE- 1117
Query: 1400 IYAKLAAKYYGPYPVVARVGAVAYQLKLPPGSKV-HPVFHVSLLKKAVGTYHEGEEL--- 1455
KL+ +Y GP+ +V RV VAY+L+LP + H VFHVS+L+K + H+ +E
Sbjct: 1118 --TKLSPRYMGPFKIVERVEPVAYRLELPDVMRAFHKVFHVSMLRKCL---HKDDEALAK 1172
Query: 1456 --PDLEGDGGILIEPTEVLATRTVQLQGQSIKQILIQWKGQQPEEATWEDVDMIKSQF 1511
DL+ + + P VL R +L+ + I I + W E TWE +K++F
Sbjct: 1173 IPEDLQPNMTLEARPVRVLERRIKELRQKKIPLIKVLWDCDGVTEETWEPEARMKARF 1230
>At2g06890 putative retroelement integrase
Length = 1215
Score = 481 bits (1239), Expect = e-135
Identities = 323/930 (34%), Positives = 480/930 (50%), Gaps = 86/930 (9%)
Query: 308 NGPRNDRQAQPEKNRSTPRDRGFTHLSYNELMERRQKGLCFKCGGAFHPMHQCPDKQLRV 367
N P +A+ + + +G +S + + + R C+KC G H ++CP+K++ +
Sbjct: 124 NKPIVSPRAESKPYAAVQDHKGKAEISTSRVRDVR----CYKCQGKGHYANECPNKRVMI 179
Query: 368 LI----MEDEEEKEGGGNLLAVEVIEEEESSEGELSSMSLSQVEQVGKDKPQTIKLLGL- 422
L+ +E EEE + L EE ++GEL + Q D+ + K L
Sbjct: 180 LLDNGEIEPEEEIPDSPSSLKEN---EELPAQGELLVARRTLSVQTKTDEQEQRKNLFHT 236
Query: 423 ---IQGLPIVILIDSGATHNFVSTSLVHKLGKTVVDTPSLRITLGDGSQARTKGKCKELM 479
+ G ++ID G+ N S ++V KLG L D + R K + +
Sbjct: 237 RCHVHGKVCSLIIDGGSCTNVASETMVKKLGLK---------WLNDSGKMRVKNQVVVPI 287
Query: 480 IIAG--NHPLCVDAQLFELGNVDMVLGIEWLRTLGDMIVNWDKKTMSFWSGHKWVTL--- 534
+I + LC D E G++ +LG W M + + + G K + +
Sbjct: 288 VIGKYEDEILC-DVLPMEAGHI--LLGRPWQSDRKVMHDGFTNRHSFEFKGGKTILVSMT 344
Query: 535 --QGHEEQEGLLVALQTMISRAGF--------SGYLGKEKVQLEKDNKGVTG------VQ 578
+ +++Q L + ++ + F S Y K+ + L + +T V
Sbjct: 345 PHEVYQDQIHLKQKKEQVVKQPNFFAKSGEVKSAYSSKQPMLLFVFKEALTSLTNFAPVL 404
Query: 579 QAELDMILERHSVVFQA--PKGLPPKRNKQHAITLKEGEGPVNVRPYRYPHHQKNEIENQ 636
+E+ +L+ + VF PKGLPP R +H I G N YR + E++ Q
Sbjct: 405 PSEMTSLLQDYKDVFPEDNPKGLPPIRGIEHQIDFVPGASLPNRPAYRTNPVETKELQRQ 464
Query: 637 VKELLEGGVIRHSTSSFSSPVILVKKKDHSWRMCVDYRALNKATIPDKFPIPIIEELLDE 696
KD SWRMC D RA+N T+ PIP ++++LDE
Sbjct: 465 --------------------------KDGSWRMCFDCRAINNVTVKYCHPIPRLDDMLDE 498
Query: 697 LHGARYFSKLDLKSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMND 756
LHG+ FSK+DLKSGYHQ+R+ E D KTAF+T G YE+LVMPFGL +APSTF LMN
Sbjct: 499 LHGSSIFSKIDLKSGYHQIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNH 558
Query: 757 IFRHLLRKRVLVFFDDILVYSKDWPSHLEHLQEVLGILREQGLVANRKKCLFGREKVEYL 816
+ R + V+V+FDDILVYS+ H+EHL VL +LR++ L AN KKC F + + +L
Sbjct: 559 VLRAFIGIFVIVYFDDILVYSESLREHIEHLDSVLNVLRKEELYANLKKCTFCTDNLVFL 618
Query: 817 GHMISGQGVEVDPSKVESVTSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKPLTELTK 876
G ++S GV+VD KV+++ WP+PK V VR F GL G+YR+F +D+ I PLTE+ K
Sbjct: 619 GFVVSADGVKVDEEKVKAIRDWPSPKTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVMK 678
Query: 877 KN-GFEWSEKAQEAFETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQEKRPIA 935
K+ GF+W + +EAF++LK KLT +PVL L +F K F IECDASG+G+GA+LMQ+++ IA
Sbjct: 679 KDVGFKWEKAQEEAFQSLKDKLTNAPVLILSEFLKTFEIECDASGIGIGAVLMQDQKLIA 738
Query: 936 YFSKALGVRNLSKSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKELLQQKVVTME 995
+FS+ LG L+ Y+KEL AL A+Q W+ YL + F + TD SLK L Q+ +
Sbjct: 739 FFSEKLGGATLNYPTYDKELYALVRALQRWQHYLWPKVFVIHTDHESLKHLKGQQKLNKR 798
Query: 996 QQNWAAKLLGFDFEISYKPGKLNKGADALSRVNETLELRQMGSHVDWLGGKDLKEEVSKD 1055
W + F + I YK GK N ADALS+ L +V +G + +KE D
Sbjct: 799 HARWVEFIETFAYVIKYKKGKDNVVADALSQRYTLLST----LNVKLMGFEQIKEVYETD 854
Query: 1056 EELQRIIKSVHEKKDSSLGYTYENGILLYEGRLVLPRESPLIHTMLTEFHTTPQGGHSGF 1115
+ Q + K+ +K +S Y ++ L YE RL +P S L + E H GH G
Sbjct: 855 HDFQEVYKAC--EKFASGRYFRQDKFLFYENRLCVPNCS-LRDLFVREAHGGGLMGHFGI 911
Query: 1116 YRTYRRLAANVYWRGMKSAVQDFVKQCDVCQRQKYLASSPGGLLQPLPIPERIWEDLSMD 1175
+T + + W MK V+ +C+ C++ K P GL PLPIP+ W D+SMD
Sbjct: 912 AKTLEVMTEHFRWPHMKCDVKRICGRCNTCKQAK-SKIQPNGLYTPLPIPKHPWNDISMD 970
Query: 1176 FITGLPKSKGFEAILVVVDRLSKYAHFIPL 1205
F+ GLP++ G ++I VV F P+
Sbjct: 971 FVMGLPRT-GKDSIFVVYSPFQIVYGFNPI 999
Score = 38.5 bits (88), Expect = 0.031
Identities = 42/171 (24%), Positives = 83/171 (47%), Gaps = 16/171 (9%)
Query: 1314 TPFEVVYGRPPPTITRWIQGETRVEAVQKELLERDEALRQLRLQLAR-AQDRMKQFA--- 1369
+PF++VYG P + I E V + ++ E ++Q+ R +++ K +A
Sbjct: 988 SPFQIVYGFNPISPFDLIPLPLS-ERVSIDGKKKAELVQQIHENARRNIEEKTKLYAKQA 1046
Query: 1370 DRKRSDRSFSIGEWVFVKLRAHRQKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLKL-- 1427
++ R ++ F +G+ V++ LR R + + +KL + GP+ ++ R+ AYQL L
Sbjct: 1047 NKGRREQIFEVGDMVWIHLRKERFPA---QRKSKLMPRIDGPFKIIKRINDNAYQLDLQD 1103
Query: 1428 PPGSKVHPVFHVSLLKKAVGTYHEGEELPDLE-----GDGGILIEPTEVLA 1473
P + +P F + + ++ E P+LE +G L+ E++A
Sbjct: 1104 EPDLRSNP-FQEGGDDMIMDSINDMEHEPELERELVAEEGAKLVAEDELVA 1153
>At3g31970 hypothetical protein
Length = 1329
Score = 473 bits (1217), Expect = e-133
Identities = 301/829 (36%), Positives = 432/829 (51%), Gaps = 108/829 (13%)
Query: 712 YHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMNDIFRHLLRKRVLVFFD 771
+ + + E +V KTAFRT GH+EF+VMPFGL N P+ F LMN +F+ L + V++F D
Sbjct: 570 FEDIPIAEANVRKTAFRTRYGHFEFVVMPFGLTNIPTAFMRLMNSVFQEFLDEFVIIFID 629
Query: 772 DILVYSKDWPSHLEHLQEVLGILREQGLVANRKKCLFGREKVEYLGHMISGQGVEVDPSK 831
DILVYSK H +E G E G + GV VD K
Sbjct: 630 DILVYSKSPEEHEVQSEESDG---------------------EAAGAEV---GVSVDLEK 665
Query: 832 VESVTSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKPLTELTKKN-GFEWSEKAQEAF 890
+E++ WP P N +R FLGL GYY++F++ + +A+P+T+LT K+ F WS + E F
Sbjct: 666 IEAIRDWPRPTNATEIRSFLGLAGYYKRFVKVFASMAQPMTKLTGKDVPFVWSPECDEGF 725
Query: 891 ETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQEKRPIAYFSKALGVRNLSKSA 950
+LK+ LT++PVLALP+ + + + DAS VG+ +LMQ
Sbjct: 726 MSLKEMLTSTPVLALPEHGEPYMVYTDASRVGLDCVLMQRG------------------- 766
Query: 951 YEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKELLQQKVVTMEQQNWAAKLLGFDFEI 1010
KV TD +SLK + Q + + Q+ W + +D EI
Sbjct: 767 ------------------------KVFTDHKSLKYIFTQPELNLRQRRWMELVADYDLEI 802
Query: 1011 SYKPGKLNKGADALSR----------VNETLELRQMGSHVDWLGGKDLKEEVSKDEELQR 1060
+Y GK N ADALSR V+E LR + LG E V + + L R
Sbjct: 803 AYHSGKANVVADALSRKRVGGSVEALVSEIGALRLCVMAQEPLG----LEAVDRADLLTR 858
Query: 1061 IIKSVHEKKDSSL-------GYTYE---NGILLYEGRLVLPRESPLIHTMLTEFHTTPQG 1110
+ + ++KD L G Y+ NG +L GR+ +P++ L +L+E H +
Sbjct: 859 V--RLAQEKDEGLIAASKADGSEYQFAANGTILVHGRVCVPKDEELRREILSEAHASMFS 916
Query: 1111 GHSGFYRTYRRLAANVYWRGMKSAVQDFVKQCDVCQRQKYLASSPGGLLQPLPIPERIWE 1170
H + YR L W GMK V ++V +CDVCQ K PGGLLQ LPI E W+
Sbjct: 917 IHPRATKMYRDLKRYYQWVGMKRDVANWVTECDVCQLVKAEHQVPGGLLQSLPILEWKWD 976
Query: 1171 DLSMDFITGLPKSKGFEAILVVVDRLSKYAHFIPLKHPYTAKSVAEVFGKEIVRLHGVPS 1230
++MDF+ GLP S+ +AI V+VDRL+K AHF+ ++ A +A+ + EIV LHGVP
Sbjct: 977 FITMDFVVGLPVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAVLLAKKYVSEIVELHGVPV 1036
Query: 1231 SIVSDRDPIFVSNFWRELFKLQGTKLKMSTAYHPESDGQSEVVNRCLETYLRCFIADQPK 1290
SIVSDRD F S FWR GTK++MSTAYHP++DGQSE + LE LR + D+
Sbjct: 1037 SIVSDRDSKFTSAFWRAFQGEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDRGG 1096
Query: 1291 TWVIWIPWAEYWYNTCFHASTGVTPFEVVYGRPPPTITRWIQGETRVEAVQKELLERDEA 1350
W + E+ YN + AS + PFE +YGRP T W Q R +LE E
Sbjct: 1097 HWADHLSLVEFAYNNSYQASIRMAPFEALYGRPCRTPLCWTQVGERSIYGADYVLETTER 1156
Query: 1351 LRQLRLQLARAQDRMKQFADRKRSDRSFSIGEWVFVKLRAHR--QKSVVTRIYAKLAAKY 1408
+R L+L + AQDR + +AD++R + F +G+ V++K+ R +S+ KL +Y
Sbjct: 1157 IRVLKLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISE---TKLTPRY 1213
Query: 1409 YGPYPVVARVGAVAYQLKLPPGSKV-HPVFHVSLLKKAVGTYHEGEE-----LPDLEGDG 1462
GP+ +V RVG VAY+L+LP + H VFHVS+L+K + H+ +E L DL+ +
Sbjct: 1214 MGPFRIVERVGPVAYRLELPDVMRAFHKVFHVSMLRKCL---HKDDEVLAKILEDLQPNM 1270
Query: 1463 GILIEPTEVLATRTVQLQGQSIKQILIQWKGQQPEEATWEDVDMIKSQF 1511
+ P +L R +L+ + I I + W E TWE +K+ F
Sbjct: 1271 TLEARPVRILERRIKELRRKKIPLIKVLWNCDGVTEETWEPEARMKASF 1319
Score = 34.7 bits (78), Expect = 0.45
Identities = 41/158 (25%), Positives = 66/158 (40%), Gaps = 12/158 (7%)
Query: 104 FDGDDPA---GWISRAEVYFRVQDTPPEVRASLAQLCMEGPTIHFFNSLLS--EEENLTW 158
F G P W SR E F P E R LA +EG ++ S+ + + +++W
Sbjct: 159 FGGTSPEEADSWKSRVEHNFGSSRCPAEYRVDLAVHFLEGDAHLWWRSVTARRRQAHMSW 218
Query: 159 ERFKCALLERYGGQGDGDVYE-QLTELRQ-RGTVEEYITAFEYLTAQIPR--LPEKQFLG 214
F +Y Q D E + EL Q +V EY F L R ++ +
Sbjct: 219 ADFVAEFNAKYFPQEALDRMEARFLELTQGERSVREYEREFNRLLVYAGRGMQDDQAQMR 278
Query: 215 YFLHGLKGEIRGRVRSMVTMADLSRMKILQIARAVERE 252
FL GL+ ++R R R + ++ +++ A VE +
Sbjct: 279 RFLRGLRPDLRVRCR---VLQYATKAALVETAAEVEED 313
>At4g07850 putative polyprotein
Length = 1138
Score = 395 bits (1014), Expect = e-109
Identities = 218/508 (42%), Positives = 293/508 (56%), Gaps = 54/508 (10%)
Query: 821 SGQGVEVDPSKVESVTSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKPLTELTKKN-G 879
S GV+VD KV+++ WP+PK+V VR F GL G+YR+F+RD+ +A PLTE+ KKN G
Sbjct: 533 STDGVKVDEEKVKAIREWPSPKSVGKVRSFHGLAGFYRRFVRDFSTLAAPLTEVIKKNVG 592
Query: 880 FEWSEKAQEAFETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQEKRPIAYFSK 939
F+W + ++AF+ LK+KLT +PVL+LPDF K F IECDA GVG+GA+LMQ+K+PIAYFS+
Sbjct: 593 FKWEQAPEDAFQALKEKLTHAPVLSLPDFLKTFEIECDAPGVGIGAVLMQDKKPIAYFSE 652
Query: 940 ALGVRNLSKSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKELLQQKVVTMEQQNW 999
LG L+ Y+KEL AL A+Q W+ YL + F + TD SLK L Q+ + W
Sbjct: 653 KLGGATLNYPTYDKELYALVRALQTWQHYLWPKEFVIHTDHESLKHLKGQQKLNKRHARW 712
Query: 1000 AAKLLGFDFEISYKPGKLNKGADALSRVNETLELRQMGSHVDWLGGKDLKEEVSKDEELQ 1059
+ F + I YK GK N ADALSR +E
Sbjct: 713 VEFIETFPYVIKYKKGKDNVVADALSRRHE------------------------------ 742
Query: 1060 RIIKSVHEKKDSSLGYTYENGILLYEGRLVLPRESPLIHTMLTEFHTTPQGGHSGFYRTY 1119
G L Y+ RL +P S L + E H GH G +T
Sbjct: 743 --------------------GFLFYDNRLCIPNSS-LRELFIREAHGGGLMGHFGVSKTL 781
Query: 1120 RRLAANVYWRGMKSAVQDFVKQCDVCQRQKYLASSPGGLLQPLPIPERIWEDLSMDFITG 1179
+ + + +W MK V+ ++C C++ K S P GL PLPIP W D+SMDF+ G
Sbjct: 782 KVMQDHFHWPHMKRDVERMCERCTTCKQAK-AKSQPHGLCTPLPIPLHPWNDISMDFVVG 840
Query: 1180 LPKSK-GFEAILVVVDRLSKYAHFIPLKHPYTAKSVAEVFGKEIVRLHGVPSSIVSDRDP 1238
LP+++ G ++I VVVDR SK AHFIP A +A +F +E+VRLHG+P +IVSDRD
Sbjct: 841 LPRTRTGKDSIFVVVDRFSKMAHFIPCHKTDDAMHIANLFFREVVRLHGMPKTIVSDRDT 900
Query: 1239 IFVSNFWRELFKLQGTKLKMSTAYHPESDGQSEVVNRCLETYLRCFIADQPKTWVIWIPW 1298
F+S FW+ L+ GTKL ST HP++DGQ+EVVNR L T LR I KTW +P
Sbjct: 901 KFLSYFWKTLWSKLGTKLLFSTTCHPQTDGQTEVVNRTLSTLLRALIKKNLKTWEDCLPH 960
Query: 1299 AEYWYNTCFHASTGVTPFEVVYGRPPPT 1326
E+ YN H++T +PF++VYG P T
Sbjct: 961 VEFAYNHSVHSATKFSPFQIVYGFNPIT 988
Score = 58.5 bits (140), Expect = 3e-08
Identities = 33/73 (45%), Positives = 40/73 (54%)
Query: 596 PKGLPPKRNKQHAITLKEGEGPVNVRPYRYPHHQKNEIENQVKELLEGGVIRHSTSSFSS 655
P+GLPP R +H I G N YR + E+E QV EL+E G I S S +
Sbjct: 457 PEGLPPIRGIEHQIDFVPGASLPNRPAYRTNPVETKELEKQVNELMERGHICESMSPCAV 516
Query: 656 PVILVKKKDHSWR 668
PV+LV KKD SWR
Sbjct: 517 PVLLVPKKDGSWR 529
Score = 48.5 bits (114), Expect = 3e-05
Identities = 44/169 (26%), Positives = 74/169 (43%), Gaps = 8/169 (4%)
Query: 286 VFVKGSKETNSGSGYNNSRAGGNGPRNDRQAQPEKNRSTPRDRGFTHLSYNELMERRQKG 345
+F K K ++ YN+S+ + K P+ + + + R +
Sbjct: 219 MFEKQIKRRSAKPSYNSSKPSYQREEKSGFQKEYKPFVKPKVEEISSKGKEKEVTRTRDL 278
Query: 346 LCFKCGGAFHPMHQCPDKQLRVLIMEDEEEKEGGGNLLAVEVIEEEESSEGE-LSSMSLS 404
CFKC G H +C +K R++I+ D E E E + EE+ +GE L +M +
Sbjct: 279 KCFKCHGLGHYASECSNK--RIMIIRDSGEVESEDE--KPEESDVEEAPKGELLVTMRVL 334
Query: 405 QVEQVGKDKPQTIKLLG---LIQGLPIVILIDSGATHNFVSTSLVHKLG 450
V +++ Q L LI+G ++ID G+ N S ++V KLG
Sbjct: 335 SVLNKAEEQAQRENLFHTRCLIKGKVCSLIIDGGSCTNVASETMVQKLG 383
>At4g16910 retrotransposon like protein
Length = 687
Score = 387 bits (993), Expect = e-107
Identities = 246/681 (36%), Positives = 366/681 (53%), Gaps = 43/681 (6%)
Query: 867 IAKPLTELTKKN-GFEWSEKAQEAFETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGA 925
+A+P+T+LT K+ F WSE+ + +F LK LT +PVL LP+ + +T+ DAS VG+G
Sbjct: 1 MAQPMTQLTGKDTAFNWSEECERSFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGC 60
Query: 926 ILMQEKRPIAYFSKALGVRNLSKSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKE 985
+L+Q+ IAY S+ L + + E+ A+ A++ WR YL G ++ TD +SLK
Sbjct: 61 VLIQKGSVIAYASRQLRKHEKNYPTNDLEMAAVVFALKIWRSYLYGAKVQIFTDHKSLKY 120
Query: 986 LLQQKVVTMEQQNWAAKLLGFDFEISYKPGKLNKGADALSRVNETLEL-RQMGSHVDWLG 1044
+ Q + + Q+ W + +D I+Y PGK N+ DALSR+ +E R + V+ +G
Sbjct: 121 IFTQPELNLRQRRWIKLVADYDLNIAYHPGKANQVVDALSRLRTVVEAERNQVNLVNMMG 180
Query: 1045 G---KDLKEEV-------SKDEELQRIIKSVHEKKDSSLGYTYEN---------GILLYE 1085
L +EV + +L I+S E+ + G+ N G ++
Sbjct: 181 TLHLNALSKEVEPLGLRAANQADLLSRIRSAQERDEEIKGWAQNNKTEYQSSNNGTIVVN 240
Query: 1086 GRLVLPRESPLIHTMLTEFHTTPQGGHSGFYRTYRRLAANVYWRGMKSAVQDFVKQCDVC 1145
GR+ P + L +L E H + H G + YR L +W GMK V +V
Sbjct: 241 GRVCGPNDKALKEEILKEAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVA----- 295
Query: 1146 QRQKYLASSPGGLLQPLPIPERIWEDLSMDFITGLP---KSKGFEAILVVVDRLSKYAHF 1202
K P G+LQ LPIPE W+ + MDF+TGLP KSK A+ VVVDRL+K AHF
Sbjct: 296 ---KEEHQVPSGMLQNLPIPEWKWDHIMMDFVTGLPTGIKSK-HNAVWVVVDRLTKSAHF 351
Query: 1203 IPLKHPYTAKSVAEVFGKEIVRLHGVPSSIVSDRDPIFVSNFWRELFKLQGTKLKMSTAY 1262
+ + A+ +AE + EIVRLHG+P SIVSDRD F S FW+ K+ GT++ +STAY
Sbjct: 352 MAISDKDAAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKPFQKVLGTRVNLSTAY 411
Query: 1263 HPESDGQSEVVNRCLETYLRCFIADQPKTWVIWIPWAEYWYNTCFHASTGVTPFEVVYGR 1322
HP++DGQSE + LE LR + D W ++ E+ YN F AS G++P+E +YGR
Sbjct: 412 HPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGR 471
Query: 1323 PPPTITRWIQ-GETRV--EAVQKELLERDEALRQLRLQLARAQDRMKQFADRKRSDRSFS 1379
T W GE R+ AV E ++ ++ L+++L AQDR K +A+++R + F
Sbjct: 472 AGRTPLCWTPVGERRLFGPAVVDETTKK---MKFLKIKLKEAQDRQKSYANKRRKELEFQ 528
Query: 1380 IGEWVFVKLRAHRQKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLKLPPG-SKVHPVFH 1438
+G+ V++K ++ T KL +Y GPY V+ RVGAVAY+L LPP H VFH
Sbjct: 529 VGDLVYLKAMTYKGAGRFTS-RKKLRPRYVGPYKVIERVGAVAYKLDLPPKLDAFHNVFH 587
Query: 1439 VSLLKKAVGTYHEGEE--LPDLEGDGGILIEPTEVLATRTVQLQGQSIKQILIQWKGQQP 1496
VS L+K + E E P L+ + + P ++ +G+S+ + I W
Sbjct: 588 VSQLRKCLSEQEESMEDVPPGLKENMTVEAWPVRIMDQMKKGTRGKSMDLLKILWNCGGR 647
Query: 1497 EEATWEDVDMIKSQFPSFCLE 1517
EE TWE +K+ FP + E
Sbjct: 648 EEYTWETETKMKANFPEWFKE 668
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 381 bits (978), Expect = e-105
Identities = 204/496 (41%), Positives = 297/496 (59%), Gaps = 9/496 (1%)
Query: 478 LMIIAGNHPLCVDAQLFELGNVDMVLGIEWLRTLGDMIVNWDKKTMSFWSGHKWVTLQGH 537
+ ++ + D + L D++LG++WL I + ++ + F + QG
Sbjct: 19 ISVVVNGVNMPADLIIVPLKKHDVILGMDWLGKYKGHI-DCHRERIQFERDEGMLKFQGI 77
Query: 538 EEQEGLLVALQTMISRAGFSGYLGK--EKVQLEKDNKGVTGVQQAELDMILERHSVVFQA 595
G LV IS LGK E K V + + +I+ S VF A
Sbjct: 78 RTTSGSLV-----ISAIQAERMLGKGCEAYLATITTKEVGASAELKDILIVNEFSDVFAA 132
Query: 596 PKGLPPKRNKQHAITLKEGEGPVNVRPYRYPHHQKNEIENQVKELLEGGVIRHSTSSFSS 655
G+PP R+ I L+ G P++ PYR + E++ Q++ELL G IR S+S + +
Sbjct: 133 VSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGA 192
Query: 656 PVILVKKKDHSWRMCVDYRALNKATIPDKFPIPIIEELLDELHGARYFSKLDLKSGYHQV 715
PV+ VKKKD S+R+C+DYR LNK T+ +K+P+P I+EL+D+L GA++FSK+DL SGYHQ+
Sbjct: 193 PVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQI 252
Query: 716 RVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMNDIFRHLLRKRVLVFFDDILV 775
++ DV KTAFRT GH+EF+VMPFGL NAP+ F +MN +FR L + V++F DDILV
Sbjct: 253 PIEPTDVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILV 312
Query: 776 YSKDWPSHLEHLQEVLGILREQGLVANRKKCLFGREKVEYLGHMISGQGVEVDPSKVESV 835
+SK W +H EHL+ VL LRE L A K F + V +LGH+IS QGV VDP K+ S+
Sbjct: 313 HSKSWEAHQEHLRAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSI 372
Query: 836 TSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKPLTELT-KKNGFEWSEKAQEAFETLK 894
WP P+N +R FLGL GYYR+F+ + +A+PLT LT K F WS++ +++F LK
Sbjct: 373 KEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELK 432
Query: 895 KKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQEKRPIAYFSKALGVRNLSKSAYEKE 954
L +PVL LP+ + +T+ DAS VG+G +LMQ+ IAY S+ L + ++ E
Sbjct: 433 AMLINAPVLVLPEEGEPYTVYTDASIVGLGCVLMQKGSVIAYASRQLRKHEKNYPTHDLE 492
Query: 955 LMALGLAIQHWRPYLL 970
+ A+ A++ WR YL+
Sbjct: 493 MAAVVFALKIWRSYLI 508
Score = 269 bits (687), Expect = 1e-71
Identities = 159/389 (40%), Positives = 224/389 (56%), Gaps = 13/389 (3%)
Query: 1076 TYENGILLYEGRLVLPRESPLIHTMLTEFHTTPQGGHSGFYRTYRRLAANVYWRGMKSAV 1135
T NG ++ GR+ +P + L +L E H + H G + YR L +W GM+ V
Sbjct: 531 TSNNGTIVVNGRVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMRKDV 590
Query: 1136 QDFVKQCDVCQRQKYLASSPGGLLQPLPIPERIWEDLSMDFITGLP---KSKGFEAILVV 1192
+V +C CQ K P GLLQ LPI E W+ ++MDF+T LP KSK A+ VV
Sbjct: 591 ARWVAKCPTCQLVKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSK-HNAVWVV 649
Query: 1193 VDRLSKYAHFIPLKHPYTAKSVAEVFGKEIVRLHGVPSSIVSDRDPIFVSNFWRELFKLQ 1252
VDRL+K AHF+ + A+ +AE + EI+RLHG+P SIVSDRD F S FW K
Sbjct: 650 VDRLTKSAHFMAISDKDGAEIIAEKYIDEIMRLHGIPVSIVSDRDTRFTSKFWNAFQKAL 709
Query: 1253 GTKLKMSTAYHPESDGQSEVVNRCLETYLRCFIADQPKTWVIWIPWAEYWYNTCFHASTG 1312
GT++ +STAYHP++DGQSE + LE LR + D W ++ E+ YN F AS G
Sbjct: 710 GTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLIEFAYNNSFQASIG 769
Query: 1313 VTPFEVVYGRPPPTITRWIQ-GETRV--EAVQKELLERDEALRQLRLQLARAQDRMKQFA 1369
++P+E +YGR T W GE R+ + E ER ++ L+++L AQDR K +A
Sbjct: 770 MSPYEALYGRACRTPLCWTPVGERRLFGPTIVDETTER---MKFLKIKLKEAQDRQKSYA 826
Query: 1370 DRKRSDRSFSIGEWVFVKLRAHRQKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLKLPP 1429
+++R + F +G+ V++K ++ T KL+ +Y GPY V+ RVGAVAY+L LPP
Sbjct: 827 NKRRKELEFQVGDLVYLKAMTYKGAGRFTS-RKKLSPRYVGPYKVIERVGAVAYKLDLPP 885
Query: 1430 GSKV-HPVFHVSLLKKAVGTYHEG-EELP 1456
V H VFHVS L+K + E E++P
Sbjct: 886 KLNVFHNVFHVSQLRKYLSDQEESVEDIP 914
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 319 bits (817), Expect = 9e-87
Identities = 155/320 (48%), Positives = 216/320 (67%), Gaps = 9/320 (2%)
Query: 1208 PYTAKSVAEVFGKEIVRLHGVPSSIVSDRDPIFVSNFWRELFKLQGTKLKMSTAYHPESD 1267
P ++SVA F +V+LHG+P SIVSD DPIF+S FW+E +KL TKL MSTAYHP++D
Sbjct: 628 PLLSRSVAAKFVSHVVKLHGIPRSIVSDCDPIFMSLFWQEFWKLSRTKLWMSTAYHPQTD 687
Query: 1268 GQSEVVNRCLETYLRCFIADQPKTWVIWIPWAEYWYNTCFHASTGVTPFEVVYGRPPPTI 1327
GQ+EVVNRC+E +LRCF+ PK W +IPWAEYWYNT FHASTG+TPF+ +YGRPP I
Sbjct: 688 GQTEVVNRCIEQFLRCFVHYHPKQWSSFIPWAEYWYNTTFHASTGMTPFQALYGRPPSPI 747
Query: 1328 TRWIQGETRVEAVQKELLERDEALRQLRLQLARAQDRMKQFADRKRSDRSFSIGEWVFVK 1387
+ G + +++ RDE L +L+ L A + MKQ AD + D SF +G+WV ++
Sbjct: 748 PAYELGSVVCGELNEQMAARDELLAELKQHLVTANNCMKQQADSRLRDVSFQVGDWVLLR 807
Query: 1388 LRAHRQKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLKLPPGSKVHPVFHVSLLKKAVG 1447
++ +RQK++ R KL+ ++YGP+ V ++ G VAY+L LP G+++HPVFHVSLLK VG
Sbjct: 808 IQPYRQKTLFRRSSQKLSHRFYGPFQVASKHGEVAYRLTLPEGTRIHPVFHVSLLKPWVG 867
Query: 1448 TYHEGE----ELPDLEGDGGILIEPTEVLATRTVQLQGQSIKQILIQWKGQQPEEATWED 1503
+GE +LP L +G + ++PT VL R + + +L+QW+G E+ATWE+
Sbjct: 868 ---DGEPDMGQLPPLRNNGELKLQPTAVLEVRWRSQDKKRVADLLVQWEGLHIEDATWEE 924
Query: 1504 VDMIKSQFPSFC--LEDKAR 1521
D + + FP F LEDK R
Sbjct: 925 YDQLAASFPEFVLNLEDKVR 944
Score = 220 bits (560), Expect = 6e-57
Identities = 133/386 (34%), Positives = 202/386 (51%), Gaps = 37/386 (9%)
Query: 386 EVIEEEESSEGELSSMSLSQVEQVGKDKPQTIKLLGLIQGLPIVILIDSGATHNFVSTSL 445
E +EEE + ++L +E G D TI++ I ++ LI+SG+THNF+
Sbjct: 289 EAVEEESTEAEGKPEITLYALE--GVDTTSTIRVRATIHRNRLIALINSGSTHNFIGEKA 346
Query: 446 VHKLGKTVVDTPSLRITLGDGSQARTKGKCKELMIIAGNHPLCVDAQLFELGNVDMVLGI 505
V + T + + +G + + + + ++ G V L +D+ +G+
Sbjct: 347 VRGMNLKATTTKPFTVRVVNGMPLVCRSRYEAIPVVMGGVVFPVTLYALPLMGLDLAMGV 406
Query: 506 EWLRTLGDMIVNWDKKTMSF-WSGHKWVTLQGHEEQEGLLVALQTMISRAGFSGYLGKEK 564
+WL TLG + NW ++T+ F W+G + V L G + V +T+ +A +
Sbjct: 407 QWLSTLGPTLCNWKEQTLQFHWAGDE-VRLMGIKPTGLRGVEHKTITKKARMGHTIFAIT 465
Query: 565 VQLEKDNKGVTGVQQAELDMILERHSVVFQAPKGLPPKRNKQHAITLKEGEGPVNVRPYR 624
+ N E+ ++E + +F+ P LP +R H I LK+G PVNVRPYR
Sbjct: 466 MA---HNSSDPNTPDGEIKKLIEEFAGLFETPTQLPSERPIVHRIALKKGTDPVNVRPYR 522
Query: 625 YPHHQKNEIENQVKELLEGGVIRHSTSSFSSPVILVKKKDHSWRMCVDYRALNKATIPDK 684
Y +QK+E+ G+IR S+S F SPV+L ++
Sbjct: 523 YAFYQKDEMSR-------AGIIRPSSSPFLSPVLL-----------------------ER 552
Query: 685 FPIPIIEELLDELHGARYFSKLDLKSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLM 744
FPIP ++++LDEL+GA YF+KLDL +GY QVR+ D+ K AF+TH GHYE+LVMPFGL
Sbjct: 553 FPIPTVDDMLDELNGAVYFTKLDLTAGYQQVRMHSPDIPKIAFQTHNGHYEYLVMPFGLC 612
Query: 745 NAPSTFQSLMNDIFRHLLRKRVLVFF 770
NAPSTFQ+LMN+IF LL + V F
Sbjct: 613 NAPSTFQALMNEIFWPLLSRSVAAKF 638
Score = 38.9 bits (89), Expect = 0.024
Identities = 32/132 (24%), Positives = 52/132 (39%), Gaps = 28/132 (21%)
Query: 98 KVELPMFDGDDPAGWISRAEVYFRVQDTPPEVRASLAQLCMEGPTIHFFNSLLSEEENLT 157
K++ P F G D W+S+A+ YF + P E ++G N
Sbjct: 105 KLDFPRFHGGDLTKWLSKAKQYFEYHEGPVEQAVRFTTFHLDGVA------------NGW 152
Query: 158 WERFKCALLERYGGQGDGDVYEQLTELRQRGTVEEYITAFEYLTAQIPRLPEKQFLGYFL 217
W+ A L R G Q G++ EY FE+L ++ ++ +G F+
Sbjct: 153 WQ----ATLRRSATIG------------QTGSLSEYQHEFEWLQNKVYGWTQEVLVGAFM 196
Query: 218 HGLKGEIRGRVR 229
+GL I +R
Sbjct: 197 NGLYYSISNGIR 208
>At4g07830 putative reverse transcriptase
Length = 611
Score = 296 bits (757), Expect = 8e-80
Identities = 207/629 (32%), Positives = 309/629 (48%), Gaps = 71/629 (11%)
Query: 928 MQEKRPIAYFSKALGVRNLSKSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKELL 987
MQ + IAY S+ L + ++ E+ A V TD +SLK +
Sbjct: 1 MQRGKVIAYGSRQLKKHEGNYPTHDLEMAA------------------VFTDHKSLKYIF 42
Query: 988 QQKVVTMEQQNWAAKLLGFDFEISYKPGKLNKGADALSRVN---------ETLELRQMGS 1038
Q + + + W + +D EI+Y PGK N DALSR ETL +
Sbjct: 43 TQLELNLRLRRWMKLVADYDLEIAYHPGKANVVTDALSRKRVGAAPGQSVETLVIEIGAL 102
Query: 1039 HVDWLGGKDLKEEVSKDEELQRIIKSVHEKKDSSL------GYTYE---NGILLYEGRLV 1089
+ + + L E +L ++ EK + + G+ Y+ NG +L GR+
Sbjct: 103 RLCAVAREPLGLEAVDQTDLLSRVQLAQEKDEGLIAASKAEGFEYQFAANGTILVHGRVC 162
Query: 1090 LPRESPLIHTMLTEFHTTPQGGHSGFYRTYRRLAANVYWRGMKSAVQDFVKQCDVCQRQK 1149
+P++ L +L+E H + H G + YR L W GMK V ++V++CDVCQ K
Sbjct: 163 VPKDKELRQEILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVGNWVEECDVCQLVK 222
Query: 1150 YLASSPGGLLQPLPIPERIWEDLSMDFITGLPKSKGFEAILVVVDRLSKYAHFIPLKHPY 1209
G LLQ LPIPE W+ ++MDF+ GLP S+ +AI V+VDRL+K AHF+ ++
Sbjct: 223 IEHQVSGSLLQSLPIPEWKWDFITMDFVVGLPVSRTKDAIWVIVDRLTKSAHFLAIRKTD 282
Query: 1210 TAKSVAEVFGKEIVRLHGVPSSIVSDRDPIFVSNFWRELFKLQGTKLKMSTAYHPESDGQ 1269
A +A+ + EIV+LHGVP SIVSDRD F S FWR GTK++MSTAYHP++DGQ
Sbjct: 283 GAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKVQMSTAYHPQTDGQ 342
Query: 1270 SEVVNRCLETYLRCFIADQPKTWVIWIPWAEYWYNTCFHASTGVTPFEVVYGRPPPTITR 1329
SE + LE LR + D W + E+ YN + AS G+ PFEV+YGRP T+
Sbjct: 343 SERTIQTLEDMLRMCVLDWRGHWADHLSLVEFAYNNSYQASIGMAPFEVLYGRPCRTLC- 401
Query: 1330 WIQGETRVEAVQKELLERDEALRQLRLQLARAQDRMKQFADRKRSDRSFSIGEWVFVKLR 1389
W Q R + E E +R L+L + AQ+R + +AD++R + F +G+ V
Sbjct: 402 WTQVGERSIYGADYVQEITERIRVLKLNMKEAQNRQRSYADKRRKELEFEVGDSVSQDGH 461
Query: 1390 AHRQKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLKLPPGSKV-HPVFHVSLLKKAVGT 1448
R + RVG VA++L+L + H VFHVS+L+K +
Sbjct: 462 VARSEQ---------------------RVGPVAFRLELSDVMRAFHKVFHVSMLRKCL-- 498
Query: 1449 YHEGEEL-----PDLEGDGGILIEPTEVLATRTVQLQGQSIKQILIQWKGQQPEEATWED 1503
H+ +E+ DL+ + + P VL R +L+ + I I + E TWE
Sbjct: 499 -HKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVLRNCDGVTEETWEP 557
Query: 1504 VDMIKSQFPSFCLEDKARAYGEGIDRTQG 1532
+K++F + + G+ +TQG
Sbjct: 558 EARLKARFKKWFEKKDV----SGLFQTQG 582
>At4g08100 putative polyprotein
Length = 1054
Score = 295 bits (754), Expect = 2e-79
Identities = 154/349 (44%), Positives = 212/349 (60%), Gaps = 29/349 (8%)
Query: 672 DYRALNKATIPDKFPIPIIEELLDELHGARYFSKLDLKSGYHQVRVKEEDVHKTAFRTHE 731
D RA+N T+ + PIP +++ LD+LHG+ FSK+DLKSGYHQ R+KE D KTA +T +
Sbjct: 527 DCRAINNITVKYRHPIPRLDDTLDKLHGSSIFSKIDLKSGYHQTRMKEGDEWKTAIKTKQ 586
Query: 732 GHYEFLVMPFGLMNAPSTFQSLMNDIFRHLLRKRVLVFFDDILVYSKDWPSHLEHLQEVL 791
YE+LVMPFGL NAP+TF LMN + R + V+V+FDDILVYSK+ H+ HL+ VL
Sbjct: 587 RLYEWLVMPFGLTNAPNTFMRLMNHVLRKHIGVFVIVYFDDILVYSKNLEYHVMHLKLVL 646
Query: 792 GILREQGLVANRKKCLFGREKVEYLGHMISGQGVEVDPSKVESVTSWPTPKNVKGVRGFL 851
+LR++ L AN KKC F + + +LG ++S G++VD KV+++ WP PKNV
Sbjct: 647 DLLRKEKLYANLKKCTFCTDNLVFLGFVVSADGIKVDEEKVKAIREWPNPKNVS------ 700
Query: 852 GLTGYYRKFIRDYGKIAKPLTELTKKNGFEWSEKAQEAFETLKKKLTTSPVLALPDFSKE 911
K GF+W + + AF+ LK+KLT S V LP+F K
Sbjct: 701 -----------------------EKDIGFKWEDAQENAFQALKEKLTNSSVPILPNFMKS 737
Query: 912 FTIECDASGVGVGAILMQEKRPIAYFSKALGVRNLSKSAYEKELMALGLAIQHWRPYLLG 971
F IECDASG+G+GA+LMQ+ +PIAYFS+ LG L+ Y+KEL AL A+Q W+ YL
Sbjct: 738 FEIECDASGLGIGAVLMQDHKPIAYFSEKLGGATLNYPTYDKELYALVRALQTWQHYLWP 797
Query: 972 RHFKVTTDQRSLKELLQQKVVTMEQQNWAAKLLGFDFEISYKPGKLNKG 1020
+ F + TD SLK L Q+ + W + F + I YK + + G
Sbjct: 798 KKFVIHTDHESLKHLKGQQKLNKRHARWVEFIETFPYVIKYKKREAHGG 846
Score = 47.8 bits (112), Expect = 5e-05
Identities = 36/120 (30%), Positives = 64/120 (53%), Gaps = 5/120 (4%)
Query: 1308 HASTGVTPFEVVYGRPPPTITRWIQGETRVEAVQKELLERDEALRQLRLQLARAQDRMKQ 1367
H++T +PFE+VYG P + I E + ++D+ ++Q+ L L + K+
Sbjct: 851 HSATKFSPFEIVYGFKPTSPLDLIPLPLS-ERSSLDGKKKDDLVQQV-LNLEARTKQYKK 908
Query: 1368 FADRKRSDRSFSIGEWVFVKLRAHRQKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLKL 1427
+A++ R + F+ G+ V+V LR R V + +KL + GP+ V+ R+ AY+L L
Sbjct: 909 YANKGRKEVIFNEGDQVWVHLRKKRFPEVRS---SKLMPRIDGPFKVLKRINNNAYKLDL 965
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 292 bits (748), Expect = 9e-79
Identities = 245/867 (28%), Positives = 387/867 (44%), Gaps = 124/867 (14%)
Query: 607 HAITLKEGEGPVNVRPYRYPH-HQKNEIENQVKELLEGGVIRH-STSSFSSPVILVKKKD 664
H I L E E ++ P R + + K ++ ++ +LL+ GVI S S++ SPV V KK
Sbjct: 889 HRIHL-ENESYSSIEPQRRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKG 947
Query: 665 H------------------SWRMCVDYRALNKATIPDKFPIPIIEELLDELHGARYFSKL 706
RMC++YR LN A+ + FP+P I+ +L+ L Y+ L
Sbjct: 948 GMTVVKNSKDELIPTRTITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCFL 1007
Query: 707 DLKSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMNDIFRHLLRKRV 766
D SG+ Q+ + D KT F G + + MPFGL NAP+TFQ M IF L+ + V
Sbjct: 1008 DSYSGFFQIPIHPNDQGKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMV 1067
Query: 767 LVFFDDILVYSKDWPSHLEHLQEVLGILREQGLVANRKKCLFGREKVEYLGHMISGQGVE 826
VF DD VY + S L +L VL E LV N +KC F + LG IS +G+E
Sbjct: 1068 EVFMDDFSVYGSSFSSCLLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKISEEGIE 1127
Query: 827 VDPSKVESVTSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKPLTE-LTKKNGFEWSEK 885
VD +K++ + PK VK +R FLG G+YR FI+D+ K+A+PLT L K+ F + ++
Sbjct: 1128 VDKAKIDVMMQLQPPKTVKDIRSFLGHAGFYRIFIKDFSKLARPLTRLLCKETEFAFDDE 1187
Query: 886 AQEAFETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQEKRPIAYFSKALGVRN 945
AF+ +K+ L T+P++ P++ F I+ + + Y
Sbjct: 1188 CLTAFKLIKEALITAPIVQAPNWDFPF------------EIITMDDAQVRY--------- 1226
Query: 946 LSKSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKELLQQKVVTMEQQNWAAKLLG 1005
+ EKEL+A+ A + +R YL+G + TD +L+ + +K W L
Sbjct: 1227 ---ATTEKELLAVVFAFEKFRSYLVGSKVTIYTDHAALRHIYAKKDTKPRLLRWILLLQE 1283
Query: 1006 FDFEISYKPGKLNKGADALSRV--------------NETLELRQM--------GSHVDWL 1043
FD EI K G N AD LSR+ + + ++Q+ HV++L
Sbjct: 1284 FDMEIVDKKGIENGVADHLSRMRIEDEVLIDDSMPEEQLMAIQQLNEKKLPWYADHVNYL 1343
Query: 1044 GGKDLKEEVSKDEELQRIIKSVHEKKDSSLGYTYENGILLYEGRLVLPRESPLIHTMLTE 1103
+ +S E+ + H D YT + R + + I +L
Sbjct: 1344 VSGEEPPNLSSYEKKKFFKDINHFYWDEPYLYTLCKDKIY---RTCVSEDE--IEGILLH 1398
Query: 1104 FHTTPQGGHSGFYRTYRR-LAANVYWRGMKSAVQDFVKQCDVCQRQKYLASSPGGLLQPL 1162
H GGH ++T + L A +W M Q+F+ +CD +
Sbjct: 1399 CHGFAYGGHFATFKTMSKILQAGFWWPSMFKDAQEFISKCDSFENFD------------- 1445
Query: 1163 PIPERIWEDLSMDFITGLPKSKGFEAILVVVDRLSKYAHFIPLKHPYTAKSVAEVFGKEI 1222
+W +DF+ P S G + ILV +D +SK+ I H A+ V ++F I
Sbjct: 1446 -----VW---GIDFMGPFPSSYGNKYILVAIDYVSKWVEAI-ASHTNDARVVLKLFKTII 1496
Query: 1223 VRLHGVPSSIVSDRDPIFVSNFWRELFKLQGTKLKMSTAYHPESDGQSEVVNRCLETYLR 1282
GVP ++SD F++ + L K G K K S GQ E+ NR ++ L
Sbjct: 1497 FPRFGVPRIVISDGGKHFINKGFENLLKKHGVKHKTS--------GQVEISNREIKAILE 1548
Query: 1283 CFIADQPKTWVIWIPWAEYWYNTCFHASTGVTPFEVVYGRP---PPTITRWIQGETRV-- 1337
+ K W + + Y T F G TPF ++YG+ P + ++
Sbjct: 1549 KTVGSTRKDWSAKLNDTLWAYRTAFKTPIGTTPFNLLYGKSCHLPVELEYKAMWAVKLLN 1608
Query: 1338 ----EAVQKELLERDEALRQLRLQLARA----QDRMKQFADRKRSDRSFSIGEWVFVKLR 1389
A +K L++ ++ L ++RL+ + ++R K F D+K R F +G+ V +
Sbjct: 1609 FDIKTAEEKRLIQLND-LNKIRLEAYESSKIYKERTKSFHDKKIVSRDFKVGDQVLL--- 1664
Query: 1390 AHRQKSVVTRIYAKLAAKYYGPYPVVA 1416
S + KL +++ GP+ V A
Sbjct: 1665 ---FNSRLRLFPGKLKSRWSGPFSVTA 1688
>At4g03650 putative reverse transcriptase
Length = 839
Score = 274 bits (700), Expect = 3e-73
Identities = 134/295 (45%), Positives = 192/295 (64%), Gaps = 1/295 (0%)
Query: 719 EEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMNDIFRHLLRKRVLVFFDDILVYSK 778
E DV KTAFRT GH+EF+VMPFGL NAP+ F LMN +F+ L + V++F DDILVYSK
Sbjct: 518 EADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSK 577
Query: 779 DWPSHLEHLQEVLGILREQGLVANRKKCLFGREKVEYLGHMISGQGVEVDPSKVESVTSW 838
H HL+ V+ LREQ L A KC F + ++ +LGH++S +GV VDP K+E++ W
Sbjct: 578 SPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDW 637
Query: 839 PTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKPLTELTKKN-GFEWSEKAQEAFETLKKKL 897
P P N +R FLGL GYYR+FI+ + +A+P+T+LT K+ F WS + +E F +LK+ L
Sbjct: 638 PRPTNATEIRSFLGLAGYYRRFIKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEML 697
Query: 898 TTSPVLALPDFSKEFTIECDASGVGVGAILMQEKRPIAYFSKALGVRNLSKSAYEKELMA 957
T++PVLALP+ + + + DASGVG+G LMQ + IAY S+ L + ++ E+ A
Sbjct: 698 TSTPVLALPEHGEPYMVYTDASGVGLGCALMQRGKVIAYASRQLRKHEGNYPTHDLEMAA 757
Query: 958 LGLAIQHWRPYLLGRHFKVTTDQRSLKELLQQKVVTMEQQNWAAKLLGFDFEISY 1012
+ A++ WR YL G +V TD +SLK + Q + + Q+ W + +D EI+Y
Sbjct: 758 VIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYDLEIAY 812
Score = 41.2 bits (95), Expect = 0.005
Identities = 22/75 (29%), Positives = 38/75 (50%)
Query: 573 GVTGVQQAELDMILERHSVVFQAPKGLPPKRNKQHAITLKEGEGPVNVRPYRYPHHQKNE 632
GV G +++ VFQ+ +GLPP R+ I L+ P++ PYR + E
Sbjct: 448 GVCGQVAVSDIRVVQEFQYVFQSLQGLPPSRSDPFTIELEPKTAPLSKAPYRMAPAKMAE 507
Query: 633 IENQVKELLEGGVIR 647
++ Q+++LL +R
Sbjct: 508 LKKQLEDLLAEADVR 522
Score = 36.6 bits (83), Expect = 0.12
Identities = 14/19 (73%), Positives = 18/19 (94%)
Query: 1253 GTKLKMSTAYHPESDGQSE 1271
GTK++MST YHP++DGQSE
Sbjct: 818 GTKVQMSTTYHPQTDGQSE 836
>At1g20390 hypothetical protein
Length = 1791
Score = 219 bits (559), Expect = 8e-57
Identities = 135/444 (30%), Positives = 225/444 (50%), Gaps = 12/444 (2%)
Query: 581 ELDMILERHSVVF----QAPKGLPPKRNKQHAITLKEGEGPVNVRPYRYPHHQKNEIENQ 636
EL +L+R+S F + KG+ P H + + PV + + + + +
Sbjct: 738 ELIALLKRNSKTFAWSIEDMKGIDPAITA-HELNVDPTFKPVKQKRRKLGPERARAVNEE 796
Query: 637 VKELLEGG-VIRHSTSSFSSPVILVKKKDHSWRMCVDYRALNKATIPDKFPIPIIEELLD 695
V++LL+ G +I + + ++VKKK+ WR+CVDY LNKA D +P+P I+ L++
Sbjct: 797 VEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVCVDYTDLNKACPKDSYPLPHIDRLVE 856
Query: 696 ELHGARYFSKLDLKSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMN 755
G S +D SGY+Q+ + ++D KT+F T G Y + VM FGL NA +T+Q +N
Sbjct: 857 ATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGTYCYKVMSFGLKNAGATYQRFVN 916
Query: 756 DIFRHLLRKRVLVFFDDILVYSKDWPSHLEHLQEVLGILREQGLVANRKKCLFGREKVEY 815
+ + + V V+ DD+LV S H+EHL + +L G+ N KC FG E+
Sbjct: 917 KMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDVLNTYGMKLNPTKCTFGVTSGEF 976
Query: 816 LGHMISGQGVEVDPSKVESVTSWPTPKNVKGVRGFLGLTGYYRKFIRDYGKIAKPLTELT 875
LG++++ +G+E +P ++ ++ P+P+N + V+ G +FI P L
Sbjct: 977 LGYVVTKRGIEANPKQIRAILELPSPRNAREVQRLTGRIAALNRFISRSTDKCLPFYNLL 1036
Query: 876 KKNG-FEWSEKAQEAFETLKKKLTTSPVLALPDFSKEFTIECDASGVGVGAILMQ----E 930
K+ F+W + ++EAFE LK L+T P+L P+ + + S V ++L++ E
Sbjct: 1037 KRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVGETLYLYIAVSDHAVSSVLVREDRGE 1096
Query: 931 KRPIAYFSKALGVRNLSKSAYEKELMALGLAIQHWRPYLLGRHFKVTTDQRSLKELLQQK 990
+RPI Y SK+L EK +A+ + + RPY V TDQ L+ L
Sbjct: 1097 QRPIFYTSKSLVEAETRYPVIEKAALAVVTSARKLRPYFQSHTIAVLTDQ-PLRVALHSP 1155
Query: 991 VVTMEQQNWAAKLLGFDFEISYKP 1014
+ WA +L +D + +P
Sbjct: 1156 SQSGRMTKWAVELSEYDIDFRPRP 1179
Score = 87.8 bits (216), Expect = 4e-17
Identities = 90/362 (24%), Positives = 148/362 (40%), Gaps = 22/362 (6%)
Query: 1097 IHTMLTEFHTTPQGGHSGFYRTYRRLAA-NVYWRGMKSAVQDFVKQCDVCQRQKYLASSP 1155
I+ ++ E H G HSG +L YW M S + F +C+ CQR P
Sbjct: 1435 INEIMKETHEGAAGNHSGGRALALKLKKLGFYWPTMISDCKTFTAKCEQCQRHAPTIHQP 1494
Query: 1156 GGLLQP--LPIPERIWEDLSMDFITGLPKSKGFEAILVVVDRLSKYAHFIPLKHPYTAKS 1213
LL+ P P W +MD + +P S+ ILV+ D +K+ A
Sbjct: 1495 TELLRAGVAPYPFMRW---AMDIVGPMPASRQKRFILVMTDYFTKWVEAESYA-TIRAND 1550
Query: 1214 VAEVFGKEIVRLHGVPSSIVSDRDPIFVSNFWRELFKLQGTKLKMSTAYHPESDGQSEVV 1273
V K I+ HG+P I++D F+S + +L ST +P+ +GQ+E
Sbjct: 1551 VQNFVWKFIICRHGLPYEIITDNGSQFISLSFENFCASWKIRLNKSTPRYPQGNGQAEAT 1610
Query: 1274 NRCLETYLRCFIADQPKTWVIWIPWAEYWYNTCFHASTGVTPFEVVYGRPPPTITRWIQG 1333
N+ + + L+ + ++ W + + Y T ++T TPF YG
Sbjct: 1611 NKTILSGLKKRLDEKKGAWADELDGVLWSYRTTPRSATDQTPFAHAYGMEAMAPAEVGYS 1670
Query: 1334 ETRVEAVQKE-------LLERDEALRQLR-LQLARAQDRMKQFA---DRKRSDRSFSIGE 1382
R + K +L+R + L ++R L R Q+ A ++K +R F +G+
Sbjct: 1671 SLRRSMMVKNPELNDRMMLDRLDDLEEIRNAALCRIQNYQLAAAKHYNQKVHNRHFDVGD 1730
Query: 1383 WVFVKLRAHRQKSVVTRIYAKLAAKYYGPYPVVARVGAVAYQLKLPPGSKVHPVFHVSLL 1442
V K+ + + KL A + G Y V V Y+L G+ V ++ L
Sbjct: 1731 LVLRKVFENTAEINA----GKLGANWEGSYQVSKIVRPGDYELLTMSGTAVPRTWNSMHL 1786
Query: 1443 KK 1444
K+
Sbjct: 1787 KR 1788
>At4g04230 putative transposon protein
Length = 315
Score = 191 bits (486), Expect = 2e-48
Identities = 94/169 (55%), Positives = 123/169 (72%)
Query: 644 GVIRHSTSSFSSPVILVKKKDHSWRMCVDYRALNKATIPDKFPIPIIEELLDELHGARYF 703
G IR S S + PV+LV KKD +WRMC+D RA+N TI + PIP + ++LDEL GA F
Sbjct: 128 GYIRESLSPCAVPVLLVPKKDGTWRMCLDCRAINNITIKYRHPIPRLYDMLDELSGAIIF 187
Query: 704 SKLDLKSGYHQVRVKEEDVHKTAFRTHEGHYEFLVMPFGLMNAPSTFQSLMNDIFRHLLR 763
SK+DL+SGYHQVR++E D KTAF+T +G YE LVMPFGL NAPSTF LMN + R +
Sbjct: 188 SKVDLRSGYHQVRMREGDEWKTAFKTKQGLYECLVMPFGLTNAPSTFMRLMNQVLRSFIG 247
Query: 764 KRVLVFFDDILVYSKDWPSHLEHLQEVLGILREQGLVANRKKCLFGREK 812
K V+V+FDDIL+Y+K + H++ L+ +L LR++GL AN KKC F +K
Sbjct: 248 KFVVVYFDDILIYNKSYSDHIQPLELILKTLRKEGLYANLKKCTFCSDK 296
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,535,203
Number of Sequences: 26719
Number of extensions: 1672735
Number of successful extensions: 5560
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 71
Number of HSP's successfully gapped in prelim test: 37
Number of HSP's that attempted gapping in prelim test: 5187
Number of HSP's gapped (non-prelim): 229
length of query: 1566
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1454
effective length of database: 8,326,068
effective search space: 12106102872
effective search space used: 12106102872
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0096b.4


