

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0096b.1
(137 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
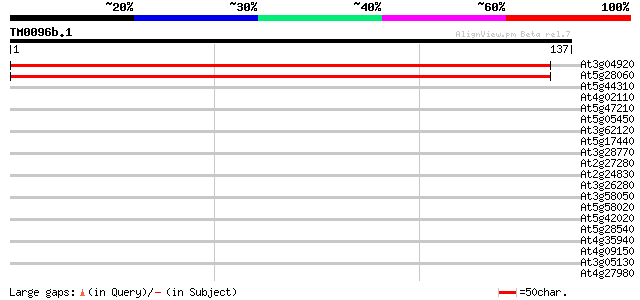
Score E
Sequences producing significant alignments: (bits) Value
At3g04920 putative ribosomal protein s19 or s24 242 4e-65
At5g28060 40S ribosomal protein S19 - like 236 2e-63
At5g44310 late embryogenesis abundant protein-like 35 0.009
At4g02110 predicted protein of unknown function 32 0.10
At5g47210 unknown protein 31 0.18
At5g05450 ATP-dependent RNA helicase-like protein 30 0.39
At3g62120 multifunctional aminoacyl-tRNA ligase-like protein 29 0.67
At5g17440 unknown protein 29 0.87
At3g28770 hypothetical protein 29 0.87
At2g27280 unknown protein 28 1.1
At2g24830 unknown protein 28 1.1
At3g26280 cytochrome P450 monooxygenase (CYP71B4) 28 1.5
At3g58050 putative protein 28 1.9
At5g58020 unknown protein 27 2.5
At5g42020 luminal binding protein (dbj|BAA13948.1) 27 2.5
At5g28540 luminal binding protein 27 2.5
At4g35940 putative protein 27 2.5
At4g09150 unknown protein 27 2.5
At3g05130 hypothetical protein 27 2.5
At4g27980 putative protein 27 3.3
>At3g04920 putative ribosomal protein s19 or s24
Length = 133
Score = 242 bits (618), Expect = 4e-65
Identities = 116/132 (87%), Positives = 129/132 (96%)
Query: 1 MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVF 60
MA+KAVTIRTRKFMTNRLLSRKQF++DVLHPGRANVSKAELKEKLAR+Y+VKDPN +FVF
Sbjct: 1 MAEKAVTIRTRKFMTNRLLSRKQFVIDVLHPGRANVSKAELKEKLARMYEVKDPNAIFVF 60
Query: 61 KFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIR 120
KFRTHFGGGKS+GFGLIYD++E+AKK+EPKYRLIRNGLDTK+EKSRKQ+KERKNRAKKIR
Sbjct: 61 KFRTHFGGGKSSGFGLIYDTVESAKKFEPKYRLIRNGLDTKIEKSRKQIKERKNRAKKIR 120
Query: 121 GVKKTKAADAAK 132
GVKKTKA DA K
Sbjct: 121 GVKKTKAGDAKK 132
>At5g28060 40S ribosomal protein S19 - like
Length = 133
Score = 236 bits (603), Expect = 2e-63
Identities = 112/132 (84%), Positives = 126/132 (94%)
Query: 1 MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVF 60
MA+KAVTIRTR FMTNRLL+RKQF++DVLHPGRANVSKAELKEKLAR+Y+VKDPN +F F
Sbjct: 1 MAEKAVTIRTRNFMTNRLLARKQFVIDVLHPGRANVSKAELKEKLARMYEVKDPNAIFCF 60
Query: 61 KFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIR 120
KFRTHFGGGKS+G+GLIYD++ENAKK+EPKYRLIRNGLDTK+EKSRKQ+KERKNRAKKIR
Sbjct: 61 KFRTHFGGGKSSGYGLIYDTVENAKKFEPKYRLIRNGLDTKIEKSRKQIKERKNRAKKIR 120
Query: 121 GVKKTKAADAAK 132
GVKKTKA D K
Sbjct: 121 GVKKTKAGDTKK 132
>At5g44310 late embryogenesis abundant protein-like
Length = 331
Score = 35.4 bits (80), Expect = 0.009
Identities = 29/106 (27%), Positives = 45/106 (42%), Gaps = 4/106 (3%)
Query: 33 RANVSKAELKEKLA-RLYDVKDPNTVFVFKFRTHFGGGKSTGFGLIYDSLENAKKYEPKY 91
RA E KEK + YDVK+ F + + G S YD E K Y +
Sbjct: 161 RAADKAYETKEKAKDKAYDVKEKTKDFAEETKEKVNEGASRAADKAYDVKEKTKNYAEQ- 219
Query: 92 RLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKTKAADAAKAGKKK 137
++ ++ ++ + +E K++AK K KA D A K+K
Sbjct: 220 --TKDKVNEGASRAADKAEETKDKAKDYAEDSKEKAEDMAHGFKEK 263
>At4g02110 predicted protein of unknown function
Length = 1293
Score = 32.0 bits (71), Expect = 0.10
Identities = 24/69 (34%), Positives = 33/69 (47%), Gaps = 3/69 (4%)
Query: 67 GGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAK--KIRGVKK 124
GG +S G E A K +P Y + +DTK K RKQ +NR + ++ K
Sbjct: 806 GGDQSLVAGETLTRKEAATK-DPSYAAAQLEVDTKKGKRRKQATVEENRLQTPSVKKAKV 864
Query: 125 TKAADAAKA 133
+K D AKA
Sbjct: 865 SKKEDGAKA 873
>At5g47210 unknown protein
Length = 357
Score = 31.2 bits (69), Expect = 0.18
Identities = 21/59 (35%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Query: 81 LENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKI---RGVKKTKAADAAKAGKK 136
LE KK ++ +DTKV +S +QL +KN ++I G K K DA + KK
Sbjct: 240 LEEKKKALQATKVEERKVDTKVFESMQQLSNKKNTDEEIFIKLGSDKEKRKDATEKAKK 298
>At5g05450 ATP-dependent RNA helicase-like protein
Length = 593
Score = 30.0 bits (66), Expect = 0.39
Identities = 29/122 (23%), Positives = 57/122 (45%), Gaps = 23/122 (18%)
Query: 36 VSKAELKEKLARLYDVKDPNTVFVFKFRTHFGGGKSTGFGLIY-DSLENAK--------- 85
V + LK ++ + K+ + F+F+++ G + G+GL+Y S+ K
Sbjct: 426 VMEKGLKAFVSFVRAYKEHHCSFIFRWKDLEIGKLAMGYGLLYLPSMSEVKQHRLSSEGF 485
Query: 86 -----------KYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKTKAADAAKAG 134
K++ KYR + + +V K ++Q +E+K + K+ R V + + D KA
Sbjct: 486 TPVEGVKFEEIKFKDKYREKQRQQNLQVRKEKRQ-EEKKEKGKRKR-VDASASNDPKKAS 543
Query: 135 KK 136
+K
Sbjct: 544 RK 545
>At3g62120 multifunctional aminoacyl-tRNA ligase-like protein
Length = 530
Score = 29.3 bits (64), Expect = 0.67
Identities = 15/35 (42%), Positives = 24/35 (67%), Gaps = 1/35 (2%)
Query: 103 EKSRKQLKERKNRAKKIRGVKKTKAADAAKAGKKK 137
E+ K +KE+K + KK + VK+ K A A+ +G+KK
Sbjct: 6 EQKEKVVKEKKEKVKKEKVVKE-KVAKASSSGQKK 39
>At5g17440 unknown protein
Length = 404
Score = 28.9 bits (63), Expect = 0.87
Identities = 30/141 (21%), Positives = 63/141 (44%), Gaps = 12/141 (8%)
Query: 1 MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARL----YDVKDPNT 56
M D+A +L +R++ +VD A+V + K +L + V D +
Sbjct: 225 MVDEAQKALEEAEALKKLTARQEPVVDSTKYTAADVRITDQKLRLCDICGAFLSVYDSDR 284
Query: 57 VFVFKFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRA 116
+ HFGG G+ LI D L A+ E K ++ + ++ + +S+++ +ER++
Sbjct: 285 ----RLADHFGGKLHLGYMLIRDKL--AELQEEKNKVHKERVEER--RSKERSRERESSK 336
Query: 117 KKIRGVKKTKAADAAKAGKKK 137
+ G + + D + + +
Sbjct: 337 DRDGGDNRDRGRDVDRRSRDR 357
>At3g28770 hypothetical protein
Length = 2081
Score = 28.9 bits (63), Expect = 0.87
Identities = 20/61 (32%), Positives = 29/61 (46%), Gaps = 8/61 (13%)
Query: 85 KKYEPKYRLIRNGLDTKVEKSRKQLKERK--------NRAKKIRGVKKTKAADAAKAGKK 136
KK E + ++K+++ K KE+K NR KK KK+K + AK KK
Sbjct: 972 KKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKK 1031
Query: 137 K 137
K
Sbjct: 1032 K 1032
Score = 26.2 bits (56), Expect = 5.6
Identities = 16/56 (28%), Positives = 27/56 (47%)
Query: 82 ENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKTKAADAAKAGKKK 137
+N +K E + + + + K EK + Q K+R+ + + R KK K KKK
Sbjct: 1008 KNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKK 1063
>At2g27280 unknown protein
Length = 764
Score = 28.5 bits (62), Expect = 1.1
Identities = 23/96 (23%), Positives = 41/96 (41%)
Query: 42 KEKLARLYDVKDPNTVFVFKFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTK 101
+E+L L + +D T +F GK+ FG + A+K E + + + K
Sbjct: 155 EERLRELREERDDVTKKKDLSDFYFNIGKNVAFGAREVEAKEAEKLEEQRKAEKLEEQRK 214
Query: 102 VEKSRKQLKERKNRAKKIRGVKKTKAADAAKAGKKK 137
EK + KE KK + +K + D+ + G +
Sbjct: 215 AEKLEELRKEVTRVEKKRKSPEKEVSPDSGEFGSSR 250
>At2g24830 unknown protein
Length = 497
Score = 28.5 bits (62), Expect = 1.1
Identities = 15/29 (51%), Positives = 18/29 (61%)
Query: 104 KSRKQLKERKNRAKKIRGVKKTKAADAAK 132
KS KQ K+R K+ RG K +AA AAK
Sbjct: 354 KSEKQKKKRSRGGKRKRGKKFAEAAKAAK 382
>At3g26280 cytochrome P450 monooxygenase (CYP71B4)
Length = 504
Score = 28.1 bits (61), Expect = 1.5
Identities = 18/79 (22%), Positives = 33/79 (40%), Gaps = 5/79 (6%)
Query: 36 VSKAELKEKLARLYDVKDPNTVFVFKFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIR 95
+S +E E+ + +D++ + R GK GFG+ D +K +R
Sbjct: 79 ISSSEAAEEALKTHDLECCSRPITMASRVFSRNGKDIGFGVYGDEWRELRKLS-----VR 133
Query: 96 NGLDTKVEKSRKQLKERKN 114
K +S K ++E +N
Sbjct: 134 EFFSVKKVQSFKYIREEEN 152
>At3g58050 putative protein
Length = 1209
Score = 27.7 bits (60), Expect = 1.9
Identities = 13/37 (35%), Positives = 20/37 (53%)
Query: 99 DTKVEKSRKQLKERKNRAKKIRGVKKTKAADAAKAGK 135
+ + E+ RK+ K K R KK+R ++ K D K K
Sbjct: 538 EKREEEERKEKKRSKEREKKLRKKERLKEKDKGKEKK 574
>At5g58020 unknown protein
Length = 354
Score = 27.3 bits (59), Expect = 2.5
Identities = 16/48 (33%), Positives = 31/48 (64%), Gaps = 3/48 (6%)
Query: 92 RLIRNGLDTKVEKSRKQLKERKNRAKKIRGV-KKTK--AADAAKAGKK 136
+++ NG + +V+ R++++E K + ++ +GV KK+K AA A G K
Sbjct: 244 KIVINGTEEEVDLLRERMEEEKAKLREKKGVSKKSKNGAAVVADTGAK 291
>At5g42020 luminal binding protein (dbj|BAA13948.1)
Length = 668
Score = 27.3 bits (59), Expect = 2.5
Identities = 17/51 (33%), Positives = 25/51 (48%), Gaps = 1/51 (1%)
Query: 82 ENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKTKAADAAK 132
E KK + K RN L+T V + Q+ ++ A K+ G +K K A K
Sbjct: 558 EEDKKVKEKIDA-RNALETYVYNMKNQVSDKDKLADKLEGDEKEKIEAATK 607
>At5g28540 luminal binding protein
Length = 669
Score = 27.3 bits (59), Expect = 2.5
Identities = 17/51 (33%), Positives = 25/51 (48%), Gaps = 1/51 (1%)
Query: 82 ENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKTKAADAAK 132
E KK + K RN L+T V + Q+ ++ A K+ G +K K A K
Sbjct: 558 EEDKKVKEKIDA-RNALETYVYNMKNQVNDKDKLADKLEGDEKEKIEAATK 607
>At4g35940 putative protein
Length = 451
Score = 27.3 bits (59), Expect = 2.5
Identities = 16/56 (28%), Positives = 29/56 (51%)
Query: 82 ENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKTKAADAAKAGKKK 137
E AKK + + + D K +K RK+ KE+K + +K R K+ + + ++K
Sbjct: 32 EKAKKEQRRKDRRSDKKDKKDKKERKEKKEKKEKKRKEREGKEVGSEKRSHKRRRK 87
>At4g09150 unknown protein
Length = 1097
Score = 27.3 bits (59), Expect = 2.5
Identities = 17/69 (24%), Positives = 31/69 (44%), Gaps = 9/69 (13%)
Query: 76 LIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQ---------LKERKNRAKKIRGVKKTK 126
++ L K + + +NGL+ +VEK R + LK KNR + + + +
Sbjct: 105 ILEKELARLAKMDEARQAAKNGLEQRVEKERDELESKVEERVLKAEKNRMLLFKAMAQRR 164
Query: 127 AADAAKAGK 135
AA +A +
Sbjct: 165 AAKRQRAAQ 173
>At3g05130 hypothetical protein
Length = 634
Score = 27.3 bits (59), Expect = 2.5
Identities = 17/58 (29%), Positives = 30/58 (51%), Gaps = 4/58 (6%)
Query: 73 GFGLIYDSLENAKKYEPKYRLIRNGLDTKVEK----SRKQLKERKNRAKKIRGVKKTK 126
GF L ++ + K+ + + L+ + K + + +KERK R ++I GVKK K
Sbjct: 181 GFDLQHEEVNRLKECVVRLEEKESNLEIVIGKLESENERLVKERKVREEEIEGVKKEK 238
>At4g27980 putative protein
Length = 565
Score = 26.9 bits (58), Expect = 3.3
Identities = 13/49 (26%), Positives = 26/49 (52%)
Query: 82 ENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKTKAADA 130
E A+K + L+R GL+ K + K+LKE + + ++ + + +A
Sbjct: 210 EEAEKLREETELMRKGLEIKEKTLEKRLKELELKQMELEETSRPQLVEA 258
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,749,684
Number of Sequences: 26719
Number of extensions: 111425
Number of successful extensions: 522
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 33
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 479
Number of HSP's gapped (non-prelim): 57
length of query: 137
length of database: 11,318,596
effective HSP length: 89
effective length of query: 48
effective length of database: 8,940,605
effective search space: 429149040
effective search space used: 429149040
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0096b.1


