

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
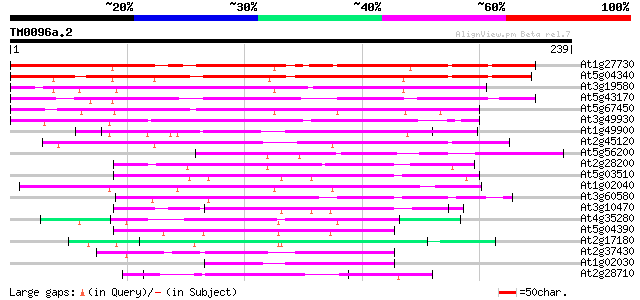
Query= TM0096a.2
(239 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g27730 salt-tolerance zinc finger protein like 218 2e-57
At5g04340 putative c2h2 zinc finger transcription factor 198 2e-51
At3g19580 zinc finger protein, putative 184 5e-47
At5g43170 Cys2/His2-type zinc finger protein 3 (dbj|BAA85109.1) 170 5e-43
At5g67450 Cys2/His2-type zinc finger protein 1 (dbj|BAA85108.1) 167 5e-42
At3g49930 zinc-finger-like protein 163 9e-41
At1g49900 Cys2/His2-type zinc finger protein, putative 97 6e-21
At2g45120 putative C2H2-type zinc finger protein 93 1e-19
At5g56200 unknown protein 91 5e-19
At2g28200 putative zinc-finger protein 90 9e-19
At5g03510 unknown protein 89 2e-18
At1g02040 hypothetical protein 89 3e-18
At3g60580 zinc finger protein - like 87 8e-18
At3g10470 unknown protein 83 1e-16
At4g35280 putative zinc-finger protein 82 3e-16
At5g04390 zinc finger transcription factor-like protein 77 8e-15
At2g17180 putative C2H2-type zinc finger protein 76 1e-14
At2g37430 putative C2H2-type zinc finger protein 72 3e-13
At1g02030 unknown protein 71 4e-13
At2g28710 putative C2H2-type zinc finger protein 70 8e-13
>At1g27730 salt-tolerance zinc finger protein like
Length = 227
Score = 218 bits (555), Expect = 2e-57
Identities = 132/236 (55%), Positives = 153/236 (63%), Gaps = 30/236 (12%)
Query: 1 MAMEALNSPTTTAPSFTFNDHPTLRHPAEPWAKRKRSKRSRS------CSEEEYLALCLI 54
MA+EAL SP +P + ++ H E W K KRSKRSRS +EEEYLA CL+
Sbjct: 1 MALEALTSPRLASPIPPLFEDSSVFHGVEHWTKGKRSKRSRSDFHHQNLTEEEYLAFCLM 60
Query: 55 MLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSG--- 111
+LAR PP PA +LSYKCSVCDK F SYQALGGHKASHRK+
Sbjct: 61 LLARDNRQ-----PPPPPAV-----EKLSYKCSVCDKTFSSYQALGGHKASHRKNLSQTL 110
Query: 112 -GGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCHYEGGG- 169
GGG+DHS TSSA TTTS+ TGSG K+H C+IC+KSFP+GQALGGHKRCHYEG
Sbjct: 111 SGGGDDHS---TSSA--TTTSAVTTGSG-KSHVCTICNKSFPSGQALGGHKRCHYEGNNN 164
Query: 170 -GSSSATASEGVGSTHTVSHSHHRDFDLNIPALPEFPSTKAGEDEVESPHPVMKKK 224
+SS + SEG GST VS S HR FDLNIP +PEF S G+DEV SP P K +
Sbjct: 165 INTSSVSNSEGAGSTSHVS-SSHRGFDLNIPPIPEF-SMVNGDDEVMSPMPAKKPR 218
>At5g04340 putative c2h2 zinc finger transcription factor
Length = 238
Score = 198 bits (503), Expect = 2e-51
Identities = 127/243 (52%), Positives = 150/243 (61%), Gaps = 35/243 (14%)
Query: 1 MAMEALNSPTTTAPSFT-FNDHPTLRHPAEPWAKRKRSKRSRS------CSEEEYLALCL 53
MA+E L SP ++P T F D H + K KRSKRSRS +E+EY+ALCL
Sbjct: 1 MALETLTSPRLSSPMPTLFQDSALGFHGS----KGKRSKRSRSEFDRQSLTEDEYIALCL 56
Query: 54 IMLARGG-------AATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASH 106
++LAR G +++++PPL P YKCSVCDKAF SYQALGGHKASH
Sbjct: 57 MLLARDGDRNRDLDLPSSSSSPPLLPPLPTP-----IYKCSVCDKAFSSYQALGGHKASH 111
Query: 107 RKH----SGGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKR 162
RK GG++ S TSSA+ T+ S G K+H CSICHKSF TGQALGGHKR
Sbjct: 112 RKSFSLTQSAGGDELS---TSSAITTSGISGGGGGSVKSHVCSICHKSFATGQALGGHKR 168
Query: 163 CHYE---GGGGSSSATASEGVGSTHTVSHSHHRDFDLNIPALPEFPSTKAGEDEVESPHP 219
CHYE GGG SSS + SE VGST VS S HR FDLNIP +PEF S G++EV SP P
Sbjct: 169 CHYEGKNGGGVSSSVSNSEDVGSTSHVS-SGHRGFDLNIPPIPEF-SMVNGDEEVMSPMP 226
Query: 220 VMK 222
K
Sbjct: 227 AKK 229
>At3g19580 zinc finger protein, putative
Length = 273
Score = 184 bits (466), Expect = 5e-47
Identities = 117/237 (49%), Positives = 131/237 (54%), Gaps = 39/237 (16%)
Query: 1 MAMEALNSPTTTAPSFT-FNDHPTLRHPA---EPWAKRKRSKRSRSCS------------ 44
MA+EA+N+PT+ SFT L + A EPW KRKRSKR RS S
Sbjct: 1 MALEAMNTPTS---SFTRIETKEDLMNDAVFIEPWLKRKRSKRQRSHSPSSSSSSPPRSR 57
Query: 45 ---------EEEYLALCLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPS 95
EEEYLALCL+MLA+ + T Q T P S L YKC+VC+KAFPS
Sbjct: 58 PKSQNQDLTEEEYLALCLLMLAKDQPSQTRFHQQSQSLTPPPESKNLPYKCNVCEKAFPS 117
Query: 96 YQALGGHKASHRKHSG---GGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFP 152
YQALGGHKASHR D S T S VA + GK HECSICHK FP
Sbjct: 118 YQALGGHKASHRIKPPTVISTTADDSTAPTISIVA--GEKHPIAASGKIHECSICHKVFP 175
Query: 153 TGQALGGHKRCHYE------GGGGSSSATASEGVGSTHTVSHSHHRDFDLNIPALPE 203
TGQALGGHKRCHYE GGGGS S + S V ST + SH DLN+PALPE
Sbjct: 176 TGQALGGHKRCHYEGNLGGGGGGGSKSISHSGSVSSTVSEERSHRGFIDLNLPALPE 232
>At5g43170 Cys2/His2-type zinc finger protein 3 (dbj|BAA85109.1)
Length = 193
Score = 170 bits (431), Expect = 5e-43
Identities = 109/232 (46%), Positives = 126/232 (53%), Gaps = 49/232 (21%)
Query: 1 MAMEALNSPTTTAPSFTFNDHPTLRHPAEPWAK-RKRSKRSRS-------CSEEEYLALC 52
MA+EALNSP FN E W K +KRSKRSRS +EEEYLA C
Sbjct: 1 MALEALNSPRLVEDPLRFNG-------VEQWTKCKKRSKRSRSDLHHNHRLTEEEYLAFC 53
Query: 53 LIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGG 112
L++LAR G + T +P SYKC VC K F SYQALGGHKASHR GG
Sbjct: 54 LMLLARDGGDLDSVTVAEKP----------SYKCGVCYKTFSSYQALGGHKASHRSLYGG 103
Query: 113 GGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCHYEGGGGSS 172
G D S +T+ K+H CS+C KSF TGQALGGHKRCHY+GG
Sbjct: 104 GENDKSTPSTAV---------------KSHVCSVCGKSFATGQALGGHKRCHYDGG---- 144
Query: 173 SATASEGVGSTHTVSHSHHRDFDLNIPALPEFPSTKAGEDEVESPHPVMKKK 224
+ SEGVGST VS S HR FDLNI + F + +DEV SP K +
Sbjct: 145 -VSNSEGVGSTSHVSSSSHRGFDLNIIPVQGF----SPDDEVMSPMATKKPR 191
>At5g67450 Cys2/His2-type zinc finger protein 1 (dbj|BAA85108.1)
Length = 245
Score = 167 bits (423), Expect = 5e-42
Identities = 108/244 (44%), Positives = 129/244 (52%), Gaps = 51/244 (20%)
Query: 1 MAMEALNSPTTTAPSFTFNDHPTLRHPAE-------PWAKRKRSKRSRSC---------- 43
MA+E LNSPT T + P LR+ E WAKRKR+KR R
Sbjct: 1 MALETLNSPTATTTA-----RPLLRYREEMEPENLEQWAKRKRTKRQRFDHGHQNQETNK 55
Query: 44 ---SEEEYLALCLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALG 100
SEEEYLALCL+MLARG A + PPL +PS YKC+VC K+F SYQALG
Sbjct: 56 NLPSEEEYLALCLLMLARGSAVQSPPLPPLPSRASPS--DHRDYKCTVCGKSFSSYQALG 113
Query: 101 GHKASHRKHSG-----GGGEDHSAGATSSAVATTTSSANTGSG----GKAHECSICHKSF 151
GHK SHRK + G E + ++S + NTG+G GK H CSIC KSF
Sbjct: 114 GHKTSHRKPTNTSITSGNQELSNNSHSNSGSVVINVTVNTGNGVSQSGKIHTCSICFKSF 173
Query: 152 PTGQALGGHKRCHYEG-----GGGSSSATASEGVGS----------THTVSHSHHRDFDL 196
+GQALGGHKRCHY+G G GSSS + GS + + HR FDL
Sbjct: 174 ASGQALGGHKRCHYDGGNNGNGNGSSSNSVELVAGSDVSDVDNERWSEESAIGGHRGFDL 233
Query: 197 NIPA 200
N+PA
Sbjct: 234 NLPA 237
>At3g49930 zinc-finger-like protein
Length = 215
Score = 163 bits (412), Expect = 9e-41
Identities = 97/211 (45%), Positives = 120/211 (55%), Gaps = 29/211 (13%)
Query: 1 MAMEALNSPTTTA-----PSFTFNDHPTLRHPAEPWAKRKRSKRSR------SCSEEEYL 49
MA++ LNSPT+T P F T E W KRKR+KR R SEEEYL
Sbjct: 1 MALDTLNSPTSTTTTTAPPPFLRCLDETEPENLESWTKRKRTKRHRIDQPNPPPSEEEYL 60
Query: 50 ALCLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKH 109
ALCL+MLARG ++ + P + +P + YKCSVC K+FPSYQALGGHK SHRK
Sbjct: 61 ALCLLMLARG-SSDHHSPPSDHHSLSPLSDHQKDYKCSVCGKSFPSYQALGGHKTSHRKP 119
Query: 110 SGGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCHYEGGG 169
+ + T++ S+ G GK H CSIC KSFP+GQALGGHKRCHY+GG
Sbjct: 120 VSVDVNNSNGTVTNNG---NISNGLVGQSGKTHNCSICFKSFPSGQALGGHKRCHYDGGN 176
Query: 170 GSSSATASEGVGSTHTVSHSHHRDFDLNIPA 200
G+S+ +SH FDLN+PA
Sbjct: 177 GNSNG------------DNSH--KFDLNLPA 193
>At1g49900 Cys2/His2-type zinc finger protein, putative
Length = 917
Score = 97.4 bits (241), Expect = 6e-21
Identities = 70/180 (38%), Positives = 82/180 (44%), Gaps = 34/180 (18%)
Query: 29 EPWAKRKRSKRSR-----------SCSEEEYLALCLIMLA--RGGAATTTAT-------- 67
+PW R KR R S SE + AL L+ + R T T T
Sbjct: 110 KPWLDSFRLKRQRLGDDEDPSSSSSPSEVDIAALTLLQFSCDRRHPQTQTLTRPQPQTHK 169
Query: 68 -------PPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAG 120
P LQ T + +KCS+C+K F SYQALGGHKASH S + +AG
Sbjct: 170 TQLQRPPPQLQSQTQTAPPKSDLFKCSICEKVFTSYQALGGHKASH---SIKAAQLENAG 226
Query: 121 ATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGV 180
A + T S GK H+C ICH FPTGQALGGHKR HYEG G E V
Sbjct: 227 ADAGE---KTRSKMLSPSGKIHKCDICHVLFPTGQALGGHKRRHYEGLLGGHKRGNDEAV 283
Score = 93.2 bits (230), Expect = 1e-19
Identities = 60/162 (37%), Positives = 79/162 (48%), Gaps = 13/162 (8%)
Query: 40 SRSCSEEEYLALCLIMLARGGAATTTATPPL-QPATAPSGSSRLSYKCSVCDKAFPSYQA 98
S S S++E ALC ++ + T + QP T S SY+C+VC + PSYQA
Sbjct: 706 SSSLSKDETAALCPTPHSQTQPQSQTQLQKVTQPQTQMLPKSD-SYQCNVCGRELPSYQA 764
Query: 99 LGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALG 158
LGGHKASHR + +A GK H+CSICH+ F TGQ+LG
Sbjct: 765 LGGHKASHR----------TKPPVENATGEKMRPKKLAPSGKIHKCSICHREFSTGQSLG 814
Query: 159 GHKRCHYEGG-GGSSSATASEGVGSTHTVSHSHHRDFDLNIP 199
GHKR HYEG G + E V +S S + ++P
Sbjct: 815 GHKRLHYEGVLRGHKRSQEKEAVSQGDKLSPSGNGSVVTHVP 856
>At2g45120 putative C2H2-type zinc finger protein
Length = 314
Score = 92.8 bits (229), Expect = 1e-19
Identities = 73/213 (34%), Positives = 90/213 (41%), Gaps = 29/213 (13%)
Query: 15 SFTFN-DHPTLRHPAEPWAKRKRSKRSRSCSEEEYLALCLIMLARGG--------AATTT 65
SF F+ + T P+E A+ + + + EE LA CLIML+R
Sbjct: 116 SFDFDFEKLTTSQPSELVAEPEHHSSASDTTTEEDLAFCLIMLSRDKWKQQKKKKQRVEE 175
Query: 66 ATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAGATSSA 125
S SR +KC C K F SYQALGGH+ASH+K + A
Sbjct: 176 DETDHDSEDYKSSKSRGRFKCETCGKVFKSYQALGGHRASHKK--------------NKA 221
Query: 126 VATTTSSANTG-----SGGKAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGV 180
T T T K HEC IC + F +GQALGGHKR H G + S+ V
Sbjct: 222 CMTKTEQVETEYVLGVKEKKVHECPICFRVFTSGQALGGHKRSHGSNIGAGRGLSVSQIV 281
Query: 181 GSTHTVSHSHHRDFDLNIPALPEFPSTKAGEDE 213
VS R DLN+PA E T DE
Sbjct: 282 QIEEEVS-VKQRMIDLNLPAPNEEDETSLVFDE 313
Score = 34.3 bits (77), Expect = 0.060
Identities = 13/23 (56%), Positives = 16/23 (69%)
Query: 142 HECSICHKSFPTGQALGGHKRCH 164
++C C KSF G+ALGGH R H
Sbjct: 4 YKCRFCFKSFINGRALGGHMRSH 26
Score = 32.0 bits (71), Expect = 0.30
Identities = 13/23 (56%), Positives = 16/23 (69%)
Query: 84 YKCSVCDKAFPSYQALGGHKASH 106
YKC C K+F + +ALGGH SH
Sbjct: 4 YKCRFCFKSFINGRALGGHMRSH 26
>At5g56200 unknown protein
Length = 493
Score = 90.9 bits (224), Expect = 5e-19
Identities = 62/170 (36%), Positives = 81/170 (47%), Gaps = 35/170 (20%)
Query: 80 SRLSYKCSVCDKAFPSYQALGGHKASHRK------HSGGGGEDHSAGAT-------SSAV 126
+R + C C+K+F SYQALGGH+ASH K H + S T +SA
Sbjct: 338 AREKHVCVTCNKSFSSYQALGGHRASHNKVKILENHQARANAEASLLGTEAIITGLASAQ 397
Query: 127 ATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTHTV 186
T TS +++ +G H C+ICHKSF TGQALGGHKRCH+ TA + TV
Sbjct: 398 GTNTSLSSSHNGD--HVCNICHKSFSTGQALGGHKRCHW---------TAPVSTVAPTTV 446
Query: 187 SHSHHRDFDLNIPALPEFPSTKAGEDEVESPHPVMKKKQRVFMIPKIEIP 236
A P P+T + E+ V K K+RV E+P
Sbjct: 447 P-----------TAAPTVPATASSSQVTETVQEVKKLKRRVLEFDLNELP 485
Score = 37.7 bits (86), Expect = 0.005
Identities = 29/87 (33%), Positives = 37/87 (42%), Gaps = 15/87 (17%)
Query: 86 CSVCDKAFPSYQALGGHKASH----RKHSGGGGEDHSAGATSSAVATTTSSANTGSGGKA 141
C C K F S +ALGGHK H RK S V+ + G A
Sbjct: 81 CCECGKRFVSGKALGGHKRIHVLETRKFSM---------MRPKMVSGMVGRSERGDLEVA 131
Query: 142 HECSICHKSFPTGQALGGHKRCHYEGG 168
C +C+K F + +AL GH R H + G
Sbjct: 132 --CCVCYKKFTSMKALYGHMRFHPDRG 156
Score = 36.2 bits (82), Expect = 0.016
Identities = 15/25 (60%), Positives = 17/25 (68%)
Query: 140 KAHECSICHKSFPTGQALGGHKRCH 164
K H C C K F +G+ALGGHKR H
Sbjct: 77 KKHICCECGKRFVSGKALGGHKRIH 101
Score = 35.4 bits (80), Expect = 0.027
Identities = 15/33 (45%), Positives = 21/33 (63%)
Query: 74 TAPSGSSRLSYKCSVCDKAFPSYQALGGHKASH 106
T+ S S + C++C K+F + QALGGHK H
Sbjct: 401 TSLSSSHNGDHVCNICHKSFSTGQALGGHKRCH 433
>At2g28200 putative zinc-finger protein
Length = 284
Score = 90.1 bits (222), Expect = 9e-19
Identities = 62/179 (34%), Positives = 89/179 (49%), Gaps = 35/179 (19%)
Query: 45 EEEYLALCLIMLARGGAATTTATPPLQPAT------APSGSSRLSYKCSVCDKAFPSYQA 98
EEE +A+CLIMLARG T +P L+ + + SS Y+C C++ F S+QA
Sbjct: 71 EEEDMAICLIMLARG---TVLPSPDLKNSRKIHQKISSENSSFYVYECKTCNRTFSSFQA 127
Query: 99 LGGHKASHRK------------------HSGGGGEDHSAGATSSAVATTTSSANTGSGGK 140
LGGH+ASH+K + G++ + SA+A+ S+ K
Sbjct: 128 LGGHRASHKKPRTSTEEKTRLPLTQPKSSASEEGQNSHFKVSGSALASQASNI-INKANK 186
Query: 141 AHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTHTVS-HSHHRDFDLNI 198
HECSIC F +GQALGGH R H ++ T V +T VS +S + ++NI
Sbjct: 187 VHECSICGSEFTSGQALGGHMRRH------RTAVTTISPVAATAEVSRNSTEEEIEINI 239
>At5g03510 unknown protein
Length = 292
Score = 89.0 bits (219), Expect = 2e-18
Identities = 66/192 (34%), Positives = 90/192 (46%), Gaps = 39/192 (20%)
Query: 45 EEEYLALCLIMLARGGAATTTATPPLQPATA------PSGSSRLS----YKCSVCDKAFP 94
E+E +A CLI+L++G A ++ P S L Y+C CDK+F
Sbjct: 69 EDEDMANCLILLSQGHQAKSSDDHLSMQRMGFFSNKKPVASLGLGLDGVYQCKTCDKSFH 128
Query: 95 SYQALGGHKASHRKHSGGGG---------------EDHSAGATSSAVA-TTTSSANTGSG 138
S+QALGGH+ASH+K G E AGA S ++ TSS +
Sbjct: 129 SFQALGGHRASHKKPKLGASVFKCVEKKTASASTVETVEAGAVGSFLSLQVTSSDGSKKP 188
Query: 139 GKAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTHTVSHSHHRD----- 193
K HECSIC F +GQALGGH R H G + +A A+ + + + S HH +
Sbjct: 189 EKTHECSICKAEFSSGQALGGHMRRH---RGLTINANATSAIKTAISSSSHHHHEESIRP 245
Query: 194 -----FDLNIPA 200
DLN+PA
Sbjct: 246 KNFLQLDLNLPA 257
>At1g02040 hypothetical protein
Length = 324
Score = 88.6 bits (218), Expect = 3e-18
Identities = 69/224 (30%), Positives = 96/224 (42%), Gaps = 33/224 (14%)
Query: 5 ALNSPTTTAPSFTFNDHPTLRHPAEPWAKRKRSKRSR------SCSEEEYLALCLIMLAR 58
A+NSP F+ H P KR+RS ++ S +E+E LA CL++L+
Sbjct: 61 AMNSPCIDFVMFSRGQHDEDNMSRRPPWKRERSMSTQHHHLNLSPNEDEELANCLVLLSN 120
Query: 59 GGAATTTATPPL----QPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSG--- 111
G A + T + ++C C K F S+QALGGH+ASH+K G
Sbjct: 121 SGDAHGGDQHKQHGHGKGKTVKKQKTAQVFQCKACKKVFTSHQALGGHRASHKKVKGCFA 180
Query: 112 -------------GGGEDHSAGATSSAVATTTSSANTG-SGGKAHECSICHKSFPTGQAL 157
+D+ S+A+ AHEC+ICH+ F +GQAL
Sbjct: 181 SQDKEEEEEEEYKEDDDDNDEDEDEEEDEEDKSTAHIARKRSNAHECTICHRVFSSGQAL 240
Query: 158 GGHKRCHYEGGGGSSSATASEGVGSTHTVSHSHHRDFDLNIPAL 201
GGHKRCH+ T S H HS R L+ P+L
Sbjct: 241 GGHKRCHWLTPSNYLRMT------SLHDHHHSVGRPQPLDQPSL 278
>At3g60580 zinc finger protein - like
Length = 288
Score = 87.0 bits (214), Expect = 8e-18
Identities = 61/180 (33%), Positives = 81/180 (44%), Gaps = 31/180 (17%)
Query: 46 EEYLALCLIMLARG----GAATTTATPPLQPATAPSGSSRLS-------YKCSVCDKAFP 94
EE LA CL+ML+R + ++ G ++++ YKC C K F
Sbjct: 124 EEDLAFCLMMLSRDKWKKNKSNKEVVEEIETEEESEGYNKINRATTKGRYKCETCGKVFK 183
Query: 95 SYQALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTG 154
SYQALGGH+ASH+K+ + T + + HEC IC + F +G
Sbjct: 184 SYQALGGHRASHKKNRVSNNKTEQRSETEYDNVVVVAK-------RIHECPICLRVFASG 236
Query: 155 QALGGHKRCHYEGGGGSSSATASEGVGSTHTVSHSHHRDFDLNIPALPEFPSTKAGEDEV 214
QALGGHKR H G G+ S V +V R DLN+PA E EDEV
Sbjct: 237 QALGGHKRSH---GVGNLSVNQQRRVHRNESVK---QRMIDLNLPAPTE-------EDEV 283
Score = 37.7 bits (86), Expect = 0.005
Identities = 31/113 (27%), Positives = 47/113 (41%), Gaps = 25/113 (22%)
Query: 140 KAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTHTVSHSHHRDFDLNIP 199
++++C +C KSF G+ALGGH R H + + VS S
Sbjct: 2 ESYKCRVCFKSFVNGKALGGHMRSHMSNSHEEEQRPSQLSYETESDVSSSD--------- 52
Query: 200 ALPEFPSTKA-----GEDEVESPHPVM----KKKQRV-----FMIPKIEIPQL 238
P+F T + GE E ES V+ K+ +R F+ K++ QL
Sbjct: 53 --PKFAFTSSVLLEDGESESESSRNVINLTRKRSKRTRKLDSFVTKKVKTSQL 103
Score = 37.4 bits (85), Expect = 0.007
Identities = 16/28 (57%), Positives = 20/28 (71%)
Query: 83 SYKCSVCDKAFPSYQALGGHKASHRKHS 110
SYKC VC K+F + +ALGGH SH +S
Sbjct: 3 SYKCRVCFKSFVNGKALGGHMRSHMSNS 30
>At3g10470 unknown protein
Length = 398
Score = 82.8 bits (203), Expect = 1e-16
Identities = 50/125 (40%), Positives = 60/125 (48%), Gaps = 28/125 (22%)
Query: 84 YKCSVCDKAFPSYQALGGHKASHRKHSGGGG----EDHSAGATSSAVA-------TTTSS 132
Y+C CD+ FPS+QALGGH+ASH+K G DH AV+ TTT +
Sbjct: 182 YQCKTCDRTFPSFQALGGHRASHKKPKAAMGLHSNHDHKKSNYDDAVSLHLNNVLTTTPN 241
Query: 133 ANT----------GSGGKAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGS 182
N+ GS K HEC IC F +GQALGGH R H A + S
Sbjct: 242 NNSNHRSLVVYGKGSNNKVHECGICGAEFTSGQALGGHMRRH-------RGAVVAAAAAS 294
Query: 183 THTVS 187
T TVS
Sbjct: 295 TATVS 299
Score = 55.5 bits (132), Expect = 3e-08
Identities = 43/149 (28%), Positives = 68/149 (44%), Gaps = 26/149 (17%)
Query: 45 EEEYLALCLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKA 104
E++ +A CLI+LA+G + P LQP P +R + SYQ G +
Sbjct: 104 EDQDMANCLILLAQGHSL-----PHLQPQPHPQQQTR---------QLMMSYQDSGNNNN 149
Query: 105 SHRKHSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCH 164
+ + S + TSS+ TTT+ +G ++C C ++FP+ QALGGH+ H
Sbjct: 150 NAYRSSSRRFLE-----TSSSNGTTTNGGGGRAGYYVYQCKTCDRTFPSFQALGGHRASH 204
Query: 165 YEGGGGSSSATASEGVGSTHTVSHSHHRD 193
A+ G+ S H S++ D
Sbjct: 205 -------KKPKAAMGLHSNHDHKKSNYDD 226
Score = 38.9 bits (89), Expect = 0.002
Identities = 21/58 (36%), Positives = 31/58 (53%), Gaps = 3/58 (5%)
Query: 78 GSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSANT 135
GS+ ++C +C F S QALGGH R+H G +A + +VA ++ANT
Sbjct: 255 GSNNKVHECGICGAEFTSGQALGGHM---RRHRGAVVAAAAASTATVSVAAIPATANT 309
>At4g35280 putative zinc-finger protein
Length = 284
Score = 81.6 bits (200), Expect = 3e-16
Identities = 49/129 (37%), Positives = 67/129 (50%), Gaps = 24/129 (18%)
Query: 44 SEEEY-LALCLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGH 102
SEE++ +A CL+ML+ G PS SS ++C C K F S+QALGGH
Sbjct: 136 SEEDHEVASCLLMLSNG---------------TPSSSSIERFECGGCKKVFGSHQALGGH 180
Query: 103 KASHRKHSGGGGEDHSAGATSSAVATTTSSANTGSG-----GKAHECSICHKSFPTGQAL 157
+ASH+ G T + +TSS + G H+C+IC + F +GQAL
Sbjct: 181 RASHKNVKGCFA---ITNVTDDPMTVSTSSGHDHQGKILTFSGHHKCNICFRVFSSGQAL 237
Query: 158 GGHKRCHYE 166
GGH RCH+E
Sbjct: 238 GGHMRCHWE 246
Score = 41.2 bits (95), Expect = 5e-04
Identities = 46/213 (21%), Positives = 76/213 (35%), Gaps = 52/213 (24%)
Query: 14 PSFTFNDHPTLRHPA----EPWAKRKRSKRSRSCSEEEYLALCLIMLARGGAATTTATPP 69
PS T N +P L+ + K+KR+K S S ++ +A+ P
Sbjct: 18 PSNTSNPNPNLQFALSSSYDHSPKKKRTKTVASSSS---------------SSPKSASKP 62
Query: 70 LQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGG---------------- 113
+ +++ C+ C + F S++AL GH H + G
Sbjct: 63 KYTKKPDPNAPKITRPCTECGRKFWSWKALFGHMRCHPERQWRGINPPPNYRVPTAASSK 122
Query: 114 --------------GEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGG 159
EDH + ++ T S+ S + EC C K F + QALGG
Sbjct: 123 QLNQILPNWVSFMSEEDHEVASCLLMLSNGTPSS---SSIERFECGGCKKVFGSHQALGG 179
Query: 160 HKRCHYEGGGGSSSATASEGVGSTHTVSHSHHR 192
H+ H G + ++ + T S H+
Sbjct: 180 HRASHKNVKGCFAITNVTDDPMTVSTSSGHDHQ 212
>At5g04390 zinc finger transcription factor-like protein
Length = 362
Score = 77.0 bits (188), Expect = 8e-15
Identities = 52/156 (33%), Positives = 71/156 (45%), Gaps = 36/156 (23%)
Query: 45 EEEYLALCLIMLARGGAATT----------------TATPPLQPATAPSGSSR--LSYKC 86
E++ +A CLI+LA+G + T+ L+ +++ SG Y+C
Sbjct: 96 EDQDIANCLILLAQGHSLPHNNHHLPNSNNNNTYRFTSRRFLETSSSNSGGKAGYYVYQC 155
Query: 87 SVCDKAFPSYQALGGHKASHRKHSGGG--------GEDHSAGATSSAVATTTSSANT--- 135
CD+ FPS+QALGGH+ASH+K ++ A S TTT N
Sbjct: 156 KTCDRTFPSFQALGGHRASHKKPKAASFYSNLDLKKNTYANDAVSLVHTTTTVFKNNNSR 215
Query: 136 -------GSGGKAHECSICHKSFPTGQALGGHKRCH 164
S K HEC IC F +GQALGGH R H
Sbjct: 216 SLVVYGKASKNKVHECGICGAEFTSGQALGGHMRRH 251
Score = 33.5 bits (75), Expect = 0.10
Identities = 25/78 (32%), Positives = 33/78 (42%), Gaps = 15/78 (19%)
Query: 84 YKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHE 143
++C +C F S QALGGH HR G + V T++ANT E
Sbjct: 229 HECGICGAEFTSGQALGGHMRRHR------GAVVVPAVIAPTVTVATAAANT-------E 275
Query: 144 CSICHKSFPTGQALGGHK 161
S+ SF Q GH+
Sbjct: 276 LSLSSMSF--DQISDGHQ 291
>At2g17180 putative C2H2-type zinc finger protein
Length = 270
Score = 76.3 bits (186), Expect = 1e-14
Identities = 58/202 (28%), Positives = 78/202 (37%), Gaps = 49/202 (24%)
Query: 26 HPAEPWA--------KRKRSKRSRSCS-------EEEYLALCLIMLARGGAATTTATPPL 70
HP W KR+ + + S S EE +A CL+M+A G T
Sbjct: 87 HPERQWRGINPPSNFKRRINSNAASSSSSWDPSEEEHNIASCLLMMANGDVPTR------ 140
Query: 71 QPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGGG-----EDHSAGATSSA 125
S ++C C K F S+QALGGH+A+H+ G ED
Sbjct: 141 ------SSEVEERFECDGCKKVFGSHQALGGHRATHKDVKGCFANKNITEDPPPPPPQEI 194
Query: 126 VATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTHT 185
V + G H C+IC + F +GQALGGH RCH+E
Sbjct: 195 VDQDKGKSVKLVSGMNHRCNICSRVFSSGQALGGHMRCHWE-----------------KD 237
Query: 186 VSHSHHRDFDLNIPALPEFPST 207
+ R DLN+PA +T
Sbjct: 238 QEENQVRGIDLNVPAATSSDTT 259
Score = 45.8 bits (107), Expect = 2e-05
Identities = 34/149 (22%), Positives = 59/149 (38%), Gaps = 26/149 (17%)
Query: 56 LARGGAATTTATPPLQPATAPS-GSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGG- 113
L +++++P +P T P +S+++ C+ C K F S +AL GH H + G
Sbjct: 36 LTNNEVGSSSSSPRPKPVTQPDPDASQIARPCTECGKQFGSLKALFGHMRCHPERQWRGI 95
Query: 114 ------------------------GEDHSAGATSSAVATTTSSANTGSGGKAHECSICHK 149
E+H+ + +A + + EC C K
Sbjct: 96 NPPSNFKRRINSNAASSSSSWDPSEEEHNIASCLLMMANGDVPTRSSEVEERFECDGCKK 155
Query: 150 SFPTGQALGGHKRCHYEGGGGSSSATASE 178
F + QALGGH+ H + G ++ +E
Sbjct: 156 VFGSHQALGGHRATHKDVKGCFANKNITE 184
>At2g37430 putative C2H2-type zinc finger protein
Length = 178
Score = 71.6 bits (174), Expect = 3e-13
Identities = 48/131 (36%), Positives = 64/131 (48%), Gaps = 20/131 (15%)
Query: 38 KRSRSCSEEEY----LALCLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAF 93
KR RS EE +A CL++LA+ L T S++ ++C C+K F
Sbjct: 2 KRERSDFEESLKNIDIAKCLMILAQTSMVKQIG---LNQHTESHTSNQ--FECKTCNKRF 56
Query: 94 PSYQALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPT 153
S+QALGGH+ASH+K + +N G H+CSIC +SF T
Sbjct: 57 SSFQALGGHRASHKKPK-----------LTVEQKDVKHLSNDYKGNHFHKCSICSQSFGT 105
Query: 154 GQALGGHKRCH 164
GQALGGH R H
Sbjct: 106 GQALGGHMRRH 116
Score = 35.0 bits (79), Expect = 0.035
Identities = 13/24 (54%), Positives = 18/24 (74%)
Query: 84 YKCSVCDKAFPSYQALGGHKASHR 107
+KCS+C ++F + QALGGH HR
Sbjct: 94 HKCSICSQSFGTGQALGGHMRRHR 117
>At1g02030 unknown protein
Length = 267
Score = 71.2 bits (173), Expect = 4e-13
Identities = 35/81 (43%), Positives = 42/81 (51%), Gaps = 9/81 (11%)
Query: 84 YKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHE 143
++C C+K F SYQALGGH+ASH+K T + S HE
Sbjct: 160 FECETCEKVFKSYQALGGHRASHKK---------KIAETDQLGSDELKKKKKKSTSSHHE 210
Query: 144 CSICHKSFPTGQALGGHKRCH 164
C IC K F +GQALGGHKR H
Sbjct: 211 CPICAKVFTSGQALGGHKRSH 231
Score = 38.1 bits (87), Expect = 0.004
Identities = 14/23 (60%), Positives = 17/23 (73%)
Query: 142 HECSICHKSFPTGQALGGHKRCH 164
H+C +C KSF G+ALGGH R H
Sbjct: 5 HKCKLCWKSFANGRALGGHMRSH 27
Score = 36.2 bits (82), Expect = 0.016
Identities = 15/28 (53%), Positives = 19/28 (67%)
Query: 79 SSRLSYKCSVCDKAFPSYQALGGHKASH 106
S+ ++C +C K F S QALGGHK SH
Sbjct: 204 STSSHHECPICAKVFTSGQALGGHKRSH 231
Score = 32.3 bits (72), Expect = 0.23
Identities = 17/52 (32%), Positives = 23/52 (43%)
Query: 140 KAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTHTVSHSHH 191
K EC C K F + QALGGH+ H + + + E + SHH
Sbjct: 158 KWFECETCEKVFKSYQALGGHRASHKKKIAETDQLGSDELKKKKKKSTSSHH 209
Score = 31.6 bits (70), Expect = 0.39
Identities = 12/23 (52%), Positives = 17/23 (73%)
Query: 84 YKCSVCDKAFPSYQALGGHKASH 106
+KC +C K+F + +ALGGH SH
Sbjct: 5 HKCKLCWKSFANGRALGGHMRSH 27
>At2g28710 putative C2H2-type zinc finger protein
Length = 156
Score = 70.5 bits (171), Expect = 8e-13
Identities = 49/135 (36%), Positives = 62/135 (45%), Gaps = 32/135 (23%)
Query: 49 LALCLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRK 108
+A CLI+L++ T SR+ + C C+K FPS+QALGGH+ASHR+
Sbjct: 14 MANCLILLSKAHQNDT--------------KSRV-FACKTCNKEFPSFQALGGHRASHRR 58
Query: 109 HSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCH---Y 165
+ G + V HEC IC F GQALGGH R H
Sbjct: 59 SAALEGHAPPSPKRVKPV--------------KHECPICGAEFAVGQALGGHMRKHRGGS 104
Query: 166 EGGGGSSSATASEGV 180
GGGG S A A+ V
Sbjct: 105 GGGGGRSLAPATAPV 119
Score = 45.8 bits (107), Expect = 2e-05
Identities = 31/87 (35%), Positives = 37/87 (41%), Gaps = 9/87 (10%)
Query: 58 RGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDH 117
R AA PP +P + ++C +C F QALGGH HR SGGGG
Sbjct: 57 RRSAALEGHAPP-----SPKRVKPVKHECPICGAEFAVGQALGGHMRKHRGGSGGGGGRS 111
Query: 118 SAGATSSAVATTTSSANTGSGGKAHEC 144
A AT A T + G GK C
Sbjct: 112 LAPAT----APVTMKKSGGGNGKRVLC 134
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.126 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,183,405
Number of Sequences: 26719
Number of extensions: 287327
Number of successful extensions: 1504
Number of sequences better than 10.0: 135
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 52
Number of HSP's that attempted gapping in prelim test: 1078
Number of HSP's gapped (non-prelim): 304
length of query: 239
length of database: 11,318,596
effective HSP length: 96
effective length of query: 143
effective length of database: 8,753,572
effective search space: 1251760796
effective search space used: 1251760796
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0096a.2


