

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0095c.5
(423 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
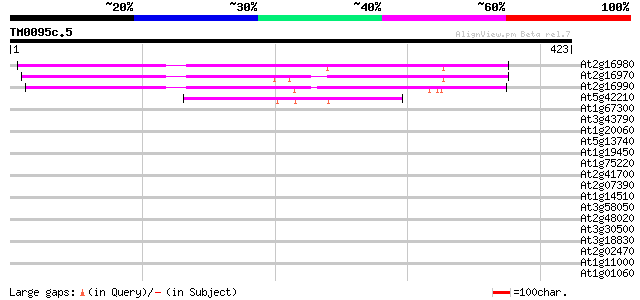
Score E
Sequences producing significant alignments: (bits) Value
At2g16980 putative tetracycline transporter protein 194 1e-49
At2g16970 putative tetracycline transporter protein 186 3e-47
At2g16990 putative tetracycline transporter protein 180 2e-45
At5g42210 putative protein 79 6e-15
At1g67300 transport protein like protein 33 0.23
At3g43790 transporter-like protein 33 0.30
At1g20060 unknown protein 32 0.86
At5g13740 transporter-like protein 31 1.1
At1g19450 integral membrane protein, putative 30 1.9
At1g75220 integral membrane protein, putative 30 2.5
At2g41700 ATP-binding cassette transporter ABCA1 30 3.3
At2g07390 hypothetical protein 29 4.3
At1g14510 unknown protein 29 4.3
At3g58050 putative protein 29 5.6
At2g48020 sugar transporter like protein 29 5.6
At3g30500 hypothetical protein 28 7.3
At3g18830 sugar transporter like protein 28 7.3
At2g02470 putative PHD-type zinc finger protein 28 7.3
At1g11000 membrane protein Mlo4 28 7.3
At1g01060 DNA-binding protein, putative 28 7.3
>At2g16980 putative tetracycline transporter protein
Length = 415
Score = 194 bits (492), Expect = 1e-49
Identities = 129/397 (32%), Positives = 201/397 (50%), Gaps = 41/397 (10%)
Query: 7 LIELRPLFHLLFPLSIHWIAEEMTVSVLVDVTTSALCPG-GSTCPKVIYINGLQQTIVGI 65
+ L L HLL + + +AE + V+ DVT +A+C G +C +Y+ G+QQ VG+
Sbjct: 3 VFRLGELRHLLVTVFLSGLAEYLIRPVMTDVTVAAVCSGLDDSCSLAVYLTGVQQVTVGM 62
Query: 66 FKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQ 125
MV++P++G LSD +G K +L + M ++ A+L + + F YA+YV++T
Sbjct: 63 GTMVMMPVIGNLSDRYGIKAMLTLPMCLSVLPPAILGYRRDTNFFYAFYVIKT------- 115
Query: 126 GSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLTF 185
+ +A V+ +KR ++F + G+ S S V ++ ARFL F V+ L
Sbjct: 116 -------LFDMAKNVHGTKRISMFGILAGVSSISGVCASLSARFLSIASTFQVAAISLFI 168
Query: 186 CPVYMQFFLVETVTPAPKKNQV-SGFCTKVVDVLHKRYKSMRNAAEILDNLIIA------ 238
VYM+ FL E + A ++ SG C +V + M + D
Sbjct: 169 GLVYMRVFLKERLQDADDDDEADSGGCRSHQEVHNGGDLKMLTEPILRDAPTKTHVFNSK 228
Query: 239 -----VQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLAS 293
Y+LKA FGFNKN F+EL ++V I SQ+ +LP L+ +GE+ +LS LL
Sbjct: 229 YSSWKDMYFLKARFGFNKNDFAELFLLVTIIGSISQLFILPTLSSTIGERKVLSTGLLME 288
Query: 294 IAYAWLYGLAWAPWVPYFSASFGIIYVLVKPS--------------GKAQTFIAGAQSIS 339
A +AW+PWVPY + V PS GK Q I+G ++ +
Sbjct: 289 FFNATCLSVAWSPWVPYAMTMLVPGAMFVMPSVCGIASRQVGSSEQGKVQGCISGVRAFA 348
Query: 340 DLLSPIAMSPLTSWFLSSNAPFDCKGFSIICASVSMV 376
+++P SPLT+ FLS NAPF GFSI+C ++S++
Sbjct: 349 QVVAPFVYSPLTALFLSENAPFYFPGFSILCIAISLM 385
>At2g16970 putative tetracycline transporter protein
Length = 414
Score = 186 bits (471), Expect = 3e-47
Identities = 129/406 (31%), Positives = 198/406 (47%), Gaps = 65/406 (16%)
Query: 10 LRPLFHLLFPLSIHWIAEEMTVSVLVDVTTSALCPG-GSTCPKVIYINGLQQTIVGIFKM 68
L L HLL + + +E + V+ DVT +A+C G TC +Y+ G++Q VG+ M
Sbjct: 6 LGELRHLLTTVFLSGFSEFLVKPVMTDVTVAAVCSGLNETCSLAVYLTGVEQVTVGLGTM 65
Query: 69 VVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSI 128
V++P++G LSD +G K LL + M +I A+LA+ + F YA+Y+ +
Sbjct: 66 VMMPVIGNLSDRYGIKTLLTLPMCLSILPPAILAYRRDTNFFYAFYITKI---------- 115
Query: 129 FCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLTFCPV 188
+ +A V KR ++F + G+ S S V AR LP IF V+ F V
Sbjct: 116 ----LFDMAKNVCGRKRISMFGVLAGVRSISGVCATFSARLLPIASIFQVAAISFFFGLV 171
Query: 189 YMQFFLVETV-----TPAPKKNQVSG-------------------FCTKVVDVLHKRYKS 224
YM+ FL E + + + SG TK+ VL+ +Y S
Sbjct: 172 YMRVFLKERLHDDDEDDCDEDDNTSGRNHHDGGDLTMLAEPILRDAPTKIHIVLNTKYSS 231
Query: 225 MRNAAEILDNLIIAVQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKV 284
+++ Y+LKA FGFNKN F+EL+++V I SQ+ +LP L +GE+
Sbjct: 232 LKD------------MYFLKARFGFNKNDFAELILLVTIIGSISQLFILPKLVSAIGERR 279
Query: 285 ILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKPS--------------GKAQT 330
+LS LL A ++W+ WVPY + + + V PS GK Q
Sbjct: 280 VLSTGLLMDSVNAACLSVSWSAWVPYATTVLVPVTMFVMPSVCGIASRQVGPGEQGKVQG 339
Query: 331 FIAGAQSISDLLSPIAMSPLTSWFLSSNAPFDCKGFSIICASVSMV 376
I+G +S S +++P SPLT+ FLS APF GFS++C + S++
Sbjct: 340 CISGVKSFSGVVAPFIYSPLTALFLSEKAPFYFPGFSLLCVTFSLM 385
>At2g16990 putative tetracycline transporter protein
Length = 418
Score = 180 bits (456), Expect = 2e-45
Identities = 119/398 (29%), Positives = 196/398 (48%), Gaps = 54/398 (13%)
Query: 13 LFHLLFPLSIHWIAEEMTVSVLVDVTTSALCPG-GSTCPKVIYINGLQQTIVGIFKMVVL 71
L H+L + + A M V V+ DVT +A+C G +C +Y+ G QQ +G+ M+++
Sbjct: 8 LRHMLATVFLSAFAGFMVVPVITDVTVAAVCSGPDDSCSLAVYLTGFQQVAIGMGTMIMM 67
Query: 72 PLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCI 131
P++G LSD +G K +L + M +I +L + + +F Y +Y+ +
Sbjct: 68 PVIGNLSDRYGIKTILTLPMCLSIVPPVILGYRRDIKFFYVFYISKI------------- 114
Query: 132 SVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLTFCPVYMQ 191
+ +A ++ S R + F + G+ + + + G ++ARFLP F VS VYM+
Sbjct: 115 -LTSMAVNIHGSTRISAFGILAGIKTIAGLFGTLVARFLPIALTFQVSAISFFVGLVYMR 173
Query: 192 FFLVETVTPAPKKNQVSGFCTK---------------------VVDVLHKRYKSMRNAAE 230
FL E + + G + V HK+Y S+++
Sbjct: 174 VFLKEKLNDDEDDDLHHGTYHQEDHDSINTTMLAEPILNDRPIKTQVFHKKYSSLKD--- 230
Query: 231 ILDNLIIAVQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAAL 290
+ +L+ Y+LKA FGF+K QF++LL+++ I SQ+ +LP +GE +LS L
Sbjct: 231 -MISLMKTRYYFLKARFGFDKKQFADLLLLITIVGSISQLFVLPRFASAIGECKLLSTGL 289
Query: 291 LASIAYAWLYGLAWAPWVPYFSASF--GIIYVL----------VKP--SGKAQTFIAGAQ 336
+ ++WAPWVPY + F G ++V+ V P GK Q I+G +
Sbjct: 290 FMEFINMAIVSISWAPWVPYLTTVFVPGALFVMPSVCGIASRQVGPGEQGKVQGCISGVR 349
Query: 337 SISDLLSPIAMSPLTSWFLSSNAPFDCKGFSIICASVS 374
S +++P SPLT+ FLS NAPF GFS++C S+S
Sbjct: 350 SFGKVVAPFVFSPLTALFLSKNAPFYFPGFSLLCISLS 387
>At5g42210 putative protein
Length = 204
Score = 78.6 bits (192), Expect = 6e-15
Identities = 53/195 (27%), Positives = 94/195 (48%), Gaps = 30/195 (15%)
Query: 132 SVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLTFCPVYMQ 191
S + AD V E +RA+ F +TG+ S + V N+ ARFL F V+ A+ +YM+
Sbjct: 5 SADFQADNVPEGRRASAFGILTGITSCAFVCANLSARFLSIGVTFQVAAAMGILSTLYMR 64
Query: 192 FFLVETVTP--------------APKKNQVSGFCTKV---VDVLHKRYKSMRNAAEILDN 234
FL +++ +P G ++ + ++H+ MR++ +
Sbjct: 65 LFLPDSIRDNSLGAPIVINEKLSSPLLEDCPGHRNRIFRAIRLVHEMASLMRSSVPLFQV 124
Query: 235 LIIAV-------------QYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVG 281
+++ YYLKA F FNK+QF++L+++VG SQ++ +P+L P +
Sbjct: 125 AMVSFFSSLAEAGLHASSMYYLKAKFHFNKDQFADLMIIVGASGSISQLLFMPVLVPALK 184
Query: 282 EKVILSAALLASIAY 296
E+ +LS L A+
Sbjct: 185 EERLLSIGLFFGCAH 199
>At1g67300 transport protein like protein
Length = 493
Score = 33.5 bits (75), Expect = 0.23
Identities = 33/113 (29%), Positives = 53/113 (46%), Gaps = 19/113 (16%)
Query: 243 LKAVFGFNKNQFSELLMMVGIGSIFSQM------VLLPILNPKVGEKVILSAALLASIAY 296
+ AVF F+ F + +G+IF + V+ +L KVG K++L + + A
Sbjct: 309 INAVFYFSSTVFKSAGVPSDLGNIFVGVSNLLGSVIAMVLMDKVGRKLLLLWSFIGMAAA 368
Query: 297 AWLYGLAWAPWVPYFSA---SFG--IIYVLVKPSGKAQTFIAGAQSISDLLSP 344
L A + ++P+FSA S G +++VL TF GA + LL P
Sbjct: 369 MALQVGATSSYLPHFSALCLSVGGTLVFVL--------TFALGAGPVPGLLLP 413
>At3g43790 transporter-like protein
Length = 478
Score = 33.1 bits (74), Expect = 0.30
Identities = 61/284 (21%), Positives = 110/284 (38%), Gaps = 72/284 (25%)
Query: 75 GQLSDEHGRKPLLLI-TMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISV 133
G+L+D +GRKP++LI T S IF+ + ++L F+ ++ +
Sbjct: 97 GKLADRYGRKPIILIGTFSVIIFNTLFGLSTSFWLAISVRFLLGCFNCLLG------VIR 150
Query: 134 AYVADVVNESKRAAVFSWIT-------------------------GLFSASHVVGNVLAR 168
AY ++VV+E A S ++ +FS S V G
Sbjct: 151 AYASEVVSEEYNALSLSVVSTSRGIGLILGPAIGGYLAQPAEKYPNIFSQSSVFGR-FPY 209
Query: 169 FLPEEYIFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVD------------ 216
FLP I V + A+L C ++L ET+ + Q T++ D
Sbjct: 210 FLPSLVISVYATAVLIAC-----WWLPETLHTRCRIAQGRLNPTELNDDESRGGGLDEQK 264
Query: 217 VLHKRYKSMRNAAEILDNLIIAVQYYLKAV------------------FGFNKNQFSELL 258
+++K S+ ++ +I+ + L+ + F+ E+L
Sbjct: 265 IINK--PSLLRNRPLMAIIIVYCVFSLQEIAYNEIFSLWAVSDRSYGGLSFSSQDVGEVL 322
Query: 259 MMVGIGSIFSQMVLLPILNPKVGEKVI--LSAALLASIAYAWLY 300
+ G+G + Q+++ P L VG + LSA LL + + Y
Sbjct: 323 AISGLGLLVFQLLVYPPLEKSVGLLAVIRLSAVLLIPLLSCYPY 366
>At1g20060 unknown protein
Length = 951
Score = 31.6 bits (70), Expect = 0.86
Identities = 16/40 (40%), Positives = 25/40 (62%)
Query: 370 CASVSMVAETENEKLKENLQDEKNKNIELEKQMIKLKAQC 409
C VAE E LK++L+ E+ K++ LE ++I LK+ C
Sbjct: 538 CNEKLKVAEGEIFTLKDSLRREQLKSLGLETELISLKSSC 577
>At5g13740 transporter-like protein
Length = 486
Score = 31.2 bits (69), Expect = 1.1
Identities = 21/99 (21%), Positives = 47/99 (47%), Gaps = 7/99 (7%)
Query: 75 GQLSDEHGRKPLLLI-TMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISV 133
G ++D +GRKP++L+ T+S IF+ + ++L +F+ ++
Sbjct: 99 GIVADRYGRKPIILLGTISIAIFNALFGLSSNFWMAIGTRFLLGSFNCLLG------TMK 152
Query: 134 AYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPE 172
AY +++ + +A S ++ + ++G L FL +
Sbjct: 153 AYASEIFRDEYQATAMSAVSTAWGIGLIIGPALGGFLAQ 191
>At1g19450 integral membrane protein, putative
Length = 488
Score = 30.4 bits (67), Expect = 1.9
Identities = 23/106 (21%), Positives = 48/106 (44%), Gaps = 4/106 (3%)
Query: 75 GQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVA 134
GQ+++ GRK L+I I + +++ + F+Y +L F G I
Sbjct: 107 GQIAEYVGRKGSLMIAAIPNIIGWLSISFAKDTSFLYMGRLLEGFGV----GIISYTVPV 162
Query: 135 YVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSI 180
Y+A++ ++ R A+ S + ++ +L F+P + V+ +
Sbjct: 163 YIAEIAPQTMRGALGSVNQLSVTIGIMLAYLLGLFVPWRILAVLGV 208
>At1g75220 integral membrane protein, putative
Length = 487
Score = 30.0 bits (66), Expect = 2.5
Identities = 30/140 (21%), Positives = 58/140 (41%), Gaps = 5/140 (3%)
Query: 75 GQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVA 134
GQ+++ GRK L+I I + +++ + F+Y +L F G I
Sbjct: 106 GQIAEYIGRKGSLMIAAIPNIIGWLCISFAKDTSFLYMGRLLEGFGV----GIISYTVPV 161
Query: 135 YVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLTFCPVYMQFFL 194
Y+A++ ++ R + S + ++ +L F+P + V+ I T + FF+
Sbjct: 162 YIAEIAPQNMRGGLGSVNQLSVTIGIMLAYLLGLFVPWRILAVLGILPCTLL-IPGLFFI 220
Query: 195 VETVTPAPKKNQVSGFCTKV 214
E+ K F T +
Sbjct: 221 PESPRWLAKMGMTDEFETSL 240
>At2g41700 ATP-binding cassette transporter ABCA1
Length = 1884
Score = 29.6 bits (65), Expect = 3.3
Identities = 23/99 (23%), Positives = 49/99 (49%), Gaps = 11/99 (11%)
Query: 196 ETVTPAPKKNQVSGFCTKVVDVLHKRYKSMRNAAEILDNLIIAVQYYLKAVFGFNKNQFS 255
E+++ ++ ++ G C +V + LHK Y S R +++L + + +NQ
Sbjct: 497 ESISLEMRQQELDGRCIQVRN-LHKVYASRRGNCCAVNSLQLTLY----------ENQIL 545
Query: 256 ELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLASI 294
LL G G + +L+ +L P G+ +IL +++ ++
Sbjct: 546 SLLGHNGAGKSTTISMLVGLLPPTSGDALILGNSIITNM 584
>At2g07390 hypothetical protein
Length = 137
Score = 29.3 bits (64), Expect = 4.3
Identities = 15/60 (25%), Positives = 29/60 (48%), Gaps = 2/60 (3%)
Query: 346 AMSPLTSWFLSSNAPFDCKGFSIICASVSMVAETENEKLKENLQDEKNKNIELEKQMIKL 405
++ P+T+ F S+ P++ +C M+ E E K+ + NK +E + + I L
Sbjct: 61 SLDPVTTRFFFSSCPYEISDTVALCDEFDMIKEETTEMKKD--VEATNKRVESQAEKIFL 118
>At1g14510 unknown protein
Length = 252
Score = 29.3 bits (64), Expect = 4.3
Identities = 12/29 (41%), Positives = 18/29 (61%)
Query: 233 DNLIIAVQYYLKAVFGFNKNQFSELLMMV 261
D+ +I+V +Y A FGF KN+ L M+
Sbjct: 99 DSWLISVAFYFGARFGFGKNERKRLFQMI 127
>At3g58050 putative protein
Length = 1209
Score = 28.9 bits (63), Expect = 5.6
Identities = 17/55 (30%), Positives = 27/55 (48%)
Query: 350 LTSWFLSSNAPFDCKGFSIICASVSMVAETENEKLKENLQDEKNKNIELEKQMIK 404
LT L + CK + V ++ E E EK +E + EK ++ E EK++ K
Sbjct: 506 LTLKLLEQHLHVACKEIITLEKQVKLLEEEEKEKREEEERKEKKRSKEREKKLRK 560
>At2g48020 sugar transporter like protein
Length = 463
Score = 28.9 bits (63), Expect = 5.6
Identities = 20/72 (27%), Positives = 32/72 (43%), Gaps = 7/72 (9%)
Query: 58 LQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIF-------SFAVLAWDQSEEFV 110
L I + ++V+ L + D GRKPLLL++ + + SF + D + E V
Sbjct: 299 LGMIIYAVLQVVITALNAPIVDRAGRKPLLLVSATGLVIGCLIAAVSFYLKVHDMAHEAV 358
Query: 111 YAYYVLRTFSYI 122
V+ YI
Sbjct: 359 PVLAVVGIMVYI 370
>At3g30500 hypothetical protein
Length = 257
Score = 28.5 bits (62), Expect = 7.3
Identities = 8/25 (32%), Positives = 20/25 (80%)
Query: 218 LHKRYKSMRNAAEILDNLIIAVQYY 242
LH++++S RN + ++DN+ +A+ ++
Sbjct: 193 LHRKFRSFRNDSIVIDNIFVALTWW 217
>At3g18830 sugar transporter like protein
Length = 539
Score = 28.5 bits (62), Expect = 7.3
Identities = 20/58 (34%), Positives = 28/58 (47%), Gaps = 5/58 (8%)
Query: 58 LQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAW-----DQSEEFV 110
L VG+ K + + L D GR+PLLL ++ + S A L DQSE+ V
Sbjct: 334 LATVAVGVVKTSFILVATFLLDRIGRRPLLLTSVGGMVLSLAALGTSLTIIDQSEKKV 391
>At2g02470 putative PHD-type zinc finger protein
Length = 256
Score = 28.5 bits (62), Expect = 7.3
Identities = 11/29 (37%), Positives = 18/29 (61%)
Query: 233 DNLIIAVQYYLKAVFGFNKNQFSELLMMV 261
D+ +++V +Y A FGF KN+ L M+
Sbjct: 99 DSWLLSVAFYFGARFGFGKNERKRLFQMI 127
>At1g11000 membrane protein Mlo4
Length = 573
Score = 28.5 bits (62), Expect = 7.3
Identities = 14/59 (23%), Positives = 26/59 (43%)
Query: 78 SDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVAYV 136
SD H +K ++ ST +F A W + ++ LR F I + F + + ++
Sbjct: 191 SDIHAKKTKVMKRQSTFVFHHASHPWSNNRFLIWMLCFLRQFRGSIRKSDYFALRLGFL 249
>At1g01060 DNA-binding protein, putative
Length = 645
Score = 28.5 bits (62), Expect = 7.3
Identities = 22/101 (21%), Positives = 42/101 (40%), Gaps = 10/101 (9%)
Query: 319 YVLVKPSGKAQTFIAGAQSISDLLSPIAMSPLTSWFLS-------SNAPFDCKGFSIICA 371
Y V SG + T ++ + ++ ++ T+W+ S + AP C FS +
Sbjct: 324 YASVGNSGDSSTPMSSSPPSITAIAAATVAAATAWWASHGLLPVCAPAPITCVPFSTVAV 383
Query: 372 SVSMVAETENEKLKENLQDEKNKNIELEKQMIKLKAQCTCS 412
+ E + EN Q + +N L+ Q + K+ + S
Sbjct: 384 PTPAMTEMDT---VENTQPFEKQNTALQDQNLASKSPASSS 421
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,758,732
Number of Sequences: 26719
Number of extensions: 341877
Number of successful extensions: 1398
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 1371
Number of HSP's gapped (non-prelim): 28
length of query: 423
length of database: 11,318,596
effective HSP length: 102
effective length of query: 321
effective length of database: 8,593,258
effective search space: 2758435818
effective search space used: 2758435818
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0095c.5


