

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0094b.11
(1170 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
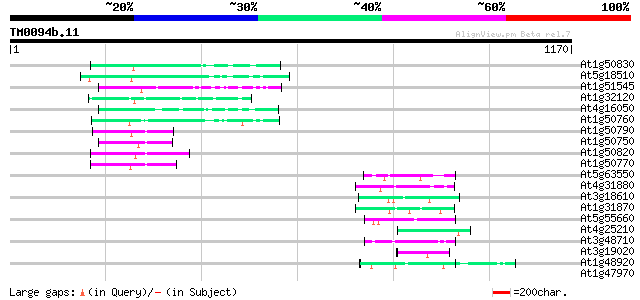
Score E
Sequences producing significant alignments: (bits) Value
At1g50830 hypothetical protein 102 2e-21
At5g18510 putative protein 92 2e-18
At1g51545 unknown protein 84 4e-16
At1g32120 hypothetical protein 76 1e-13
At4g16050 hypothetical protein 68 3e-11
At1g50760 hypothetical protein 68 3e-11
At1g50790 hypothetical protein 62 2e-09
At1g50750 hypothetical protein 62 2e-09
At1g50820 hypothetical protein 60 6e-09
At1g50770 hypothetical protein 60 6e-09
At5g63550 unknown protein 56 1e-07
At4g31880 unknown protein 52 2e-06
At3g18610 unknown protein 50 1e-05
At1g31870 unknown protein 45 2e-04
At5g55660 putative protein 45 2e-04
At4g25210 unknown protein 45 2e-04
At3g48710 unknown protein 45 3e-04
At3g19020 hypothetical protein 45 3e-04
At1g48920 unknown protein 45 3e-04
At1g47970 Unknown protein (T2J15.12) 43 0.001
>At1g50830 hypothetical protein
Length = 768
Score = 102 bits (253), Expect = 2e-21
Identities = 109/418 (26%), Positives = 163/418 (38%), Gaps = 57/418 (13%)
Query: 168 DKAYLKWLDRVEEDKRQHWKDVGILDLIQLSRSPISYNPAMLLSALYFWERSTNCLHVPF 227
D ++ WL ++E Q W+ GI + I++S I+ NP+++LS W T P+
Sbjct: 79 DAPFVSWLAKMEALHAQTWRKAGIFEAIKVSTYSITKNPSLILSVSEKWCPETKSFVFPW 138
Query: 228 GMITPTLLDVAAITGLWPIGDDYHSAPAT---------VKRISIPTDNISFSRFIKDHYV 278
G T TL DV + G +G + T ++ + I N S R +
Sbjct: 139 GEATITLEDVMVLLGFSVLGSPVFAPLETSETRDSVKKLENVRIQHMNSSTDRRVSQKSW 198
Query: 279 ES---GEVSDAEHVAFLLYWLSAYVFCTKSLR-IPAKLLPLANLLHEGRQLAMARLVLGN 334
S G D EHVAFL+ WLS +VF KS R I + P+A L G ++A+A +L
Sbjct: 199 VSTFLGRGGDMEHVAFLVLWLSLFVFPVKSRRNISNHVFPIAVRLARGERIALAPAILAI 258
Query: 335 LYQMLNEAVEDIRNTKTVSLNAAGPLWLFQLWLNAVFESLPPAQDNPPTVSNTRIDAHRL 394
LY+ L+ E R + L Q+W F ++ P + P H L
Sbjct: 259 LYRDLDRIHEVSREDCVDKFHLESLFKLVQVWTWERFRNIRPKASDIPKGEPRIAQWHGL 318
Query: 395 ETLTPAYDASSFEADFRKYFTMFLELKHYRSSFSPYSKAVRG----PFWLRNSYPNAYDS 450
+ DA DF + PY+KA+ F+L + D
Sbjct: 319 HRRSK--DAWFCFDDF---------------EWRPYTKALNNWNPFRFYLEEAIWVTVDE 361
Query: 451 ALPKEHHITLWRTLLSPRVLTVGFASSDYTLCGYNPQLVSRQFGLSQVLPNTLFDKSLVL 510
++ E R +TV D + Y P V+RQFGL Q LP
Sbjct: 362 SIDDEF-------ASFARCVTVSQIVGDGFVEDYFPNRVARQFGLDQDLPGL-------- 406
Query: 511 YPGAIKRAPAFDTTVQFYNKKLLNLSPFTYAPSYY----ATNAFKAWWSEYWAQISKP 564
+R YNK L+ L+ Y PS T ++ WW + ++ P
Sbjct: 407 --ATCQRNSTEKEAWNDYNKSLIGLN--LYMPSRLDQGSVTARYRVWWLKSVSKFLSP 460
>At5g18510 putative protein
Length = 702
Score = 92.0 bits (227), Expect = 2e-18
Identities = 115/462 (24%), Positives = 178/462 (37%), Gaps = 58/462 (12%)
Query: 147 MPFLSDPKRAFRSAPPNPS-----ANDKAYLKWLDRVEEDKRQHWKDVGILDLIQLSRSP 201
+P L D +R S+ N S A+ +L WL +++ WK GI + I+ S
Sbjct: 31 LPRLDDQQRLSDSSLHNTSSGFWAADHHFFLSWLGKMQALYEPIWKKAGIFEAIKASTYK 90
Query: 202 ISYNPAMLLSALYFWERSTNCLHVPFGMITPTLLDVAAITGLWPIGDDYHS-------AP 254
I + + +LS W T P+G T TL DV + G +G S
Sbjct: 91 IIKDTSSILSIAEKWCSETKSFIFPWGEATITLEDVMVLLGFSVLGSPVFSPLECSEMRD 150
Query: 255 ATVKRISIPTDNISFSRFIKDHYVES---GEVSDAEHVAFLLYWLSAYVFCTKSLR-IPA 310
+ K + D++ ++ + S G EH AFL+ WLS +VF K R I
Sbjct: 151 SAEKLEKVRRDSLGKAKRVSQRSWTSSFMGRGGQMEHEAFLVLWLSLFVFPGKFCRSIST 210
Query: 311 KLLPLANLLHEGRQLAMARLVLGNLYQMLNEAVEDIRNTKTVSLNAAGPLWLFQLWLNAV 370
++P+A L G ++A+A VL LY+ L+ + R +N L Q+W
Sbjct: 211 NVIPIAVRLARGERIALAPAVLAFLYKDLDRICDFSRGKCAGKVNLKSLFKLVQVWTWER 270
Query: 371 FESLPPAQDNPPTVSNTRIDAHRLETLTPAYDASSFEADFRKYFTMFLELKHYRSSFSPY 430
F ++ P P L+ ++ S ++R Y +++P
Sbjct: 271 FSNIRPKAKEIPKGEPRIAQWDGLQQISKNVKLSFDVFEWRPYTKPL-------KNWNPL 323
Query: 431 SKAVRGPFWLRNSYPNAYDSALPKEHHITLWRTLLSPRVLTVGFASSDYTLCGYNPQLVS 490
V WL + ++ D A R + V + + + + Y P V+
Sbjct: 324 RFYVDEAMWL--TVDDSVDDAFAS-----------FARCVKVSYLAGNGFVEDYFPYRVA 370
Query: 491 RQFGLSQVLPNTLFDKSLVLYPGAIKRAPAFDTTVQFYNKKLLNLSPFTYAPSY----YA 546
RQFGLSQ LP LV I A+D Y+ L L+ Y PS Y
Sbjct: 371 RQFGLSQDLP------GLVTRRRKITEKDAWDD----YSNSLEGLN--LYLPSQLDRGYV 418
Query: 547 TNAFKAWW----SEYWA--QISKPLVDCLQCMTDAFLLQKQD 582
T ++ WW SEY+ +I K + L K D
Sbjct: 419 TARYQDWWFKSASEYFGSEEIQKESTEAFDARNTFDHLDKDD 460
>At1g51545 unknown protein
Length = 629
Score = 84.3 bits (207), Expect = 4e-16
Identities = 106/397 (26%), Positives = 162/397 (40%), Gaps = 52/397 (13%)
Query: 186 WKDVGILDLIQLSRSPISYNPAMLLSALYFWERSTNCLHVPFGMITPTLLDVAAITGLWP 245
W+ GI + I+ S I N ++LL+ + W T P+G T TL DV + G
Sbjct: 9 WRKAGIFEAIKASMYKIRKNQSLLLALVEKWCPETKSFLFPWGEATITLEDVLVLLGFSV 68
Query: 246 IGDDYHSAPATVKRISIPTDNISFSRFI---------KDHYVES--GEVSDAEHVAFLLY 294
G AP + + + +R ++ +V S G EH AFL +
Sbjct: 69 QGSPVF-APLESSEMRDSVEKLEKARLENRGQDGLVRQNLWVSSFLGRGDQMEHEAFLAF 127
Query: 295 WLSAYVF---CTKSLRIPAKLLPLANLLHEGRQLAMARLVLGNLYQMLNEAVEDIRNTKT 351
WLS +VF C +S I K+LP+A L G ++A A VL LY+ L + R T
Sbjct: 128 WLSQFVFPDMCRRS--ISTKVLPMAVRLARGERIAFAPAVLARLYRDLGQIQASAREKST 185
Query: 352 VSLNAAGPLWLFQLWLNAVFESLPPAQDNPPTVSNTRIDAHRLETLTPAYDASSFEADFR 411
++ L QLW F+S P P RI +T + + + D+R
Sbjct: 186 PNVTLKSLFKLVQLWAWERFKSTSPKARVIPK-GEPRISRWHSQT-SKNVRLNLVDFDWR 243
Query: 412 KYFTMFLELKHYRSSFSPYSKAVRGPFWLRNSYPNAYDSALPKEHHITLWRTLLSPRVLT 471
Y T L++ ++P W+ D L + ++ R + +++
Sbjct: 244 PY-TKPLQI------WNPPRFYPEEAMWM------TVDDNL-DDGFVSFARCMRVSQLVG 289
Query: 472 VGFASSDYTLCGYNPQLVSRQFGLSQVLPNTLFDKSLVLYPGAIKRAPAFDTTVQFYNKK 531
VG Y P V+ QFGL+Q LP + D S + A+D YNK
Sbjct: 290 VGIVED------YYPNRVAMQFGLAQDLPGLVTDHS------SFTEKEAWDG----YNKS 333
Query: 532 LLNLSPFTYAPSYYATNAFKAWWSEYWAQ-ISKPLVD 567
L L Y PS AT + + ++W + ISK +D
Sbjct: 334 LDGL--MLYIPSRVATTSVTERYRDWWLKSISKFFLD 368
Score = 32.0 bits (71), Expect = 2.1
Identities = 29/131 (22%), Positives = 48/131 (36%), Gaps = 8/131 (6%)
Query: 723 GQGQASRDEDDDDDDEDDDVSLADKLKSRKRKPKPVASASQKRAKGAGVSTTSGASKLAS 782
G+ +A D+ D +DDDV++A +KSR+ + +KG S S +
Sbjct: 466 GRKRAREDDVSSMDHKDDDVTIAQMIKSRRN--------DECGSKGKKSSMVQRPSNENN 517
Query: 783 DAQPEVDDKEGGGDAAIQADVHEHSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKK 842
+ P VD G D ++ + + + S E S A+K
Sbjct: 518 SSDPLVDSNGGIIDISVSPSETRQTCDDELHVDRSKKEELVHEDEEKQRSNENLCSEAEK 577
Query: 843 TSTSEGRRKVR 853
+ R K R
Sbjct: 578 EEDIDERLKQR 588
>At1g32120 hypothetical protein
Length = 1206
Score = 75.9 bits (185), Expect = 1e-13
Identities = 91/362 (25%), Positives = 145/362 (39%), Gaps = 45/362 (12%)
Query: 164 PSANDKAYLKWLDRVEEDKRQHWKDVGILDLIQLSRSPISYNPAMLLSALYFWERSTNCL 223
PS N +++W++ + + WK G+ D I SR I + ++++ + W TN
Sbjct: 80 PSLN---WIEWVNVMAKSHATVWKKSGVYDAILASRYQIKRHDDLIVALVEKWCIETNTF 136
Query: 224 HVPFGMITPTLLDVAAITGLWPIGDDYHSAPATVKR----------------ISIPTDNI 267
P+G T TL D+ + GL G++ A A VKR I + +
Sbjct: 137 VFPWGEATLTLEDMIVLGGLSVTGNN---ALAPVKRDGMKEVEEKMKEAKRYIEVSLEKK 193
Query: 268 SFSRFIKDHYVESGEVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAM 327
+ SG ++ EH AF++ WLS +VF + KL P A L +G +LA+
Sbjct: 194 CCVSMWMKEMMNSG--NEIEHEAFMVSWLSRFVFTNSGDVLREKLFPAAVQLAKGVRLAL 251
Query: 328 ARLVLGNLY---QMLNEAVEDIRNTKTVSLNAAGPLWLFQLWLNAVFESL-PPAQDNPPT 383
A VL +Y +L E + +TV + + P Q+W F +L PP Q +
Sbjct: 252 APAVLARIYGDLGVLKEFLTGYSEKETVVVKS--PFQFVQVWALERFMALQPPGQPSQLK 309
Query: 384 VSNTRIDAHRLETLTPAYDASSFEADFRKYFTMFLELKHYRSSFSPYSKAVRGPFWLRNS 443
RI R D + + R E YR PY+K V F
Sbjct: 310 TGEPRI--ARWHHYGGGQDVYGYPENIRAVLDSAKESFDYR----PYTKPVNN-FRFPKF 362
Query: 444 Y--PNAYDSALPKEHHITLWRTLLSPRVLTVGFASSDYTLCGYNPQLVSRQFGLSQVLPN 501
Y + + P E+ + R L +++ + Y P V+ QFG Q +P
Sbjct: 363 YFEDDCWVRVRPDENIVAFGRCLRFAKLVGLDCIEP------YYPHRVALQFGYDQDVPG 416
Query: 502 TL 503
+
Sbjct: 417 VV 418
Score = 34.3 bits (77), Expect = 0.43
Identities = 26/121 (21%), Positives = 44/121 (35%), Gaps = 10/121 (8%)
Query: 803 VHEHSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPR 862
++ NPPS S P + S + K + + + S G S K P
Sbjct: 758 INSRGQKNPPSPRVSKEPYMSSSLSVSSSSTRKKPPRSHEAANSRGYNHTPSLRASKEPY 817
Query: 863 RSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSPQAKGGSS 922
+S S + SS +++ P R E + N P P+ + + +P G S
Sbjct: 818 KSSSLSGSSSTRK----------KPPRSHEASSSRGYNHPPSPRVSKELNKTPSISGSPS 867
Query: 923 S 923
+
Sbjct: 868 A 868
Score = 34.3 bits (77), Expect = 0.43
Identities = 31/140 (22%), Positives = 51/140 (36%), Gaps = 2/140 (1%)
Query: 786 PEVDDKEGGGDAAIQADVHEHSTSNPPSRENSPAPNPAKSKAEGVE--KPPKAASSAKKT 843
P V D ++ H S ++PP +EN + + S E + P +S K
Sbjct: 689 PRVADLAKQSSSSPWVPRHTSSVAHPPKQENYIKHHNSTSSRVSKESNRTPSVSSYPLKR 748
Query: 844 STSEGRRKVRSKSGHKSPRRSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMP 903
TS+ + + G K+P R S S + + P R E + N P
Sbjct: 749 KTSQRSPEAINSRGQKNPPSPRVSKEPYMSSSLSVSSSSTRKKPPRSHEAANSRGYNHTP 808
Query: 904 PPKSTATVQPSPQAKGGSSS 923
+++ S G SS+
Sbjct: 809 SLRASKEPYKSSSLSGSSST 828
>At4g16050 hypothetical protein
Length = 900
Score = 68.2 bits (165), Expect = 3e-11
Identities = 91/385 (23%), Positives = 148/385 (37%), Gaps = 57/385 (14%)
Query: 186 WKDVGILDLIQLSRSPISYNPAMLLSALYFWERSTNCLHVPFGMITPTLLDVAAITGLWP 245
W GI + I+ S I NP+++LS W TN P+G T TL DV + G
Sbjct: 341 WLKSGIFEAIKASTYRIHKNPSLILSLAQNWCPETNTFVFPWGEATITLEDVNVLLGFSI 400
Query: 246 IGDDYHSA--PATVKRISIPTDNISFSRFIKDHYVESGEVSDAEHVAFLLYWLSAYVFCT 303
G ++ + +K ++ ++ S + EH AFL+ WLS +VF
Sbjct: 401 SGSSVFASLQSSEMKEAVEKLQKRCQGSMKQESWISSFVDDEMEHEAFLVLWLSKFVFPD 460
Query: 304 KSLRIPAKLLPLANLLHEGRQLAMARLVLGNLYQMLNE--AVEDIRNTKTVSLNAAGPLW 361
K G ++A A VL NLY L + I+N SL
Sbjct: 461 K-------------FCTRGERIAFAPAVLANLYNDLGHICVLASIQNVLASSL-----FK 502
Query: 362 LFQLWLNAVFESLPPAQD----NPPTVSNTRIDAHRLETLTPAYDASSFEADFRKYFTMF 417
L Q+W+ F+S+ P P ++ R + + +F D+R Y
Sbjct: 503 LVQVWIWERFKSIRPEAKVIPRGQPRIAQWSGLKQRFKNVGLIIFHGNF--DWRPY---- 556
Query: 418 LELKHYRSSFSPYSKAVRGPFWLR--NSYPNAYDSALPKEHHITLWRTLLSPRVLTVGFA 475
+++P V W+R S YD + ++ R + +++ +G
Sbjct: 557 ---SEPLENWNPPRFYVEEAKWVRIDESLDGDYDD---DDEFVSFARCVRVSKLVGIGVV 610
Query: 476 SSDYTLCGYNPQLVSRQFGLSQVLPNTLFDKSLVLYPGAIKRAPAFDTTVQFYNKKLLNL 535
+ Y P V+ QFGL+Q +P + +R + YNK L+ L
Sbjct: 611 EN------YYPNRVAMQFGLAQDVP---------VLGTNHRRNFTEEEAWDDYNKPLVGL 655
Query: 536 SPFTYAPSYYATNAFKAWWSEYWAQ 560
Y PS AT + + ++WA+
Sbjct: 656 K--LYFPSRVATASVTTRYRDWWAK 678
>At1g50760 hypothetical protein
Length = 649
Score = 68.2 bits (165), Expect = 3e-11
Identities = 93/426 (21%), Positives = 166/426 (38%), Gaps = 77/426 (18%)
Query: 171 YLKWLDRVEEDKRQHWKDVGILDLIQLSRSPISYNPAMLLSALYFWERSTNCLHVPFGMI 230
+ +W ++ W+ GI + + S I N ++L W T +G
Sbjct: 53 FKRWARKMSALHEPIWRKAGIFEAVNASTYKIHKNTDLVLGVAEKWSPDTKTFVFSWGEA 112
Query: 231 TPTLLDVAAITGLWPIG---------------DDYHSAPATVKR---ISIPTDNISFSRF 272
T TL DV ++G +G + +A T+KR +++ T RF
Sbjct: 113 TITLEDVMVLSGFSVLGSPAFATLDSSGKKIMERLKNAWQTIKREGKVNLLTQVAWMDRF 172
Query: 273 IKDHYVESGEVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARLVL 332
+ GE+ EHVAFL+ WL +V ++ + + P+A L G ++A+A VL
Sbjct: 173 MNC----GGEL---EHVAFLVLWLGYFVLPSRFHHLCETIFPIAVNLSSGTRIALAPAVL 225
Query: 333 GNLYQMLNEAVEDI-RNTKTVSLNAAGPLWLFQLWLNAVFESLPPAQDN----PPTVSNT 387
+LY L+ I ++ V + + L Q+W F L P ++ P V+
Sbjct: 226 AHLYADLSLLKTHISKSPPRVEIRLSALFMLVQVWTWERFRELQPKPNSLLAGKPRVAIW 285
Query: 388 RIDAHRLETLTPAYDASSFEADFRKYFTMFLELKHYRSSFSPYSKAVRGPFWLRNSYPNA 447
+ + D S ++ + PY+++V W +P
Sbjct: 286 NKLKQQTSNVRQILDNSKIDS----------------FEWRPYTRSVTN--W---KFPQF 324
Query: 448 YDSALPKEHHITLWRTLLSPRVLTVGFASSDYTLCG------YNPQLVSRQFGLSQVLPN 501
Y P++ L +++ D L G Y P V+ QFG+ Q +P+
Sbjct: 325 Y----PEKAMCVHVSPSLDEELISFARCIKDSELVGIEKVEHYFPNRVASQFGMLQDVPS 380
Query: 502 TLFDKSLVLYPGAIKRAPAFDTTVQFYNKKLLNLSPFTYAPSYYA----TNAFKAWWSEY 557
D++ + R A++ YNK + +L+ + PS A T+ F WW +
Sbjct: 381 PHVDRN------NLSREAAWND----YNKPIDDLA--LHIPSRSAIPRVTSTFCEWWRKT 428
Query: 558 WAQISK 563
+ ++ K
Sbjct: 429 YPELIK 434
>At1g50790 hypothetical protein
Length = 812
Score = 62.0 bits (149), Expect = 2e-09
Identities = 49/179 (27%), Positives = 80/179 (44%), Gaps = 14/179 (7%)
Query: 174 WLDRVEEDKRQHWKDVGILDLIQLSRSPISYNPAMLLSALYFWERSTNCLHVPFGMITPT 233
W ++ W+ GI + I S I N +++ W TN +G T T
Sbjct: 67 WARKMSALHEPIWRKAGIFEAILASTYKIFKNTDLVMGIAEKWCPDTNTFVFSWGEATIT 126
Query: 234 LLDVAAITGLWPIGDDYHS----------APATVKRISIPTDNISFSRFIK--DHYVESG 281
L DV + G +G + A + + I D +F I + +++SG
Sbjct: 127 LEDVMVLLGFSVLGSPVFATLDSSGKEIMAKLGKEWLKIKKDKGTFVTQIAWIERFMDSG 186
Query: 282 EVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARLVLGNLYQMLN 340
+ + EH+AFL+ WLS +VF T+ I + P+A L G ++A+A VL +LY L+
Sbjct: 187 D--ELEHLAFLVLWLSYFVFPTRYYHIYEAIWPIAIHLSNGTKMALAPAVLAHLYADLS 243
>At1g50750 hypothetical protein
Length = 816
Score = 62.0 bits (149), Expect = 2e-09
Identities = 43/166 (25%), Positives = 79/166 (46%), Gaps = 14/166 (8%)
Query: 186 WKDVGILDLIQLSRSPISYNPAMLLSALYFWERSTNCLHVPFGMITPTLLDVAAITGLWP 245
W++ GI + + S I NP ++L W T P+G TL DV ++G
Sbjct: 54 WREAGIFEAVMASIYRIPKNPDLILGIAEKWCPYTKTFVFPWGETAVTLEDVMVLSGFSV 113
Query: 246 IGDD-YHSAPATVKRISIPTDN---------ISFSRFIK--DHYVESGEVSDAEHVAFLL 293
+G + + ++ K + D ++F + + +++SG+ + EHVAFL+
Sbjct: 114 LGSPVFATLDSSGKEVKAKLDKEWKKIKKAKVNFVTQVAWMERFMDSGD--ELEHVAFLV 171
Query: 294 YWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARLVLGNLYQML 339
WL+ +VF ++ + + P+ L G ++A+A VL +LY L
Sbjct: 172 LWLNYFVFPSRLYHLYKAVFPIVVHLSTGTRIALALAVLAHLYAEL 217
Score = 30.4 bits (67), Expect = 6.2
Identities = 25/109 (22%), Positives = 46/109 (41%), Gaps = 21/109 (19%)
Query: 725 GQASRDEDDDDDDEDDDVSLADKLKSRKRK-----------PKPVASASQKRAKGAGVST 773
GQ D++DEDD++S+A L+ K+K +P+ S+ A + T
Sbjct: 493 GQCHTHVHSDNNDEDDNLSIAQFLRLNKKKYSDVRNSGRDASEPLGKISRVEADNNDLGT 552
Query: 774 TSGASKLASDAQP--EVDDKEGGGDAA--------IQADVHEHSTSNPP 812
+ + + P E+ + G D A ++ E+++S PP
Sbjct: 553 FQKLASIRDEIVPPSEIAHRNGETDEAGSRAGTNMDESPFDENNSSEPP 601
>At1g50820 hypothetical protein
Length = 528
Score = 60.5 bits (145), Expect = 6e-09
Identities = 56/223 (25%), Positives = 93/223 (41%), Gaps = 18/223 (8%)
Query: 168 DKAYLKWLDRVEEDKRQHWKDVGILDLIQLSRSPISYNPAMLLSALYFWERSTNCLHVPF 227
+K + W ++ W+ GI + + S I + ++L W T P+
Sbjct: 57 NKKFKSWARKMASLHEPIWRKAGIFEAVIASTYKIPKDTDLVLGLAEKWCPDTKTFIFPW 116
Query: 228 GMITPTLLDVAAITGLWPIG-DDYHSAPATVKRI---------SIPTDNISFSRFIK--D 275
G T TL DV + G +G + + ++ K I I D + + +
Sbjct: 117 GEATITLEDVMVLLGFSVLGLPVFATVDSSGKEIMAKLEKEWKKIKNDKVCLVTKLAWME 176
Query: 276 HYVESGEVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARLVLGNL 335
++ SG+ + EHV FL+ WLS + F + I +LP+A L G ++A+A VL +L
Sbjct: 177 RFMNSGD--ELEHVGFLVLWLSYFAFPSHLFHISEAILPVAVHLSSGTKMALAPAVLAHL 234
Query: 336 YQMLNEAVEDIR----NTKTVSLNAAGPLWLFQLWLNAVFESL 374
Y L+ IR + V L+ L Q+W F L
Sbjct: 235 YADLSLLQGHIRVFSESLIKVQLDLNALFKLVQVWAWERFREL 277
>At1g50770 hypothetical protein
Length = 632
Score = 60.5 bits (145), Expect = 6e-09
Identities = 46/193 (23%), Positives = 79/193 (40%), Gaps = 15/193 (7%)
Query: 168 DKAYLKWLDRVEEDKRQHWKDVGILDLIQLSRSPISYNPAMLLSALYFWERSTNCLHVPF 227
+K + W ++ W+ GI + + S I+ N ++L W T P+
Sbjct: 60 NKNFKSWARKMAALHEPIWRKAGIFEAVTASTYKINPNTELVLGIAEKWCPDTKTFVFPW 119
Query: 228 GMITPTLLDVAAITGLWPIGDDY------------HSAPATVKRISIPTDNISFSRFIKD 275
G T TL DV + G +G K++ N + R K+
Sbjct: 120 GETTITLEDVMLLLGFSVLGSPVFVTLDSSGEIIREKLEKEWKKVKKDKGNAT-QRTWKE 178
Query: 276 HYVESGEVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARLVLGNL 335
+++SG+ + EHVAFL+ WLS +VF ++ I +LP+A L RL + +
Sbjct: 179 RFMDSGD--ELEHVAFLVLWLSYFVFPSRYYHIYGAILPIAVHLSSDNDEVDGRLTIAQM 236
Query: 336 YQMLNEAVEDIRN 348
+ D++N
Sbjct: 237 VGPSKKKYSDVKN 249
>At5g63550 unknown protein
Length = 530
Score = 56.2 bits (134), Expect = 1e-07
Identities = 64/210 (30%), Positives = 87/210 (40%), Gaps = 48/210 (22%)
Query: 738 EDDDVSLADKLKSRKRKPKPVASASQKRAKGAGVSTTSGASK-------LASDAQPEVDD 790
E DV +AD+ K++KRK P KR K S T K L SD + D
Sbjct: 241 ETRDVIIADQEKAKKRKSTP------KRGKSGESSDTPAKRKRQTKKRDLPSDTEEGKD- 293
Query: 791 KEGGGDAAIQADVHEHSTSNPPSRENSPAPNPAKSKAE-GVEKPPKAASSAKKT----ST 845
EG D+ D HE + P + + K E VEKP K SS+KKT S
Sbjct: 294 -EGDADSEGTNDPHEEDDAAPEEESDHEKTDTDDEKDEVEVEKPSKKKSSSKKTVEESSG 352
Query: 846 SEGRRKVRS-----KSGHKSPRRSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDN 900
S+G+ K S +SG KS ++ ST SSP+K+ + +
Sbjct: 353 SKGKDKQPSAKGSARSGEKSSKQIAKST-SSPAKKQKVDHVESS---------------- 395
Query: 901 PMPPPKSTATVQPS-PQAKGGSSSHVSSEK 929
K + QPS PQAKG +++K
Sbjct: 396 -----KEKSKKQPSKPQAKGSKEKGKATKK 420
Score = 35.4 bits (80), Expect = 0.19
Identities = 30/127 (23%), Positives = 50/127 (38%), Gaps = 18/127 (14%)
Query: 726 QASRDEDDDDDDEDDDVSLADKLKSRKRKPKPVASASQKRAKGAGVSTTSGASKLASDAQ 785
+ S E D DDE D+V + K + K V +S + K D Q
Sbjct: 315 EESDHEKTDTDDEKDEVEVEKPSKKKSSSKKTVEESSGSKGK---------------DKQ 359
Query: 786 PEVDDKEGGGDAAIQADVHEHSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKK-TS 844
P G+ + + STS+P ++ +K K++ P+A S +K +
Sbjct: 360 PSAKGSARSGEKSSKQIA--KSTSSPAKKQKVDHVESSKEKSKKQPSKPQAKGSKEKGKA 417
Query: 845 TSEGRRK 851
T +G+ K
Sbjct: 418 TKKGKAK 424
>At4g31880 unknown protein
Length = 873
Score = 52.0 bits (123), Expect = 2e-06
Identities = 52/218 (23%), Positives = 91/218 (40%), Gaps = 22/218 (10%)
Query: 722 IGQGQASRDED--DDDDDEDDDVSLADKLKSRKRKPKPVASASQKRAKGAGVS-----TT 774
+ + + S+DE+ D E+D ++ K++ K + ++S K+ GAG S
Sbjct: 660 LDESELSQDEEAADQTGQEEDASTVPLTKKAKTGKQSKMDNSSAKKGSGAGSSKAKATPA 719
Query: 775 SGASKLASD---AQPEVDDKEGGGDAAIQADVHEHSTSNPPSRENSPAPNPAKSKAEGVE 831
S +SK + D A D KE + ++ E PP + +K V
Sbjct: 720 SKSSKTSQDDKTASKSKDSKEASREEEASSE-EESEEEEPPKTVGKSGSSRSKKDISSVS 778
Query: 832 KPPKA-ASSAKKTSTSEGRRKVRSKSGHKSPRRSRSSTNSSPSKESACQETQGAISPIRE 890
K K+ ASS KK S+ +SKSG ++S T +K + A +P +
Sbjct: 779 KSGKSKASSKKKEEPSKATTSSKSKSGPVKSVPAKSKTGKGKAKSGS------ASTPASK 832
Query: 891 AEPLPGKDDNPMPPPKSTATVQPSPQAKGGSSSHVSSE 928
A+ + ++ P + +P+ +AK G S S+
Sbjct: 833 AKESASESESEETPKEP----EPATKAKSGKSQGSQSK 866
>At3g18610 unknown protein
Length = 636
Score = 49.7 bits (117), Expect = 1e-05
Identities = 50/237 (21%), Positives = 89/237 (37%), Gaps = 30/237 (12%)
Query: 728 SRDEDDDDDDEDDDVSLADKLKSRKRKPKPVASASQKRAKGAGVSTTSGASKLASDAQPE 787
S+ +D+ +E+DD S +++ K++P+P+ A + + + +TS QP
Sbjct: 83 SKLKDESSSEEEDDSSSDEEIAPAKKRPEPIKKAKVE-SSSSDDDSTSDEETAPVKKQPA 141
Query: 788 V-----------DDKEGGGDAAI----------QADVHEHSTSNPPSRENSPAPNPAKSK 826
V DD + + +A + S+ + S + P K
Sbjct: 142 VLEKAKVESSSSDDDSSSDEETVPVKKQPAVLEKAKIESSSSDDDSSSDEETVP---MKK 198
Query: 827 AEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSSTNSSPS-----KESACQET 881
V + KA SS+ +S +K ++ S +SS K+
Sbjct: 199 QTAVLEKAKAESSSSDDGSSSDEEPTPAKKEPIVVKKDSSDESSSDEETPVVKKKPTTVV 258
Query: 882 QGAISPIREAEPLPGKDDNPMPPPKSTATVQPSPQAKGGSSSHVSSEKLDTLFEEDP 938
+ A + +E DD P P K T P AK SSS S++ ++ E+ P
Sbjct: 259 KDAKAESSSSEEESSSDDEPTPAKKPTVVKNAKPAAKDSSSSEEDSDEEESDDEKPP 315
Score = 41.6 bits (96), Expect = 0.003
Identities = 46/214 (21%), Positives = 89/214 (41%), Gaps = 19/214 (8%)
Query: 735 DDDEDDDVSLADKLKSRKRKPKPVASASQKRAKGAGVSTTSGASKLASDAQPEVDDKEGG 794
D +ED D+ + ++K+K + + +++A+ +S ASD+ E KE
Sbjct: 28 DAEEDLDMQV-----TKKQKKELIDVVQKEKAEKTVPKKVESSSSDASDSDEEEKTKETP 82
Query: 795 GDAAIQADVHEHSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGR----R 850
++ E S+ + PAK + E ++K +SS+ STS+ +
Sbjct: 83 SKLKDESSSEEEDDSS-----SDEEIAPAKKRPEPIKKAKVESSSSDDDSTSDEETAPVK 137
Query: 851 KVRSKSGHKSPRRSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDN-----PMPPP 905
K + S S +SS +E+ + Q A+ + E DD+ +P
Sbjct: 138 KQPAVLEKAKVESSSSDDDSSSDEETVPVKKQPAVLEKAKIESSSSDDDSSSDEETVPMK 197
Query: 906 KSTATVQPSPQAKGGSSSHVSSEKLDTLFEEDPL 939
K TA ++ + S SS++ T +++P+
Sbjct: 198 KQTAVLEKAKAESSSSDDGSSSDEEPTPAKKEPI 231
>At1g31870 unknown protein
Length = 561
Score = 45.4 bits (106), Expect = 2e-04
Identities = 59/239 (24%), Positives = 88/239 (36%), Gaps = 38/239 (15%)
Query: 722 IGQGQASRDEDDDDDDEDDDVSLADKLKSRKRKPKPVASASQKRAKGAGVSTTSGASKLA 781
+ Q Q +ED+++DD ++ L D+ KR + + +RA A SG L
Sbjct: 48 VWQKQVDPEEDENEDDSAEETPLVDEDIEVKRMRR-LEEIKARRAHNAIAEDGSGWVTLP 106
Query: 782 ---SDAQPEVD------------DKEGGGDAAIQADVHEHSTSNPP---SRENSPAPNPA 823
D Q + E G ++ V + +PP R NSP+P P
Sbjct: 107 LNREDTQSNISPPRRQRTRNDSPSPEPGPRRSVADRVD--TDMSPPRRRKRHNSPSPEPN 164
Query: 824 KSKAEGVE-----KPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSR---SSTNSSPSKE 875
+ + V PP+ + + + E K S+ SP R R S + S K
Sbjct: 165 RKHTKPVSLDSDMSPPRKRKARNDSPSPEPEAKYLSED--LSPPRRRHVHSPSRESSRKR 222
Query: 876 SACQETQGAISPIREAEPLPG-------KDDNPMPPPKSTATVQPSPQAKGGSSSHVSS 927
S E +SP R L G K N + PP+ PSP+ SS S
Sbjct: 223 SDSVELDDDLSPPRRKRDLHGSPVSDVKKKSNDLSPPRRRRYHSPSPEPARRSSKSFGS 281
>At5g55660 putative protein
Length = 759
Score = 45.1 bits (105), Expect = 2e-04
Identities = 53/204 (25%), Positives = 84/204 (40%), Gaps = 22/204 (10%)
Query: 741 DVSLADKLKSRKRKPKP------VASASQKRA-----KGAGVSTTSGASKLASDAQPEVD 789
DV + +K K KRK P S+S KR+ K + T+ S SD + E +
Sbjct: 454 DVLVNEKEKGVKRKRTPKKSSPAAGSSSSKRSAKSQKKTEEATRTNKKSVAHSDDESE-E 512
Query: 790 DKEGGGDAAIQADVHEHSTSNP---PSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTS 846
+KE + + +V E N P + AP ++S+ E VE ++ KK
Sbjct: 513 EKEDDEEEEKEQEVEEEEEENENGIPDKSEDEAPQLSESE-ENVESEEESEEETKKK--- 568
Query: 847 EGRRKVRSKSGHK-SPRRSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPP 905
+R R+ S K S +SRS + P+K S ++ S + + D +P
Sbjct: 569 --KRGSRTSSDKKESAGKSRSKKTAVPTKSSPPKKATQKRSAGKRKKSDDDSDTSPKASS 626
Query: 906 KSTATVQPSPQAKGGSSSHVSSEK 929
K T +P+ + VS EK
Sbjct: 627 KRKKTEKPAKEQAAAPLKSVSKEK 650
>At4g25210 unknown protein
Length = 368
Score = 45.1 bits (105), Expect = 2e-04
Identities = 39/162 (24%), Positives = 57/162 (35%), Gaps = 9/162 (5%)
Query: 809 SNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSST 868
S P S E + + + E P K SS K S SEG + S SG + +
Sbjct: 12 SPPVSSEEEESGSSGEESESSAEVPKKVESSQKPESDSEGESESESSSGPEPESEPAKTI 71
Query: 869 NSSPSKESACQETQGAISPIREA----EPLPGKDDNPMPPPKSTATVQPSPQAKGGSSSH 924
P ET G+ + + E+ PL + K++ T
Sbjct: 72 KLKPVGTKPIPETSGSAATVPESSTAKRPLKEAAPEAIKKQKTSDTEHVKKPITNDEVKK 131
Query: 925 VSSEKLDTLF-----EEDPLAALDGFLDGTLEWDSPPHQADS 961
+SSE +F E D +A L G +D T P D+
Sbjct: 132 ISSEDAKKMFQRLFSETDEIALLQGIIDFTSTKGDPYEDIDA 173
>At3g48710 unknown protein
Length = 462
Score = 44.7 bits (104), Expect = 3e-04
Identities = 45/190 (23%), Positives = 78/190 (40%), Gaps = 14/190 (7%)
Query: 741 DVSLADKLKSRKRKPKPVASASQKRAKGAGVSTTSGASKLASDAQPEVDDKEGGGDAAIQ 800
D+ LAD K K++ K ++ K S + + Q + + EG G++
Sbjct: 204 DILLADSEKETKKRKK----STSKNVTSGESSHVPAKRRRQAKKQEQPTETEGNGES--- 256
Query: 801 ADVHEHSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKT-STSEGRRKVRSKSGHK 859
DV T N + E+ AP +K+E E + + +KT ST + R R+K
Sbjct: 257 -DVGSEGT-NDSNGEDDVAPEEENNKSEDTETEDEKDKAKEKTKSTDKKRLSKRTKKEKP 314
Query: 860 SPRRSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSPQAKG 919
+ +S S+ S + ++ + + + + + KDD+ K T PQAKG
Sbjct: 315 AAEEEKSIKGSAKSSRKSFRQVDKSTTSSSKKQKV-DKDDSSKEKGK---TQTSKPQAKG 370
Query: 920 GSSSHVSSEK 929
S +K
Sbjct: 371 SKDQGQSRKK 380
>At3g19020 hypothetical protein
Length = 951
Score = 44.7 bits (104), Expect = 3e-04
Identities = 29/114 (25%), Positives = 47/114 (40%), Gaps = 6/114 (5%)
Query: 809 SNPPSRENSPAPNPAKSKAEGVEKP-PKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSS 867
S+ PS P P K ++ E+P PK S +++ E + + K +SP++ S
Sbjct: 449 SHEPSNPKEPKPESPKQESPKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSK 508
Query: 868 TN-----SSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSPQ 916
SP E E P ++ P P + P PP + T + SP+
Sbjct: 509 QEPPKPEESPKPEPPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPK 562
Score = 44.3 bits (103), Expect = 4e-04
Identities = 26/109 (23%), Positives = 42/109 (37%)
Query: 808 TSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSS 867
T P + SP K +A E+P S K+ S+ + K + P+ S
Sbjct: 470 TEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKPEESPKPEPPKPEESP 529
Query: 868 TNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSPQ 916
P +E+ E P ++ P P + P PP + T + SP+
Sbjct: 530 KPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPK 578
Score = 40.0 bits (92), Expect = 0.008
Identities = 30/111 (27%), Positives = 41/111 (36%), Gaps = 17/111 (15%)
Query: 805 EHSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRS 864
E S PP E SP P P K + +PPK + + S + P++
Sbjct: 505 ESSKQEPPKPEESPKPEPPKPEESPKPQPPKQETPKPEESPKP-----------QPPKQE 553
Query: 865 RSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSP 915
SP + QET P +P P K + PPK+ A SP
Sbjct: 554 TPKPEESPKPQPPKQETP---KPEESPKPQPPKQEQ---PPKTEAPKMGSP 598
Score = 37.0 bits (84), Expect = 0.066
Identities = 37/175 (21%), Positives = 61/175 (34%), Gaps = 25/175 (14%)
Query: 756 KPVASASQKRAKGAGVSTTSGASKLASDAQPEVDDKEGGGDAAIQADVHEHSTSNP--PS 813
+PV + K A G G + + PE + +++ + P P
Sbjct: 383 RPVDCSKDKCAGGGGGGSNPSPKPTPTPKAPEP-----------KKEINPPNLEEPSKPK 431
Query: 814 RENSPAPNPAKSKAE------GVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSS 867
E SP P K E K PK S +++ +E + +SP++
Sbjct: 432 PEESPKPQQPSPKPETPSHEPSNPKEPKPESPKQESPKTEQPKPKPESPKQESPKQEAPK 491
Query: 868 TN------SSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSPQ 916
SP +ES+ QE + P P + P PP + T + SP+
Sbjct: 492 PEQPKPKPESPKQESSKQEPPKPEESPKPEPPKPEESPKPQPPKQETPKPEESPK 546
Score = 35.8 bits (81), Expect = 0.15
Identities = 29/115 (25%), Positives = 43/115 (37%), Gaps = 12/115 (10%)
Query: 805 EHSTSNPPSREN------SPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGH 858
E S P ++ SP P P K + E+ PK ++T E K +
Sbjct: 526 EESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPK-Q 584
Query: 859 KSPRRSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQP 913
+ P ++ + SP ES SPI++ P P P P + T T P
Sbjct: 585 EQPPKTEAPKMGSPPLESPVPNDPYDASPIKKRRPQP-----PSPSTEETKTTSP 634
Score = 35.4 bits (80), Expect = 0.19
Identities = 30/118 (25%), Positives = 44/118 (36%), Gaps = 4/118 (3%)
Query: 809 SNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSST 868
S PP + P P+P S V PP + +TS + S +S S
Sbjct: 789 SPPPPVHSPPPPSPIYSPPPPVFSPPPKPVTPLPPATSP-MANAPTPSSSESGEISTPVQ 847
Query: 869 NSSPSKESACQETQGAISPIREAEPLPGKDDNP---MPPPKSTATVQPSPQAKGGSSS 923
+P E + SP+ ++ P P D P P P S V+ Q++ SS
Sbjct: 848 APTPDSEDIEAPSDSNHSPVFKSSPAPSPDSEPEVEAPVPSSEPEVEAPKQSEATPSS 905
>At1g48920 unknown protein
Length = 557
Score = 44.7 bits (104), Expect = 3e-04
Identities = 75/331 (22%), Positives = 128/331 (38%), Gaps = 38/331 (11%)
Query: 733 DDDDDEDDDVSLADKLKSRKRKPKPVASASQKRAKGAGVSTTSGASKLASDAQPEVDDKE 792
+ +DD D VSL K++ K V +A QK V +S SD++ E ++K
Sbjct: 26 EPEDDIDTKVSL-------KKQKKDVIAAVQKEKAVKKVPKKVESSD-DSDSESEEEEKA 77
Query: 793 GGGDAAIQADVHEHSTSNPPSRENSPAPNPAKSKAEG-VEKPPKAASSAKKTSTSEGRRK 851
A A + S S+ S ++ PAP A + G V K K SS+ +S+
Sbjct: 78 KKVPAKKAASSSDES-SDDSSSDDEPAPKKAVAATNGTVAKKSKDDSSSSDDDSSDEEVA 136
Query: 852 VRSKSGHKSPRRSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPM-------PP 904
V K + S + S S++ + E + A P + KD +
Sbjct: 137 VTKKPAAAAKNGSVKAKKESSSEDDSSSEDEPAKKPAAKIAKPAAKDSSSSDDDSDEDSE 196
Query: 905 PKSTATVQPSP-QAKGGSSSHVSSEKLDTLFEEDPLAALDGFLDGTLEWDSPPHQADSTA 963
+ AT + +P AK SSS S E D E++ A +AD+ A
Sbjct: 197 DEKPATKKAAPAAAKAASSSDSSDEDSDEESEDEKPA---------------QKKADTKA 241
Query: 964 ETAQSSGVANFQQIEESMGRLKAIVFASGFLDQIQVDPQASHAAKELLVFLLGQKLDSHQ 1023
SS ++ + +ES + S ++ + + ++ K G +
Sbjct: 242 SKKSSSDESSESEEDESEDEEETPKKKSSDVEMVDAEKSSAKQPKTPSTPAAG----GSK 297
Query: 1024 AAALANLQSFLVDASTTFHQVKDVGQAVSLK 1054
ANL SF ++ + + K+ G+ V ++
Sbjct: 298 TLFAANL-SFNIERADVENFFKEAGEVVDVR 327
Score = 43.5 bits (101), Expect = 7e-04
Identities = 43/222 (19%), Positives = 88/222 (39%), Gaps = 26/222 (11%)
Query: 730 DEDDDDDDEDDDVSLADKLKSRKR-----KPKPVASASQKRAKGAGVSTTSG-ASKLASD 783
+ DD D E ++ A K+ ++K + +S+ + A V+ T+G +K + D
Sbjct: 62 ESSDDSDSESEEEEKAKKVPAKKAASSSDESSDDSSSDDEPAPKKAVAATNGTVAKKSKD 121
Query: 784 AQPEVDDKEGGGDAAIQAD------------VHEHSTSNPPSRENSPAPNPAKSKAEGVE 831
DD + A+ E S+ + S E+ PA PA +
Sbjct: 122 DSSSSDDDSSDEEVAVTKKPAAAAKNGSVKAKKESSSEDDSSSEDEPAKKPAAK----IA 177
Query: 832 KPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSSTNSSPSKESACQETQGAISPIREA 891
KP SS+ + E + + +P ++++++S S E + +E++ ++A
Sbjct: 178 KPAAKDSSSSDDDSDEDSEDEKPATKKAAPAAAKAASSSDSSDEDSDEESEDEKPAQKKA 237
Query: 892 EPLPGK----DDNPMPPPKSTATVQPSPQAKGGSSSHVSSEK 929
+ K D++ + + +P+ K V +EK
Sbjct: 238 DTKASKKSSSDESSESEEDESEDEEETPKKKSSDVEMVDAEK 279
Score = 42.4 bits (98), Expect = 0.002
Identities = 55/207 (26%), Positives = 85/207 (40%), Gaps = 29/207 (14%)
Query: 725 GQASRDEDDDDDDEDDDVSLADKLKSRKRKPKPVASASQKRAKGAGVSTTSGASKLASDA 784
G ++ DD DDD S D+ + +KP A +AK +S +S+
Sbjct: 113 GTVAKKSKDDSSSSDDDSS--DEEVAVTKKPAAAAKNGSVKAK----KESSSEDDSSSED 166
Query: 785 QPEVDDKEGGGDAAIQADVHEHSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKT- 843
+P K+ A A + S+S+ S E+S PA KA KAASS+ +
Sbjct: 167 EPA---KKPAAKIAKPA-AKDSSSSDDDSDEDSEDEKPATKKA--APAAAKAASSSDSSD 220
Query: 844 ----STSEGRRKVRSKSGHKSPRRSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDD 899
SE + + K+ K+ ++S SS SS S+E ++ E P D
Sbjct: 221 EDSDEESEDEKPAQKKADTKASKKS-SSDESSESEEDESED--------EEETPKKKSSD 271
Query: 900 NPMPPPKSTATVQ---PSPQAKGGSSS 923
M + ++ Q PS A GGS +
Sbjct: 272 VEMVDAEKSSAKQPKTPSTPAAGGSKT 298
>At1g47970 Unknown protein (T2J15.12)
Length = 198
Score = 43.1 bits (100), Expect = 0.001
Identities = 28/106 (26%), Positives = 45/106 (42%), Gaps = 11/106 (10%)
Query: 730 DEDDDDDDEDDDVSLADKLKSRKRKPKPVASASQKRAKGAGVSTTSGASKLASDAQPEVD 789
D+DDDDDDED+DV L + +PV A + +G + + + + +
Sbjct: 103 DDDDDDDDEDEDVEDEGDL-GTEYLVRPVGRAEDEEDASDFEPEENGVEEDIDEGEDDEN 161
Query: 790 DKEGGGDAAIQADVHEHSTSNPPSRENSPAPNPAKSKAEGVEKPPK 835
D GG + PP R+ +P + S E ++PPK
Sbjct: 162 DNSGGAG----------KSEAPPKRKRAPEEDEEDSGDEDDDRPPK 197
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.132 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,173,215
Number of Sequences: 26719
Number of extensions: 1236677
Number of successful extensions: 9987
Number of sequences better than 10.0: 363
Number of HSP's better than 10.0 without gapping: 97
Number of HSP's successfully gapped in prelim test: 280
Number of HSP's that attempted gapping in prelim test: 8144
Number of HSP's gapped (non-prelim): 1287
length of query: 1170
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1060
effective length of database: 8,379,506
effective search space: 8882276360
effective search space used: 8882276360
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0094b.11


