

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0093b.6
(443 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
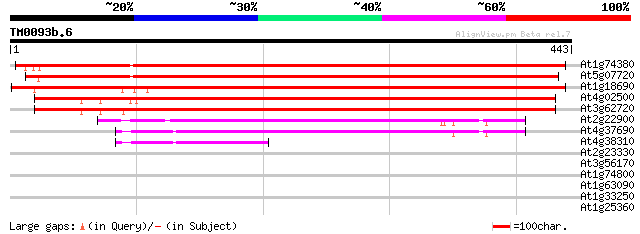
Score E
Sequences producing significant alignments: (bits) Value
At1g74380 putative alpha galactosyltransferase 755 0.0
At5g07720 alpha galactosyltransferase protein 747 0.0
At1g18690 hypothetical protein 649 0.0
At4g02500 unknown protein 555 e-158
At3g62720 alpha galactosyltransferase-like protein 539 e-153
At2g22900 unknown protein 301 4e-82
At4g37690 unknown protein 300 1e-81
At4g38310 putative protein 99 5e-21
At2g23330 putative retroelement pol polyprotein 32 0.70
At3g56170 Ca(2+)-dependent nuclease 30 2.7
At1g74800 unknown protein (At1g74800) 30 3.5
At1g63090 unknown protein 29 5.9
At1g33250 hypothetical protein 28 7.8
At1g25360 28 7.8
>At1g74380 putative alpha galactosyltransferase
Length = 457
Score = 755 bits (1950), Expect = 0.0
Identities = 362/449 (80%), Positives = 399/449 (88%), Gaps = 16/449 (3%)
Query: 5 AHKRTTT---GLPTTT-----ARGGR-----RGRQIQKTFNNVKITILCGFVTILVLRGT 51
AHKR + GLPTTT RGGR RGRQ+QKTFNN+KITILCGFVTILVLRGT
Sbjct: 8 AHKRPSGSGGGLPTTTLTNGGGRGGRGGLLPRGRQMQKTFNNIKITILCGFVTILVLRGT 67
Query: 52 IGV-NLSSSDADAVNQNVIEETNRILAEIRSDADPSDPDDAAAAETFFSPNATFTLGPKI 110
IGV NL SS ADAVNQN+IEETNRILAEIRSD+DP+D D+ + +PNAT+ LGPKI
Sbjct: 68 IGVGNLGSSSADAVNQNIIEETNRILAEIRSDSDPTDLDEPQEGD--MNPNATYVLGPKI 125
Query: 111 TGWDLQRKAWLDQNPEYPNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRL 170
T WD QRK WL+QNPE+P+ V GKARILLLTGSPPKPCDNPIGDHYLLKS+KNKIDYCRL
Sbjct: 126 TDWDSQRKVWLNQNPEFPSTVNGKARILLLTGSPPKPCDNPIGDHYLLKSVKNKIDYCRL 185
Query: 171 HGIEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKY 230
HGIEIVYN+AHLD ELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDA FTD++F++PL++Y
Sbjct: 186 HGIEIVYNMAHLDKELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDALFTDILFQIPLARY 245
Query: 231 DDYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVL 290
+NLV+HGYPDLLF+QKSWIA+NTGSFL RNCQWSLDLLDAWAPMGPKGP+R+EAGKVL
Sbjct: 246 QKHNLVIHGYPDLLFDQKSWIALNTGSFLLRNCQWSLDLLDAWAPMGPKGPIRDEAGKVL 305
Query: 291 TANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYH 350
TA LKGRPAFEADDQSALIYLLLS+KD WM+K F+EN +YLHG+W GLVDRYEEMIEKYH
Sbjct: 306 TAYLKGRPAFEADDQSALIYLLLSQKDTWMEKVFVENQYYLHGFWEGLVDRYEEMIEKYH 365
Query: 351 PGLGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSP 410
PGLGDERWPFVTHFVGCKPCGSY DY VE+CL SMERAFNFADNQVLKLYGF HRGLLSP
Sbjct: 366 PGLGDERWPFVTHFVGCKPCGSYADYAVERCLKSMERAFNFADNQVLKLYGFSHRGLLSP 425
Query: 411 KIKRIRNETVTPLEFVDQFDIRRHSSESR 439
KIKRIRNETV+PLEFVD+FDIRR E++
Sbjct: 426 KIKRIRNETVSPLEFVDKFDIRRTPVETK 454
>At5g07720 alpha galactosyltransferase protein
Length = 457
Score = 747 bits (1928), Expect = 0.0
Identities = 343/426 (80%), Positives = 388/426 (90%), Gaps = 7/426 (1%)
Query: 13 LPTTTARGG-----RRGRQIQKTFNNVKITILCGFVTILVLRGTIGVNLSSSDADAVNQN 67
LPTT A GG RGRQIQKTFNNVK+TILCGFVTILVLRGTIG+N +SDAD VNQN
Sbjct: 24 LPTTMASGGVRRPPPRGRQIQKTFNNVKMTILCGFVTILVLRGTIGINFGTSDADVVNQN 83
Query: 68 VIEETNRILAEIRSDADPSDPDDAAAAETFFSPNATFTLGPKITGWDLQRKAWLDQNPEY 127
+IEETNR+LAEIRSD+DP+D ++ ++ N T+TLGPKIT WD +RK WL QNP++
Sbjct: 84 IIEETNRLLAEIRSDSDPTDSNEPPDSD--LDLNMTYTLGPKITNWDQKRKLWLTQNPDF 141
Query: 128 PNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNLAHLDVELA 187
P+F+ GKA++LLLTGSPPKPCDNPIGDHYLLKS+KNKIDYCR+HGIEIVYN+AHLD ELA
Sbjct: 142 PSFINGKAKVLLLTGSPPKPCDNPIGDHYLLKSVKNKIDYCRIHGIEIVYNMAHLDKELA 201
Query: 188 GYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQ 247
GYWAKLPMIRRLMLSHPE+EWIWWMDSDA FTDMVFE+PLS+Y+++NLV+HGYPDLLF+Q
Sbjct: 202 GYWAKLPMIRRLMLSHPEIEWIWWMDSDALFTDMVFEIPLSRYENHNLVIHGYPDLLFDQ 261
Query: 248 KSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAFEADDQSA 307
KSWIA+NTGSFLFRNCQWSLDLLDAWAPMGPKGP+REEAGK+LTANLKGRPAFEADDQSA
Sbjct: 262 KSWIALNTGSFLFRNCQWSLDLLDAWAPMGPKGPIREEAGKILTANLKGRPAFEADDQSA 321
Query: 308 LIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPGLGDERWPFVTHFVGC 367
LIYLLLS+K+ WM+K F+EN +YLHG+W GLVD+YEEM+EKYHPGLGDERWPF+THFVGC
Sbjct: 322 LIYLLLSQKETWMEKVFVENQYYLHGFWEGLVDKYEEMMEKYHPGLGDERWPFITHFVGC 381
Query: 368 KPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKIKRIRNETVTPLEFVD 427
KPCGSY DY VE+CL SMERAFNFADNQVLKLYGF HRGLLSPKIKRIRNET PL+FVD
Sbjct: 382 KPCGSYADYAVERCLKSMERAFNFADNQVLKLYGFGHRGLLSPKIKRIRNETTFPLKFVD 441
Query: 428 QFDIRR 433
+FDIRR
Sbjct: 442 RFDIRR 447
>At1g18690 hypothetical protein
Length = 632
Score = 649 bits (1675), Expect = 0.0
Identities = 326/509 (64%), Positives = 375/509 (73%), Gaps = 71/509 (13%)
Query: 2 NGSAHKRTTTGLPTTTA-------RGGRRGRQIQKT-FNNVKITILCGFVTILVLRGTIG 53
+GS + GL TT RG RG QIQ T FNN+K ILC FVTIL+L GTI
Sbjct: 123 DGSRSSGSGRGLSTTAVSNGGWRTRGFLRGWQIQNTLFNNIKFMILCCFVTILILLGTIR 182
Query: 54 V-NLSSSDADAVNQNVIEETNRILAEIRSDADPSD------------------------P 88
V NL SS+AD+VNQ+ I+ET ILAEI SD+ +D P
Sbjct: 183 VGNLGSSNADSVNQSFIKETIPILAEIPSDSHSTDLAEPPKADVSPNATYTLEPRIAEIP 242
Query: 89 DDAAAAETF------FSPNATFTLG--------------------------------PKI 110
D + + SPNAT+TLG PKI
Sbjct: 243 SDVHSTDLVELPKADISPNATYTLGPRIAEIPSDSHLTDLLEPPKADISPNATYTLGPKI 302
Query: 111 TGWDLQRKAWLDQNPEYPNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRL 170
T WD QRK WL+QNPE+PN V GKARILLLTGS P PCD PIG++YLLK++KNKIDYCRL
Sbjct: 303 TNWDSQRKVWLNQNPEFPNIVNGKARILLLTGSSPGPCDKPIGNYYLLKAVKNKIDYCRL 362
Query: 171 HGIEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKY 230
HGIEIVYN+A+LD EL+GYW KLPMIR LMLSHPEVEWIWWMDSDA FTD++FE+PL +Y
Sbjct: 363 HGIEIVYNMANLDEELSGYWTKLPMIRTLMLSHPEVEWIWWMDSDALFTDILFEIPLPRY 422
Query: 231 DDYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVL 290
+++NLV+HGYPDLLF QKSW+A+NTG FL RNCQWSLDLLDAWAPMGPKG +R+E GK+L
Sbjct: 423 ENHNLVIHGYPDLLFNQKSWVALNTGIFLLRNCQWSLDLLDAWAPMGPKGKIRDETGKIL 482
Query: 291 TANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYH 350
TA LKGRPAFEADDQSALIYLLLS+K+KW++K ++EN +YLHG+W GLVDRYEEMIEKYH
Sbjct: 483 TAYLKGRPAFEADDQSALIYLLLSQKEKWIEKVYVENQYYLHGFWEGLVDRYEEMIEKYH 542
Query: 351 PGLGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSP 410
PGLGDERWPFVTHFVGCKPCGSY DY V++C SMERAFNFADNQVLKLYGF HRGLLSP
Sbjct: 543 PGLGDERWPFVTHFVGCKPCGSYADYAVDRCFKSMERAFNFADNQVLKLYGFSHRGLLSP 602
Query: 411 KIKRIRNETVTPLEFVDQFDIRRHSSESR 439
KIKRIRNETV+PLE VD+FDIRR E++
Sbjct: 603 KIKRIRNETVSPLESVDKFDIRRMHMETK 631
>At4g02500 unknown protein
Length = 461
Score = 555 bits (1430), Expect = e-158
Identities = 259/440 (58%), Positives = 323/440 (72%), Gaps = 28/440 (6%)
Query: 20 GGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGVN---LSSSDADAVNQNVIE----ET 72
G R R+IQ+ +K+TILC +T++VLR TIG D D + Q+ E
Sbjct: 7 GAYRCRRIQRALRQLKVTILCLLLTVVVLRSTIGAGKFGTPEQDLDEIRQHFHARKRGEP 66
Query: 73 NRILAEIRSDADPSDPDDAAAA------ETFF--------------SPNATFTLGPKITG 112
+R+L EI++ D S D + ETF PN +TLGPKI+
Sbjct: 67 HRVLEEIQTGGDSSSGDGGGNSGGSNNYETFDINKIFVDEGEEEKPDPNKPYTLGPKISD 126
Query: 113 WDLQRKAWLDQNPEYPNFVR-GKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLH 171
WD QR WL +NP +PNF+ K R+LL+TGS PKPC+NP+GDHYLLKSIKNKIDYCRLH
Sbjct: 127 WDEQRSDWLAKNPSFPNFIGPNKPRVLLVTGSAPKPCENPVGDHYLLKSIKNKIDYCRLH 186
Query: 172 GIEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYD 231
GIEI YN+A LD E+AG+WAKLP+IR+L+LSHPE+E++WWMDSDA FTDM FELP +Y
Sbjct: 187 GIEIFYNMALLDAEMAGFWAKLPLIRKLLLSHPEIEFLWWMDSDAMFTDMAFELPWERYK 246
Query: 232 DYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLT 291
DYNLV+HG+ +++++QK+WI +NTGSFL RN QW+LDLLD WAPMGPKG +REEAGKVLT
Sbjct: 247 DYNLVMHGWNEMVYDQKNWIGLNTGSFLLRNNQWALDLLDTWAPMGPKGKIREEAGKVLT 306
Query: 292 ANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHP 351
LK RP FEADDQSA++YLL +++D W +K +LE+ +YLHGYW LVDRYEEMIE YHP
Sbjct: 307 RELKDRPVFEADDQSAMVYLLATQRDAWGNKVYLESGYYLHGYWGILVDRYEEMIENYHP 366
Query: 352 GLGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPK 411
GLGD RWP VTHFVGCKPCG +GDYPVE+CL M+RAFNF DNQ+L++YGF H+ L S K
Sbjct: 367 GLGDHRWPLVTHFVGCKPCGKFGDYPVERCLKQMDRAFNFGDNQILQIYGFTHKSLASRK 426
Query: 412 IKRIRNETVTPLEFVDQFDI 431
+KR+RNET PLE D+ +
Sbjct: 427 VKRVRNETSNPLEMKDELGL 446
>At3g62720 alpha galactosyltransferase-like protein
Length = 460
Score = 539 bits (1388), Expect = e-153
Identities = 250/439 (56%), Positives = 321/439 (72%), Gaps = 27/439 (6%)
Query: 20 GGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGVN---LSSSDADAVNQNVIE-----E 71
G R R++Q+ K+TILC +T++VLRGTIG D + + ++ E
Sbjct: 7 GAHRFRRLQRFMRQGKVTILCLVLTVIVLRGTIGAGKFGTPEKDIEEIREHFFYTRKRGE 66
Query: 72 TNRILAEIRSDADPSDP------------------DDAAAAETFFSPNATFTLGPKITGW 113
+R+L E+ S S+ D+ ++ N ++LGPKI+ W
Sbjct: 67 PHRVLVEVSSKTTSSEDGGNGGNSYETFDINKLFVDEGDEEKSRDRTNKPYSLGPKISDW 126
Query: 114 DLQRKAWLDQNPEYPNFVR-GKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHG 172
D QR+ WL QNP +PNFV K R+LL+TGS PKPC+NP+GDHYLLKSIKNKIDYCR+HG
Sbjct: 127 DEQRRDWLKQNPSFPNFVAPNKPRVLLVTGSAPKPCENPVGDHYLLKSIKNKIDYCRIHG 186
Query: 173 IEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDD 232
IEI YN+A LD E+AG+WAKLP+IR+L+LSHPE+E++WWMDSDA FTDMVFELP +Y D
Sbjct: 187 IEIFYNMALLDAEMAGFWAKLPLIRKLLLSHPEIEFLWWMDSDAMFTDMVFELPWERYKD 246
Query: 233 YNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTA 292
YNLV+HG+ +++++QK+WI +NTGSFL RN QWSLDLLDAWAPMGPKG +REEAGKVLT
Sbjct: 247 YNLVMHGWNEMVYDQKNWIGLNTGSFLLRNSQWSLDLLDAWAPMGPKGKIREEAGKVLTR 306
Query: 293 NLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPG 352
LK RPAFEADDQSA++YLL ++++KW K +LE+ +YLHGYW LVDRYEEMIE + PG
Sbjct: 307 ELKDRPAFEADDQSAMVYLLATEREKWGGKVYLESGYYLHGYWGILVDRYEEMIENHKPG 366
Query: 353 LGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKI 412
GD RWP VTHFVGCKPCG +GDYPVE+CL M+RAFNF DNQ+L++YGF H+ L S ++
Sbjct: 367 FGDHRWPLVTHFVGCKPCGKFGDYPVERCLRQMDRAFNFGDNQILQMYGFTHKSLGSRRV 426
Query: 413 KRIRNETVTPLEFVDQFDI 431
K RN+T PL+ D+F +
Sbjct: 427 KPTRNQTDRPLDAKDEFGL 445
>At2g22900 unknown protein
Length = 449
Score = 301 bits (772), Expect = 4e-82
Identities = 153/377 (40%), Positives = 221/377 (58%), Gaps = 50/377 (13%)
Query: 70 EETNRILAEIRSDADPSDPDDAAAAETFFSPNATFTLGPKITGWDLQRKAWLDQNPEYPN 129
E +N+ + I DP+DP + P+ T+T+ + WD +R+ WL+ +P+
Sbjct: 66 ESSNKCSSGIDMSQDPTDP------VYYDDPDLTYTIEKPVKNWDEKRRRWLNL---HPS 116
Query: 130 FVRG-KARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNLAHLDVELAG 188
F+ G + R +++TGS PC NPIGDH LL+ KNK+DYCR+HG +I Y+ A L ++
Sbjct: 117 FIPGAENRTVMVTGSQSAPCKNPIGDHLLLRFFKNKVDYCRIHGHDIFYSNALLHPKMNS 176
Query: 189 YWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQK 248
YWAKLP ++ M++HPE EWIWW+DSDA FTDM F P +Y ++NLV+HG+P +++ +
Sbjct: 177 YWAKLPAVKAAMIAHPEAEWIWWVDSDALFTDMDFTPPWRRYKEHNLVVHGWPGVIYNDR 236
Query: 249 SWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAFEADDQSAL 308
SW A+N G FL RNCQWS++L+D W MGP P + G++ + K + E+DDQ+AL
Sbjct: 237 SWTALNAGVFLIRNCQWSMELIDTWTGMGPVSPEYAKWGQIQRSIFKDKLFPESDDQTAL 296
Query: 309 IYLLLSKKDKWMDKTFLENSFYLHGYWAGLV-------DRY---------------EEMI 346
+YLL ++ + K +LE FY GYW +V +RY E++
Sbjct: 297 LYLLYKHREVYYPKIYLEGDFYFEGYWLEIVPGLSNVTERYLEMEREDATLRRRHAEKVS 356
Query: 347 EKY------------HPGLGDERWPFVTHFVGCKPCGSYGD----YPVEKCLSSMERAFN 390
E+Y G G +R PFVTHF GC+PC GD Y + C + M +A N
Sbjct: 357 ERYAAFREERFLKGERGGKGSKRRPFVTHFTGCQPCS--GDHNKMYDGDTCWNGMIKAIN 414
Query: 391 FADNQVLKLYGFRHRGL 407
FADNQV++ YGF H L
Sbjct: 415 FADNQVMRKYGFVHSDL 431
>At4g37690 unknown protein
Length = 432
Score = 300 bits (768), Expect = 1e-81
Identities = 147/362 (40%), Positives = 212/362 (57%), Gaps = 48/362 (13%)
Query: 84 DPSDPDDAAAAETFFSPNATFTLGPKITGWDLQRKAWLDQNPEYPNFVRGKARILLLTGS 143
DPS+P + P+ ++++ IT WD +R W + +P + + RI+++TGS
Sbjct: 63 DPSEPG------FYDDPDLSYSIEKPITKWDEKRNQWFESHPSFKP--GSENRIVMVTGS 114
Query: 144 PPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNLAHLDVELAGYWAKLPMIRRLMLSH 203
PC NPIGDH LL+ KNK+DY R+HG +I Y+ + L ++ YWAKLP+++ ML+H
Sbjct: 115 QSSPCKNPIGDHLLLRCFKNKVDYARIHGHDIFYSNSLLHPKMNSYWAKLPVVKAAMLAH 174
Query: 204 PEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNC 263
PE EWIWW+DSDA FTDM F+ PL +Y +NLV+HG+P++++E++SW A+N G FL RNC
Sbjct: 175 PEAEWIWWVDSDAIFTDMEFKPPLHRYRQHNLVVHGWPNIIYEKQSWTALNAGVFLIRNC 234
Query: 264 QWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAFEADDQSALIYLLLSKKDKWMDKT 323
QWS+DL+D W MGP P ++ G + + K + E+DDQ+ALIYLL K+ + K
Sbjct: 235 QWSMDLIDTWKSMGPVSPDYKKWGPIQRSIFKDKLFPESDDQTALIYLLYKHKELYYPKI 294
Query: 324 FLENSFYLHGYWAGLVDRYEEMIEKY---------------------------------- 349
+LE +YL GYW G+ + + E+Y
Sbjct: 295 YLEAEYYLQGYWIGVFGDFANVTERYLEMEREDDTLRRRHAEKVSERYGAFREERFLKGE 354
Query: 350 HPGLGDERWPFVTHFVGCKPCGSYGD----YPVEKCLSSMERAFNFADNQVLKLYGFRHR 405
G G R F+THF GC+PC GD Y + C + M RA NFADNQV+++YG+ H
Sbjct: 355 FGGRGSRRRAFITHFTGCQPCS--GDHNPSYDGDTCWNEMIRALNFADNQVMRVYGYVHS 412
Query: 406 GL 407
L
Sbjct: 413 DL 414
>At4g38310 putative protein
Length = 120
Score = 99.0 bits (245), Expect = 5e-21
Identities = 48/122 (39%), Positives = 73/122 (59%), Gaps = 9/122 (7%)
Query: 84 DPSDPDDAAAAETFFSPNATFTLGPKITGWDLQRKAWLDQNPEYPNFVRGKARILLLTGS 143
DP +P + P+ ++++ IT WD +R W +P + + RIL++TGS
Sbjct: 4 DPPEPG------FYDDPDLSYSIEKSITNWDEKRHEWFKSHPSFKP--GSENRILMVTGS 55
Query: 144 PPKPCDNPIGDHYLL-KSIKNKIDYCRLHGIEIVYNLAHLDVELAGYWAKLPMIRRLMLS 202
PC NPIGDH LL + KNK+DY R+HG +I Y+ + L ++ YWAKLP+++ ML+
Sbjct: 56 QSSPCKNPIGDHLLLLRCFKNKVDYARIHGHDIFYSNSLLHPKMNSYWAKLPVVKAAMLA 115
Query: 203 HP 204
HP
Sbjct: 116 HP 117
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 32.0 bits (71), Expect = 0.70
Identities = 17/59 (28%), Positives = 31/59 (51%)
Query: 93 AAETFFSPNATFTLGPKITGWDLQRKAWLDQNPEYPNFVRGKARILLLTGSPPKPCDNP 151
++ T + NA+ G I+ L+ + + + E NF+R K ++ + GS PKP +P
Sbjct: 17 SSTTAYLINASDNPGALISSVVLKENNYAEWSEELQNFLRAKQKLGFIDGSIPKPAADP 75
>At3g56170 Ca(2+)-dependent nuclease
Length = 323
Score = 30.0 bits (66), Expect = 2.7
Identities = 19/59 (32%), Positives = 29/59 (48%), Gaps = 5/59 (8%)
Query: 242 DLL-FEQKSWIAVNTGSFLFRN----CQWSLDLLDAWAPMGPKGPVREEAGKVLTANLK 295
DLL FE S + GS++ + W +L+AW P+ EEA +++ A LK
Sbjct: 30 DLLNFETTSQVPEKLGSYVVSSQKAQANWYRKILEAWKQAKPRPKTPEEASRLVIAALK 88
>At1g74800 unknown protein (At1g74800)
Length = 672
Score = 29.6 bits (65), Expect = 3.5
Identities = 40/167 (23%), Positives = 63/167 (36%), Gaps = 32/167 (19%)
Query: 62 DAVNQNVIEETNRILAEIRSDADPSDPDDA-AAAETFFSPNATFTLGPKITGWDLQRKAW 120
D V V E +L+ +R D++ DP + E S + LG K+ W
Sbjct: 101 DLVQNKVREHHRGVLSSLRFDSETFDPSSKDGSVELHKSAKEAWQLGRKL--WKELESGR 158
Query: 121 LDQNPEYP--NFVRGKARILLLTGSP---------PKPCDNPIGDHYLL-----KSIKNK 164
L++ E P N + LTGS PC +G H L K+ +
Sbjct: 159 LEKLVEKPEKNKPDSCPHSVSLTGSEFMNRENKLMELPCGLTLGSHITLVGRPRKAHPKE 218
Query: 165 IDYCRL--------HGIEIVYN-----LAHLDVELAGYWAKLPMIRR 198
D+ +L G++ V + H + L G W+K P+I +
Sbjct: 219 GDWSKLVSQFVIELQGLKTVEGEDPPRILHFNPRLKGDWSKKPVIEQ 265
>At1g63090 unknown protein
Length = 289
Score = 28.9 bits (63), Expect = 5.9
Identities = 19/67 (28%), Positives = 26/67 (38%), Gaps = 6/67 (8%)
Query: 189 YWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQK 248
YW+ +P S V+ IWW D ++ F P Y Y + G P F
Sbjct: 139 YWSHIPSDDSRFASVAYVQQIWWFQVDG---EIDFPFPAGTYSVYFRLQLGKPGKRF--- 192
Query: 249 SWIAVNT 255
W V+T
Sbjct: 193 GWKVVDT 199
>At1g33250 hypothetical protein
Length = 548
Score = 28.5 bits (62), Expect = 7.8
Identities = 18/54 (33%), Positives = 21/54 (38%)
Query: 201 LSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQKSWIAVN 254
LS P V W D D F LSKYD +V G P S+ + N
Sbjct: 216 LSLPNVRWFVLGDDDTIFNVHNLLAVLSKYDPSEMVYIGNPSESHSANSYFSHN 269
>At1g25360
Length = 790
Score = 28.5 bits (62), Expect = 7.8
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 3/40 (7%)
Query: 332 HGYWAGLVDRYEEMIEKYHPGLGDERWPFVTHFVGCKPCG 371
HG+ A VD YEEM++K G+ +R +T C G
Sbjct: 496 HGHGAEAVDVYEEMLKK---GIRPDRITLLTVLTACSHAG 532
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.139 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,823,582
Number of Sequences: 26719
Number of extensions: 490454
Number of successful extensions: 1048
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1018
Number of HSP's gapped (non-prelim): 20
length of query: 443
length of database: 11,318,596
effective HSP length: 102
effective length of query: 341
effective length of database: 8,593,258
effective search space: 2930300978
effective search space used: 2930300978
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0093b.6


