

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0093b.4
(565 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
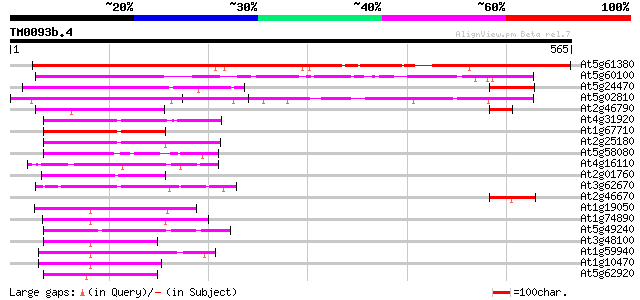
Score E
Sequences producing significant alignments: (bits) Value
At5g61380 pseudo-response regulator 1 512 e-145
At5g60100 pseudo-response regulator 3 (APRR3) 177 1e-44
At5g24470 pseudo-response regulator 5 (APRR5) 120 3e-27
At5g02810 unknown protein 105 7e-23
At2g46790 hypothetical protein 103 3e-22
At4g31920 predicted protein 93 3e-19
At1g67710 response regulator 11 (ARR11) 89 5e-18
At2g25180 putative two-component response regulator protein 89 7e-18
At5g58080 ARR2 - like protein 79 5e-15
At4g16110 hypothetical protein 75 1e-13
At2g01760 putative two-component response regulator protein 74 3e-13
At3g62670 putative protein 67 3e-11
At2g46670 hypothetical protein 64 2e-10
At1g19050 putative protein 64 3e-10
At1g74890 putative response regulator (two component phosphotran... 63 4e-10
At5g49240 putative protein 62 1e-09
At3g48100 response reactor 2 (ATRR2) 61 1e-09
At1g59940 putative protein 61 1e-09
At1g10470 putative response regulator 3 61 2e-09
At5g62920 response regulator 6 (ARR6) 59 7e-09
>At5g61380 pseudo-response regulator 1
Length = 618
Score = 512 bits (1319), Expect = e-145
Identities = 315/627 (50%), Positives = 386/627 (61%), Gaps = 107/627 (17%)
Query: 24 KSGDAFIDRSKVRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQYIDI 83
K GD FIDRS+VRILLCD+D S EVFTLL +CSYQVT V+SARQVIDALNAEG IDI
Sbjct: 8 KGGDGFIDRSRVRILLCDNDSTSLGEVFTLLSECSYQVTAVKSARQVIDALNAEGPDIDI 67
Query: 84 ILAEVDLPIKKGMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKPLRT 143
ILAE+DLP+ KGMK+L+Y++RDK+LRRIPVIMMS +DEVP+VVKCL+LGAADYLVKPLRT
Sbjct: 68 ILAEIDLPMAKGMKMLRYITRDKDLRRIPVIMMSRQDEVPVVVKCLKLGAADYLVKPLRT 127
Query: 144 NELLNLWTHMWRRRRMLGLAEKNILSYDFDMVASDPSDANTNSTTLFSDDTDDRSKRSTN 203
NELLNLWTHMWRRRRMLGLAEKN+LSYDFD+V SD SD NTNST LFSDDTDDRS RSTN
Sbjct: 128 NELLNLWTHMWRRRRMLGLAEKNMLSYDFDLVGSDQSDPNTNSTNLFSDDTDDRSLRSTN 187
Query: 204 PE---AAQQEPEVNV----------------SISKAAAVIEELPGAQISECRPNVPGISD 244
P+ + QE E +V + + + AV P + P +
Sbjct: 188 PQRGNLSHQENEWSVATAPVHARDGGLGADGTATSSLAVTAIEPPLDHLAGSHHEPMKRN 247
Query: 245 RRTGNFSSGPKKSELRIGESSAFFTYVKATTLKSNVQEIAHVDNHATTQ----------- 293
FSS PKKS L+IGESSAFFTYVK+T L++N Q+ VD + +
Sbjct: 248 SNPAQFSSAPKKSRLKIGESSAFFTYVKSTVLRTNGQDPPLVDGNGSLHLHRGLAEKFQV 307
Query: 294 VRVEDMN-----------------------------------------QACARQRVNDLK 312
V E +N Q A + +N+ K
Sbjct: 308 VASEGINNTKQARRATPKSTVLRTNGQDPPLVNGNGSHHLHRGAAEKFQVVASEGINNTK 367
Query: 313 -THENQEMFESRSQ-EDLPSSNSFPDSFSIERSCTPPASVEVSQQKNFREEYYPQGSLYP 370
H ++ + SQ E L + S+P S+ERS T P S+E S +N++E +
Sbjct: 368 QAHRSRGTEQYHSQGETLQNGASYP--HSLERSRTLPTSME-SHGRNYQEGNMNIPQVAM 424
Query: 371 SNGSRGSKLEPSGMPAQHAYPYYYNSGVVNHVMLPSSAQLYQKNMQDIHSHANSSMIAQY 430
+ S+++ SG A +AYPYY + GV+N VM+ S+A M+ QY
Sbjct: 425 NRSKDSSQVDGSGFSAPNAYPYYMH-GVMNQVMMQSAA-----------------MMPQY 466
Query: 431 NH-LAQCSP-HANGMTSFPYY--PMNICLQPGQVS--------TPHSWPSLGSSGSCEVN 478
H + C P H NGMT +PYY PMN LQ Q+S HSW G+ S EV
Sbjct: 467 GHQIPHCQPNHPNGMTGYPYYHHPMNTSLQHSQMSLQNGQMSMVHHSWSPAGNPPSNEVR 526
Query: 479 LSKADRREAALMKFKQKRKDRCFDKKIRYVNRKRLAERRPRVRGQFVRKLSGVDVYLNGQ 538
++K DRRE AL+KF++KR RCFDKKIRYVNRKRLAERRPRV+GQFVRK++GV+V LNGQ
Sbjct: 527 VNKLDRREEALLKFRRKRNQRCFDKKIRYVNRKRLAERRPRVKGQFVRKMNGVNVDLNGQ 586
Query: 539 PASADY-DEEEEEEEDNHVARDSSLED 564
P SADY DEEEEEEE+ RDSS +D
Sbjct: 587 PDSADYDDEEEEEEEEEEENRDSSPQD 613
>At5g60100 pseudo-response regulator 3 (APRR3)
Length = 495
Score = 177 bits (450), Expect = 1e-44
Identities = 155/512 (30%), Positives = 227/512 (44%), Gaps = 94/512 (18%)
Query: 27 DAFIDRSKVRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQYIDIILA 86
+ ++ +++LL ++D + V LL CSY+VT V + L E ID++L
Sbjct: 56 ERYLPVRSLKVLLVENDDSTRHIVTALLKNCSYEVTAVPDVLEAWRILEDEKSCIDLVLT 115
Query: 87 EVDLPIKKGMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKPLRTNEL 146
EVD+P+ G LL + K L+ IPVIMMS+ D + +V KCL GA D+LVKP+R NEL
Sbjct: 116 EVDMPVHSGTGLLSKIMSHKTLKNIPVIMMSSHDSMVLVFKCLSNGAVDFLVKPIRKNEL 175
Query: 147 LNLWTHMWRRRRMLGLAEKNILSYDFDMVASDPSDANTNSTTLFSDDTDDRSKRSTNPEA 206
NLW H+WRR +S++ ++ K+S PE+
Sbjct: 176 KNLWQHVWRR---------------------------CHSSSGSGSESGIHDKKSVKPES 208
Query: 207 AQQEPEVNVSISKAAAVIEELPGAQISECRPNVPGISDRRTGNFSSGPKKSELRIGESSA 266
Q A IS+ N G S G S+ S+ G S+
Sbjct: 209 TQGSEN----------------DASISDEHRNESGSS----GGLSNQDGGSDNGSGTQSS 248
Query: 267 FFTYVKATTLKSNVQEIAHVDNHATTQVRVEDMNQACARQRVNDLKTHENQ-EMFESRSQ 325
+ T S + N T CA VN LK E+Q E + SQ
Sbjct: 249 WTKRASDTKSTSPSNQFPDAPNKKGT------YENGCA--HVNRLKEAEDQKEQIGTGSQ 300
Query: 326 EDLPSSNSFPDSFSIERSCTPPASVEVSQQKNFREEYYPQGSLYPSNGSRGSKLEPSGMP 385
+ S + +E++ SV+ ++ N +L N S G+ S P
Sbjct: 301 TGMSMSKKAEEPGDLEKNA--KYSVQALERNN-------DDTL---NRSSGNSQVESKAP 348
Query: 386 AQHAYPYYYNSGVVNHVMLPSSAQLYQKNMQDIHSHANSSMIAQYNHLAQCSPHANGMTS 445
+ N L S Q +K +D + ++++L+ S + NG TS
Sbjct: 349 SS------------NREDLQSLEQTLKKTREDRDYKVGDRSVLRHSNLSAFSKYNNGATS 396
Query: 446 FPYYPMNICLQPGQVSTPHSWP---SLGSSGSCEVNL----SKADR---REAALMKFKQK 495
P + + +PH P LGSS S + L S +DR REAALMKF+ K
Sbjct: 397 AKKAPE----ENVESCSPHDSPIAKLLGSSSSSDNPLKQQSSGSDRWAQREAALMKFRLK 452
Query: 496 RKDRCFDKKIRYVNRKRLAERRPRVRGQFVRK 527
RK+RCF+KK+RY +RK+LAE+RP V+GQF+RK
Sbjct: 453 RKERCFEKKVRYHSRKKLAEQRPHVKGQFIRK 484
>At5g24470 pseudo-response regulator 5 (APRR5)
Length = 667
Score = 120 bits (300), Expect = 3e-27
Identities = 76/229 (33%), Positives = 114/229 (49%), Gaps = 11/229 (4%)
Query: 14 RENNCGGANSKSGDAFIDRSKVRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDA 73
R+ + G + F+ + +R+LL + D + + + LL KCSY+V V + +
Sbjct: 138 RKKDAGVDGLVKWERFLPKIALRVLLVEADDSTRQIIAALLRKCSYRVAAVPDGLKAWEM 197
Query: 74 LNAEGQYIDIILAEVDLPIKKGMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGA 133
L + + +D+IL EVDLP G LL + + IPVIMMS +D V V KC+ GA
Sbjct: 198 LKGKPESVDLILTEVDLPSISGYALLTLIMEHDICKNIPVIMMSTQDSVNTVYKCMLKGA 257
Query: 134 ADYLVKPLRTNELLNLWTHMWRRRRMLGLAEKNILSYDFDMVASDPSDANTNSTT----- 188
ADYLVKPLR NEL NLW H+WRR+ L + ++ + A+ N++
Sbjct: 258 ADYLVKPLRRNELRNLWQHVWRRQTSLA---PDSFPWNESVGQQKAEGASANNSNGKRDD 314
Query: 189 -LFSDDTDDRSKRSTNPEAAQQEPEVNVSISKAAAVIEELPGAQISECR 236
+ S + D T PE + +V VS A + E +Q +E R
Sbjct: 315 HVVSGNGGDAQSSCTRPEMEGESADVEVSARDAVQM--ECAKSQFNETR 361
Score = 72.0 bits (175), Expect = 8e-13
Identities = 30/45 (66%), Positives = 42/45 (92%)
Query: 484 RREAALMKFKQKRKDRCFDKKIRYVNRKRLAERRPRVRGQFVRKL 528
+REAAL KF+ KRKDRC++KK+RY +RK+LAE+RPR++GQFVR++
Sbjct: 617 QREAALTKFRMKRKDRCYEKKVRYESRKKLAEQRPRIKGQFVRQV 661
>At5g02810 unknown protein
Length = 727
Score = 105 bits (262), Expect = 7e-23
Identities = 75/260 (28%), Positives = 116/260 (43%), Gaps = 21/260 (8%)
Query: 2 ESEAEEILNLNSRENNCGG---------ANSKSGDAFIDRSKVRILLCDDDPKSSEEVFT 52
E E + ++ R + GG A + + F+ +R+LL ++D + V
Sbjct: 36 EEEKTNGITMDVRNGSSGGLQIPLSQQTAATVCWERFLHVRTIRVLLVENDDCTRYIVTA 95
Query: 53 LLVKCSYQVTPVRSARQVIDALNAEGQYIDIILAEVDLPIKKGMKLLKYLSRDKELRRIP 112
LL CSY+V + Q L +IDI+L EV +P G+ LL + K R IP
Sbjct: 96 LLRNCSYEVVEASNGIQAWKVLEDLNNHIDIVLTEVIMPYLSGIGLLCKILNHKSRRNIP 155
Query: 113 VIMMSAKDEVPIVVKCLRLGAADYLVKPLRTNELLNLWTHMWRRRRMLG---------LA 163
VIMMS+ D + +V KCL GA D+LVKP+R NEL LW H+WRR +
Sbjct: 156 VIMMSSHDSMGLVFKCLSKGAVDFLVKPIRKNELKILWQHVWRRCQSSSGSGSESGTHQT 215
Query: 164 EKNILSYDFDMVASDPSDANTNSTTLFSDDTDDRSKRSTNPEAAQQEPEVNVSISKAAAV 223
+K++ S D ++ N + D S + +++ + V+V S A
Sbjct: 216 QKSVKSKSIKKSDQDSGSSDENENGSIGLNASDGSSDGSGAQSSWTKKAVDVDDSPRAVS 275
Query: 224 I---EELPGAQISECRPNVP 240
+ + AQ+ P P
Sbjct: 276 LWDRVDSTCAQVVHSNPEFP 295
Score = 85.9 bits (211), Expect = 6e-17
Identities = 101/373 (27%), Positives = 154/373 (41%), Gaps = 48/373 (12%)
Query: 175 VASDPSDANTNSTTL--FSDDTDDRSKRSTNPEAAQQEPEVNVSISKAAAVIEELPGAQI 232
V P D ++ S + +D K ++ +EPE + K E +I
Sbjct: 367 VGKGPLDLSSESPSSKQMHEDGGSSFKAMSSHLQDNREPEAPNTHLKTLDTNEA--SVKI 424
Query: 233 SECRPNVPGISDRRTGNFSSGP----KKSELRIGESSAFFTYVKATTLKS----NVQEIA 284
SE +V S R G G ++ LR E SAF Y A+ N+ +
Sbjct: 425 SEELMHVEHSSKRHRGTKDDGTLVRDDRNVLRRSEGSAFSRYNPASNANKISGGNLGSTS 484
Query: 285 HVDNHATTQVRVEDMNQACARQRVNDLKTHENQEMFESRSQEDLPSSNSFPDSFSIERSC 344
DN++ ++ + C H N + SN+F S + E +
Sbjct: 485 LQDNNSQDLIKKTEAAYDC----------HSNMNESLPHNHRSHVGSNNFDMSSTTENNA 534
Query: 345 TPPASVEVSQQKNFREEYYPQGSLYPSNGSRGSKLEPSGMPAQHAYPYYYNSGVVNHVML 404
F + P+ S S+ + S +P + + Y V L
Sbjct: 535 -------------FTKPGAPKVSSAGSSSVKHSSFQPLPCDHHNNHASYNLVHVAERKKL 581
Query: 405 P---SSAQLYQKNMQDIHSHANSSMIAQYNHLAQCS--PH--ANGMTSFPYYPMNICLQP 457
P S+ +Y + ++ ++ N S+ + S P+ +NGM + MN+
Sbjct: 582 PPQCGSSNVYNETIEGNNNTVNYSVNGSVSGSGHGSNGPYGSSNGMNAGG---MNMGSDN 638
Query: 458 GQVSTPHSWPSLGSSGSCEVNLS---KADRREAALMKFKQKRKDRCFDKKIRYVNRKRLA 514
G + S SGS NL+ K +REAAL KF+QKRK+RCF KK+RY +RK+LA
Sbjct: 639 GAGKNGNGDGSGSGSGSGSGNLADENKISQREAALTKFRQKRKERCFRKKVRYQSRKKLA 698
Query: 515 ERRPRVRGQFVRK 527
E+RPRVRGQFVRK
Sbjct: 699 EQRPRVRGQFVRK 711
>At2g46790 hypothetical protein
Length = 454
Score = 103 bits (256), Expect = 3e-22
Identities = 52/140 (37%), Positives = 80/140 (57%), Gaps = 10/140 (7%)
Query: 27 DAFIDRSKVRILLCDDDPKSSEEVFTLLVKCSYQ----------VTPVRSARQVIDALNA 76
+ ++ ++ +R+LL + D + + + LL KC Y+ V + L
Sbjct: 29 EKYLPKTVLRVLLVESDYSTRQIITALLRKCCYKGRLVFPFISLFVAVSDGLAAWEVLKE 88
Query: 77 EGQYIDIILAEVDLPIKKGMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADY 136
+ ID+IL E+DLP G LL + + + IPVIMMS++D + +V+KC+ GAADY
Sbjct: 89 KSHNIDLILTELDLPSISGFALLALVMEHEACKNIPVIMMSSQDSIKMVLKCMLRGAADY 148
Query: 137 LVKPLRTNELLNLWTHMWRR 156
L+KP+R NEL NLW H+WRR
Sbjct: 149 LIKPMRKNELKNLWQHVWRR 168
Score = 44.7 bits (104), Expect = 1e-04
Identities = 19/23 (82%), Positives = 22/23 (95%)
Query: 484 RREAALMKFKQKRKDRCFDKKIR 506
+REAALMKF+ KRKDRCFDKK+R
Sbjct: 426 QREAALMKFRLKRKDRCFDKKVR 448
>At4g31920 predicted protein
Length = 552
Score = 93.2 bits (230), Expect = 3e-19
Identities = 53/179 (29%), Positives = 94/179 (51%), Gaps = 9/179 (5%)
Query: 35 VRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQYIDIILAEVDLPIKK 94
+R+L DDD + TLL +C Y VT A+ ++ L D+++++VD+P
Sbjct: 17 MRVLAVDDDQTCLRILQTLLQRCQYHVTTTNQAQTALELLRENKNKFDLVISDVDMPDMD 76
Query: 95 GMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKPLRTNELLNLWTHMW 154
G KLL+ + + +L PVIM+SA + V+K ++ GA DYL+KP+R EL N+W H+
Sbjct: 77 GFKLLELVGLEMDL---PVIMLSAHSDPKYVMKGVKHGACDYLLKPVRIEELKNIWQHVV 133
Query: 155 RRRRMLGLAEKNILSYDFDMVASDPSDANTNSTTLFSDDTDDRSKRSTNPEAAQQEPEV 213
R+ ++ +KN + + + AN + ++ ++ + AQ++P V
Sbjct: 134 RKSKL----KKN--KSNVSNGSGNCDKANRKRKEQYEEEEEEERGNDNDDPTAQKKPRV 186
>At1g67710 response regulator 11 (ARR11)
Length = 521
Score = 89.4 bits (220), Expect = 5e-18
Identities = 47/123 (38%), Positives = 76/123 (61%), Gaps = 3/123 (2%)
Query: 35 VRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQYIDIILAEVDLPIKK 94
+R+L+ DDDP + + +L KCSY+VT AR+ + L DI++++V++P
Sbjct: 11 LRVLVVDDDPTWLKILEKMLKKCSYEVTTCGLAREALRLLRERKDGYDIVISDVNMPDMD 70
Query: 95 GMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKPLRTNELLNLWTHMW 154
G KLL+++ + +L PVIMMS E V+K ++ GA DYL+KP+R EL +W H+
Sbjct: 71 GFKLLEHVGLELDL---PVIMMSVDGETSRVMKGVQHGACDYLLKPIRMKELKIIWQHVL 127
Query: 155 RRR 157
R++
Sbjct: 128 RKK 130
>At2g25180 putative two-component response regulator protein
Length = 573
Score = 89.0 bits (219), Expect = 7e-18
Identities = 55/183 (30%), Positives = 93/183 (50%), Gaps = 8/183 (4%)
Query: 35 VRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQYIDIILAEVDLPIKK 94
+R+L DDD + + +LL C Y VT A++ ++ L D+++++VD+P
Sbjct: 17 MRVLAVDDDQTCLKILESLLRHCQYHVTTTNQAQKALELLRENKNKFDLVISDVDMPDMD 76
Query: 95 GMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKPLRTNELLNLWTHMW 154
G KLL+ + + +L PVIM+SA + V+K + GA DYL+KP+R EL N+W H+
Sbjct: 77 GFKLLELVGLEMDL---PVIMLSAHSDPKYVMKGVTHGACDYLLKPVRIEELKNIWQHVV 133
Query: 155 R-----RRRMLGLAEKNILSYDFDMVASDPSDANTNSTTLFSDDTDDRSKRSTNPEAAQQ 209
R R +K S + + SDP++ N + D+ R N ++ Q
Sbjct: 134 RSRFDKNRGSNNNGDKRDGSGNEGVGNSDPNNGKGNRKRKDQYNEDEDEDRDDNDDSCAQ 193
Query: 210 EPE 212
+ +
Sbjct: 194 KKQ 196
>At5g58080 ARR2 - like protein
Length = 632
Score = 79.3 bits (194), Expect = 5e-15
Identities = 55/183 (30%), Positives = 92/183 (50%), Gaps = 25/183 (13%)
Query: 35 VRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQYIDIILAEVDLPIKK 94
+R+L DD+P ++ LL++C Y VT +R+ ++ L D+++++V++P
Sbjct: 18 MRVLAVDDNPTCLRKLEELLLRCKYHVTKTMESRKALEMLRENSNMFDLVISDVEMPDTD 77
Query: 95 GMKLLKY-LSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKPLRTNELLNLWTHM 153
G KLL+ L D +PVI S D V+K + GA DYLVKP+ EL N+W H
Sbjct: 78 GFKLLEIGLEMD-----LPVITHSDYDS---VMKGIIHGACDYLVKPVGLKELQNIWHH- 128
Query: 154 WRRRRMLGLAEKNILSYDFDMVASDPSDANTNSTTLFSD------DTDDRSKRSTNPEAA 207
+ +KNI SY ++ SD+ +++ D D DD + + E +
Sbjct: 129 --------VVKKNIKSY-AKLLPPSESDSVPSASRKRKDKVNDSGDEDDSDREEDDGEGS 179
Query: 208 QQE 210
+Q+
Sbjct: 180 EQD 182
>At4g16110 hypothetical protein
Length = 644
Score = 75.1 bits (183), Expect = 1e-13
Identities = 61/209 (29%), Positives = 99/209 (47%), Gaps = 33/209 (15%)
Query: 19 GGANSKSGDAFIDRSKVRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEG 78
GG+NS D F + +R+L+ DDDP + +L+ C Y R R
Sbjct: 17 GGSNS---DPF--PANLRVLVVDDDPTCLMILERMLMTCLY-----REQRAHCLCFGRTK 66
Query: 79 QYIDIILAEVDLPIKKGMKLLKYLSRDKELRRIP-------VIMMSAKDEVPIVVKCLRL 131
DI++++V +P G KLL+++ + +L I VI+MSA D +V+K +
Sbjct: 67 NGFDIVISDVHMPDMDGFKLLEHVGLEMDLPVINLNVLKPLVIVMSADDSKSVVLKGVTH 126
Query: 132 GAADYLVKPLRTNELLNLWTHMWRRRRMLGLAEKNILSY----------DFDMVASDPSD 181
GA DYL+KP+R L N+W H+ R++R E N+ + D D D
Sbjct: 127 GAVDYLIKPVRIEALKNIWQHVVRKKR----NEWNVSEHSGGSIEDTGGDRDRQQQHRED 182
Query: 182 ANTNSTTLFSDDTDDRSKRSTNPEAAQQE 210
A+ NS+++ ++ + RS R E +
Sbjct: 183 ADNNSSSV--NEGNGRSSRKRKEEEVDDQ 209
>At2g01760 putative two-component response regulator protein
Length = 382
Score = 73.6 bits (179), Expect = 3e-13
Identities = 43/125 (34%), Positives = 66/125 (52%), Gaps = 2/125 (1%)
Query: 33 SKVRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQYIDIILAEVDLPI 92
S +RIL+ DDD + +L++ YQVT A + L D++L++V +P
Sbjct: 9 SGLRILVVDDDTSCLFILEKMLLRLMYQVTICSQADVALTILRERKDSFDLVLSDVHMPG 68
Query: 93 KKGMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKPLRTNELLNLWTH 152
G LL+ + + +PVIMMS V+ + GA DYL+KP+R EL N+W H
Sbjct: 69 MNGYNLLQQVGLLE--MDLPVIMMSVDGRTTTVMTGINHGACDYLIKPIRPEELKNIWQH 126
Query: 153 MWRRR 157
+ RR+
Sbjct: 127 VVRRK 131
>At3g62670 putative protein
Length = 426
Score = 66.6 bits (161), Expect = 3e-11
Identities = 54/215 (25%), Positives = 102/215 (47%), Gaps = 22/215 (10%)
Query: 27 DAFIDRSKVRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQYIDIILA 86
+ F+ +S R+LL D SS + L+ + SYQVT S + + L ID+++
Sbjct: 34 EEFLTKSN-RVLLVGADSNSSLK--NLMTQYSYQVTKYESGEEAMAFLMKNKHEIDLVIW 90
Query: 87 EVDLPIKKGMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKPLRTNEL 146
+ +P G+ L + + +L PV++MS + + V++ ++ GA D+LVKP+ +
Sbjct: 91 DFHMPDINGLDALNIIGKQMDL---PVVIMSHEYKKETVMESIKYGACDFLVKPVSKEVI 147
Query: 147 LNLWTHMWRRRRM---------LGLAEKNILSYDFDMVASDPSDANTNSTTLFSDDTDDR 197
LW H++R+R G E + YD D+ + ++N + D +++
Sbjct: 148 AVLWRHVYRKRMSKSGLDKPGESGTVESDPDEYD-DLEQDNLYESNEEGSKNTCDHKEEK 206
Query: 198 SKRSTNPEAAQQEPEVN----VSISKAAAVIEELP 228
S T Q PE++ V++ K ++ + P
Sbjct: 207 S--PTKKPRMQWTPELHHKFEVAVEKMGSLEKAFP 239
>At2g46670 hypothetical protein
Length = 203
Score = 64.3 bits (155), Expect = 2e-10
Identities = 34/66 (51%), Positives = 43/66 (64%), Gaps = 20/66 (30%)
Query: 484 RREAALMKFKQKRKDRCFDKK--------------------IRYVNRKRLAERRPRVRGQ 523
+REAALMKF+ KRKDRCFDKK +RY +RK+LAE+RPRV+GQ
Sbjct: 131 QREAALMKFRLKRKDRCFDKKHLKQIQDQTDSMEMYKNGLQVRYQSRKKLAEQRPRVKGQ 190
Query: 524 FVRKLS 529
FVR ++
Sbjct: 191 FVRTVN 196
>At1g19050 putative protein
Length = 206
Score = 63.5 bits (153), Expect = 3e-10
Identities = 41/176 (23%), Positives = 84/176 (47%), Gaps = 13/176 (7%)
Query: 26 GDAFIDRSKVRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQY----- 80
GD + ++ +L DD + + LL S +VT V S + + L +G
Sbjct: 15 GDLTVTTPELHVLAVDDSIVDRKVIERLLRISSCKVTTVESGTRALQYLGLDGGKGASNL 74
Query: 81 ----IDIILAEVDLPIKKGMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADY 136
+++I+ + +P G LLK + R +PV++MS+++ +P + +CL+ GA ++
Sbjct: 75 KDLKVNLIVTDYSMPGLSGYDLLKKIKESSAFREVPVVIMSSENILPRIQECLKEGAEEF 134
Query: 137 LVKPLRTNELLNLWTHMWRRR----RMLGLAEKNILSYDFDMVASDPSDANTNSTT 188
L+KP++ ++ + + R ++L + K L D D +S D + ++
Sbjct: 135 LLKPVKLADVKRIKQLIMRNEAEECKILSHSNKRKLQEDSDTSSSSHDDTSIKDSS 190
>At1g74890 putative response regulator (two component
phosphotransfer signaling)
Length = 206
Score = 63.2 bits (152), Expect = 4e-10
Identities = 48/183 (26%), Positives = 89/183 (48%), Gaps = 16/183 (8%)
Query: 34 KVRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQY---------IDII 84
++ +L DD + + LL + +VT V S + + L +G +++I
Sbjct: 17 ELHVLAVDDSFVDRKVIERLLKISACKVTTVESGTRALQYLGLDGDNGSSGLKDLKVNLI 76
Query: 85 LAEVDLPIKKGMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKPLRTN 144
+ + +P G +LLK + LR IPV++MS+++ P + +C+ GA ++L+KP++
Sbjct: 77 VTDYSMPGLTGYELLKKIKESSALREIPVVIMSSENIQPRIEQCMIEGAEEFLLKPVKLA 136
Query: 145 ELLNLWTHMWR-----RRRMLGLAEKNILSYDFDMVASDPSDANTNSTTLFS--DDTDDR 197
++ L + R + L+ K IL D D S S ++++S+ S DD
Sbjct: 137 DVKRLKELIMRGGEAEEGKTKKLSPKRILQNDIDSSPSSSSTSSSSSSHDVSSLDDDTPS 196
Query: 198 SKR 200
SKR
Sbjct: 197 SKR 199
>At5g49240 putative protein
Length = 292
Score = 61.6 bits (148), Expect = 1e-09
Identities = 48/188 (25%), Positives = 91/188 (47%), Gaps = 11/188 (5%)
Query: 35 VRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQYIDIILAEVDLPIKK 94
+R+L+ D+DP + L K YQVT + + L D+ + +V+
Sbjct: 42 LRVLVFDEDPSYLLILERHLQKFQYQVTICNEVNKAMHTLRNHRNRFDLAMIQVN---NA 98
Query: 95 GMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKPLRTNELLNLWTHMW 154
+ ++LS +P+I++S D V V K + GAADYL+KP+R +L ++ H+
Sbjct: 99 EGDIFRFLSEIGSEMDLPIIIISEDDSVKSVKKWMINGAADYLIKPIRPEDLRIVFKHLV 158
Query: 155 RRRRMLGLAEKNILSYDFDMVASDPSDANTNSTTLFSDDTDDRSKRSTNPEAAQQEPEVN 214
++ R +++++ + + A + S + +ST ++SKRS+ EA E + +
Sbjct: 159 KKMR----ERRSVVTGEAEKAAGEKSSSVGDSTI----RNPNKSKRSSCLEAEVNEEDRH 210
Query: 215 VSISKAAA 222
+A A
Sbjct: 211 DHNDRACA 218
>At3g48100 response reactor 2 (ATRR2)
Length = 184
Score = 61.2 bits (147), Expect = 1e-09
Identities = 36/125 (28%), Positives = 66/125 (52%), Gaps = 10/125 (8%)
Query: 35 VRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQY----------IDII 84
+ +L DD + + LL S +VT V SA + + L +G+ I++I
Sbjct: 25 LHVLAVDDSMVDRKFIERLLRVSSCKVTVVDSATRALQYLGLDGENNSSVGFEDLKINLI 84
Query: 85 LAEVDLPIKKGMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKPLRTN 144
+ + +P G +LLK + R IPV++MS+++ +P + +CL GA D+L+KP++
Sbjct: 85 MTDYSMPGMTGYELLKKIKESSAFREIPVVIMSSENILPRIDRCLEEGAEDFLLKPVKLA 144
Query: 145 ELLNL 149
++ L
Sbjct: 145 DVKRL 149
>At1g59940 putative protein
Length = 231
Score = 61.2 bits (147), Expect = 1e-09
Identities = 46/195 (23%), Positives = 89/195 (45%), Gaps = 22/195 (11%)
Query: 30 IDRSKVRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQY--------- 80
+D +V +L DD + LL S +VT V S + ++ L +
Sbjct: 28 LDSDQVHVLAVDDSLVDRIVIERLLRITSCKVTAVDSGWRALEFLGLDDDKAAVEFDRLK 87
Query: 81 IDIILAEVDLPIKKGMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKP 140
+D+I+ + +P G +LLK + + +PV++MS+++ + + +CL GA D+L+KP
Sbjct: 88 VDLIITDYCMPGMTGYELLKKIKESTSFKEVPVVIMSSENVMTRIDRCLEEGAEDFLLKP 147
Query: 141 LRTNELLNLWTHMWRRRRMLGLAEKNILSYDFDMVASDPSDANTNSTTLFSDDT------ 194
++ ++ L +++ R ++ K L+ P + S+ SD T
Sbjct: 148 VKLADVKRLRSYLTRDVKVAAEGNKRKLT-----TPPPPPPLSATSSMESSDSTVESPLS 202
Query: 195 --DDRSKRSTNPEAA 207
DD + +PE+A
Sbjct: 203 MVDDEDSLTMSPESA 217
>At1g10470 putative response regulator 3
Length = 259
Score = 60.8 bits (146), Expect = 2e-09
Identities = 35/133 (26%), Positives = 69/133 (51%), Gaps = 9/133 (6%)
Query: 30 IDRSKVRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNAEGQY--------- 80
+D +V +L DD + LL S +VT V S + ++ L + +
Sbjct: 29 LDLDEVHVLAVDDSLVDRIVIERLLRITSCKVTAVDSGWRALEFLGLDNEKASAEFDRLK 88
Query: 81 IDIILAEVDLPIKKGMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKP 140
+D+I+ + +P G +LLK + R +PV++MS+++ + + +CL GA D+L+KP
Sbjct: 89 VDLIITDYCMPGMTGYELLKKIKESSNFREVPVVIMSSENVLTRIDRCLEEGAQDFLLKP 148
Query: 141 LRTNELLNLWTHM 153
++ ++ L +H+
Sbjct: 149 VKLADVKRLRSHL 161
>At5g62920 response regulator 6 (ARR6)
Length = 186
Score = 58.9 bits (141), Expect = 7e-09
Identities = 34/124 (27%), Positives = 65/124 (52%), Gaps = 9/124 (7%)
Query: 35 VRILLCDDDPKSSEEVFTLLVKCSYQVTPVRSARQVIDALNA---------EGQYIDIIL 85
+ +L DD + + LL S +VT V SA + + L E +++I+
Sbjct: 25 LHVLAVDDSHVDRKFIERLLRVSSCKVTVVDSATRALQYLGLDVEEKSVGFEDLKVNLIM 84
Query: 86 AEVDLPIKKGMKLLKYLSRDKELRRIPVIMMSAKDEVPIVVKCLRLGAADYLVKPLRTNE 145
+ +P G +LLK + R +PV++MS+++ +P + +CL GA D+L+KP++ ++
Sbjct: 85 TDYSMPGMTGYELLKKIKESSAFREVPVVIMSSENILPRIDRCLEEGAEDFLLKPVKLSD 144
Query: 146 LLNL 149
+ L
Sbjct: 145 VKRL 148
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.129 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,936,638
Number of Sequences: 26719
Number of extensions: 568052
Number of successful extensions: 2374
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 71
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 2258
Number of HSP's gapped (non-prelim): 113
length of query: 565
length of database: 11,318,596
effective HSP length: 104
effective length of query: 461
effective length of database: 8,539,820
effective search space: 3936857020
effective search space used: 3936857020
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0093b.4


