

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0093a.1
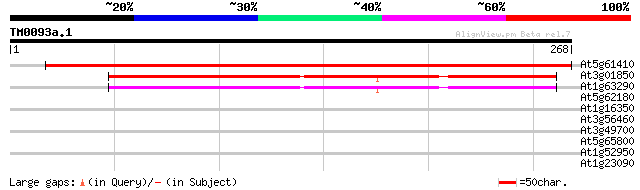
(268 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g61410 ribulose-5-phosphate-3-epimerase 463 e-131
At3g01850 D-ribulose-5-phosphate 3-epimerase like protein 158 3e-39
At1g63290 D-ribulose-5-phosphate like protein 154 4e-38
At5g62180 putative protein 32 0.27
At1g16350 inosine-5'-monophosphate dehydrogenase 32 0.46
At3g56460 quinone reductase-like protein 28 3.9
At3g49700 1-aminocyclopropane-1-carboxylate synthase -like protein 28 5.1
At5g65800 1-aminocyclopropane-1-carboxylate synthase ACS5 (pir||... 28 6.7
At1g52950 replication protein A1, putative 28 6.7
At1g23090 putative sulphate transporter protein 27 8.7
>At5g61410 ribulose-5-phosphate-3-epimerase
Length = 281
Score = 463 bits (1192), Expect = e-131
Identities = 233/251 (92%), Positives = 242/251 (95%)
Query: 18 PRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHV 77
P S +FSRR+ IVKASSRVD+FSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHV
Sbjct: 30 PTSFSFSRRRTHGIVKASSRVDRFSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHV 89
Query: 78 DVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQS 137
DVMDGRFVPNITIGPL+VDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVS+HCEQ
Sbjct: 90 DVMDGRFVPNITIGPLVVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSVHCEQQ 149
Query: 138 STIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQ 197
STIHLHRTVNQ+KSLGAKAGVVLNPGTPLSAIEYVLD+VDLVLIMSVNPGFGGQSFIESQ
Sbjct: 150 STIHLHRTVNQIKSLGAKAGVVLNPGTPLSAIEYVLDMVDLVLIMSVNPGFGGQSFIESQ 209
Query: 198 VKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKG 257
VKKISDLR++CAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKG
Sbjct: 210 VKKISDLRKMCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKG 269
Query: 258 IKTSKRPEPVA 268
IK SKRP VA
Sbjct: 270 IKASKRPAAVA 280
>At3g01850 D-ribulose-5-phosphate 3-epimerase like protein
Length = 225
Score = 158 bits (400), Expect = 3e-39
Identities = 85/217 (39%), Positives = 132/217 (60%), Gaps = 8/217 (3%)
Query: 48 VSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLD 107
++PS+LS++FA L + + G +W+H+D+MDG FVPN+TIG ++++LR T+ LD
Sbjct: 6 IAPSMLSSDFANLAAEANRMIDLGANWLHMDIMDGHFVPNLTIGAPVIESLRKHTNAYLD 65
Query: 108 VHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLS 167
HLM+ P V KAGA + H E + + + V ++KS G + GV L PGTP+
Sbjct: 66 CHLMVTNPMDYVAQMAKAGASGFTFHVEVAQD-NWQQLVEKIKSTGMRPGVALKPGTPVE 124
Query: 168 AIEYVLD---VVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVT 224
+ +++ V++VL+M+V PGFGGQ F+ + K+ LR +K I+VDGG+
Sbjct: 125 QVYPLVEGTTPVEMVLVMTVEPGFGGQKFMPDMMDKVRALR----QKYPTLDIQVDGGLG 180
Query: 225 PANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTS 261
P+ AGAN +VAGS+VFGA + + I ++TS
Sbjct: 181 PSTIDTAAAAGANCIVAGSSVFGAPEPGDVISLLRTS 217
>At1g63290 D-ribulose-5-phosphate like protein
Length = 227
Score = 154 bits (390), Expect = 4e-38
Identities = 85/217 (39%), Positives = 129/217 (59%), Gaps = 8/217 (3%)
Query: 48 VSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLD 107
++PS+LS++FA L + K + G +W+H+D+MDG FV N+TIG ++++LR T+ LD
Sbjct: 8 IAPSMLSSDFANLAAEAKRMIDLGANWLHMDIMDGHFVSNLTIGAPVIESLRKHTNAYLD 67
Query: 108 VHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLS 167
HLM+ P V KAGA + H E + + V ++K+ G + GV L PGTP+
Sbjct: 68 CHLMVTNPMDYVDQMAKAGASGFTFHVEVAQE-NWQELVKKIKAAGMRPGVALKPGTPVE 126
Query: 168 AIEYVLD---VVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVT 224
+ +++ V++VL+M+V PGFGGQ F+ S + K+ LR K IEVDGG+
Sbjct: 127 QVYPLVEGTNPVEMVLVMTVEPGFGGQKFMPSMMDKVRALR----NKYPTLDIEVDGGLG 182
Query: 225 PANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTS 261
P+ AGAN +VAGS+VFGA + I ++ S
Sbjct: 183 PSTIDAAAAAGANCIVAGSSVFGAPKPGDVISLLRAS 219
>At5g62180 putative protein
Length = 327
Score = 32.3 bits (72), Expect = 0.27
Identities = 24/92 (26%), Positives = 43/92 (46%), Gaps = 3/92 (3%)
Query: 115 PEQRVPDFIKAGADIVS-IHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVL 173
PE R+P G + + I I H + V +G AG L L +++ V
Sbjct: 125 PEHRLPAAYDDGVEALDWIKTSDDEWIKSHADFSNVFLMGTSAGGNLAYNVGLRSVDSVS 184
Query: 174 DVVDLVL--IMSVNPGFGGQSFIESQVKKISD 203
D+ L + ++ +P FGG+ ES+++ ++D
Sbjct: 185 DLSPLQIRGLILHHPFFGGEERSESEIRLMND 216
>At1g16350 inosine-5'-monophosphate dehydrogenase
Length = 502
Score = 31.6 bits (70), Expect = 0.46
Identities = 34/146 (23%), Positives = 63/146 (42%), Gaps = 18/146 (12%)
Query: 116 EQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVV-LNPGTPLSAIEYVLD 174
++R+ +KAGA++V + Q ++I+ + VK+ + VV N T A +
Sbjct: 247 KERLEHLVKAGANVVVLDSSQGNSIYQLEMIKYVKNTYPELDVVGGNVVTMYQAENLIKA 306
Query: 175 VVDLVLIMSVNPGFGGQSFI---------ESQVKKISDLRRLCAEKGVNPWIEVDGGVT- 224
VD + G G S Q + + L A+ GV + DGG++
Sbjct: 307 GVD-----GLRVGMGSGSICTTQEVCAVGRGQATAVYKVSTLAAQHGVP--VIADGGISN 359
Query: 225 PANAYKVIEAGANALVAGSAVFGAKD 250
+ K + GA+ ++ GS + G+ +
Sbjct: 360 SGHIVKALVLGASTVMMGSFLAGSTE 385
>At3g56460 quinone reductase-like protein
Length = 348
Score = 28.5 bits (62), Expect = 3.9
Identities = 39/134 (29%), Positives = 57/134 (42%), Gaps = 13/134 (9%)
Query: 121 DFIKAGADIVSIHCEQSSTIHLHR-TVNQVKS-LGAKAGVVLNP-------GTPLSAIEY 171
D + A A V+ + +H R T QV LGA GV L G + A+
Sbjct: 124 DMVAAAALPVAFGTSHVALVHRARLTSGQVLLVLGAAGGVGLAAVQIGKVCGAIVIAVAR 183
Query: 172 VLDVVDLVLIMSVNPGFG-GQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYK 230
+ + L+ M V+ G + S VK+ R+L KGV+ + GG + K
Sbjct: 184 GTEKIQLLKSMGVDHVVDLGTENVISSVKEFIKTRKL---KGVDVLYDPVGGKLTKESMK 240
Query: 231 VIEAGANALVAGSA 244
V++ GA LV G A
Sbjct: 241 VLKWGAQILVIGFA 254
>At3g49700 1-aminocyclopropane-1-carboxylate synthase -like protein
Length = 470
Score = 28.1 bits (61), Expect = 5.1
Identities = 18/53 (33%), Positives = 24/53 (44%), Gaps = 7/53 (13%)
Query: 124 KAGADIVSIHCEQSSTIH-----LHRTVNQVKSLGAKAGVVL--NPGTPLSAI 169
+ GA+IV IHC S+ L + Q + L K VL NP PL +
Sbjct: 155 RTGAEIVPIHCSSSNGFQITESALQQAYQQAQKLDLKVKGVLVTNPSNPLGTM 207
>At5g65800 1-aminocyclopropane-1-carboxylate synthase ACS5
(pir||S71174)
Length = 470
Score = 27.7 bits (60), Expect = 6.7
Identities = 18/50 (36%), Positives = 23/50 (46%), Gaps = 7/50 (14%)
Query: 124 KAGADIVSIHCEQSSTIH-----LHRTVNQVKSLGAKAGVVL--NPGTPL 166
+ GA+IV IHC S+ L + Q + L K VL NP PL
Sbjct: 155 RTGAEIVPIHCSSSNGFQITESALQQAYQQAQKLDLKVKGVLVTNPSNPL 204
>At1g52950 replication protein A1, putative
Length = 566
Score = 27.7 bits (60), Expect = 6.7
Identities = 16/53 (30%), Positives = 28/53 (52%), Gaps = 3/53 (5%)
Query: 74 WIHVDVMDGRFVPNITIGPLIVDAL--RPVTDLPLDVHLMIVEPEQRVPDFIK 124
W+H+D+MD + + V+ + P T+L LDV ++ Q +PD +K
Sbjct: 347 WLHLDIMDNTGESKLMLFDSFVEPIIGCPATEL-LDVTNEQIDEPQPLPDVVK 398
>At1g23090 putative sulphate transporter protein
Length = 631
Score = 27.3 bits (59), Expect = 8.7
Identities = 10/31 (32%), Positives = 19/31 (61%)
Query: 84 FVPNITIGPLIVDALRPVTDLPLDVHLMIVE 114
+ PN+ +G +IV A+ + DLP H+ ++
Sbjct: 407 YTPNVVLGAIIVTAVIGLIDLPAACHIWKID 437
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,645,927
Number of Sequences: 26719
Number of extensions: 226822
Number of successful extensions: 556
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 546
Number of HSP's gapped (non-prelim): 10
length of query: 268
length of database: 11,318,596
effective HSP length: 98
effective length of query: 170
effective length of database: 8,700,134
effective search space: 1479022780
effective search space used: 1479022780
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0093a.1


