

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
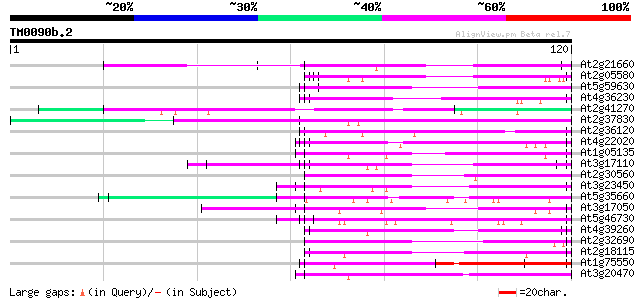
Query= TM0090b.2
(120 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g21660 glycine-rich RNA binding protein 57 1e-09
At2g05580 unknown protein 57 2e-09
At5g59630 glycine-rich protein - like 56 2e-09
At4g36230 putative glycine-rich cell wall protein 56 2e-09
At2g41270 pseudogene 56 2e-09
At2g37830 hypothetical protein 55 4e-09
At2g36120 unknown protein 55 4e-09
At4g22020 glycine-rich protein 55 7e-09
At1g05135 unknown protein 55 7e-09
At3g17110 hypothetical protein 54 2e-08
At2g30560 putative glycine-rich protein 54 2e-08
At3g23450 hypothetical protein 53 2e-08
At5g35660 unknown protein 53 3e-08
At3g17050 unknown protein 51 1e-07
At5g46730 unknown protein 50 2e-07
At4g39260 glycine-rich protein (clone AtGRP8) 50 2e-07
At2g32690 unknown protein 50 2e-07
At2g18115 putative protein 48 7e-07
At1g75550 hypothetical protein 48 7e-07
At3g20470 glycine-rich protein, putative 47 1e-06
>At2g21660 glycine-rich RNA binding protein
Length = 176
Score = 57.4 bits (137), Expect = 1e-09
Identities = 40/101 (39%), Positives = 45/101 (43%), Gaps = 34/101 (33%)
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSS-GG 79
GGG SGGGG RR +GGGG GGG G SS GG
Sbjct: 108 GGGYSGGGGSYGGGGGRR------------------------EGGGGYSGGGGGYSSRGG 143
Query: 80 GARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G ++ G RR+ GGG G +GGG GG GGGGG
Sbjct: 144 GGGSYGGGRRE---------GGGGYGGGEGGGYGGSGGGGG 175
Score = 46.6 bits (109), Expect = 2e-06
Identities = 26/57 (45%), Positives = 28/57 (48%), Gaps = 10/57 (17%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG G S GGG+ G RR +GGG S GGG GGGGG
Sbjct: 100 GGGYRSGGGGGYSGGGGSYGGGGGRR----------EGGGGYSGGGGGYSSRGGGGG 146
Score = 39.7 bits (91), Expect = 2e-04
Identities = 25/65 (38%), Positives = 28/65 (42%), Gaps = 19/65 (29%)
Query: 54 AVVVEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG 113
++ V G GG GGG G GGG R+ GGG G GGGS
Sbjct: 78 SITVNEAQSRGSGG--GGGHRGGGGGGYRS-----------------GGGGGYSGGGGSY 118
Query: 114 GGGGG 118
GGGGG
Sbjct: 119 GGGGG 123
Score = 31.2 bits (69), Expect = 0.086
Identities = 18/52 (34%), Positives = 19/52 (35%), Gaps = 17/52 (32%)
Query: 68 NDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
N+ G GGG R GGG G GGG G GGGG
Sbjct: 82 NEAQSRGSGGGGGHRG-----------------GGGGGYRSGGGGGYSGGGG 116
Score = 28.5 bits (62), Expect = 0.56
Identities = 11/23 (47%), Positives = 12/23 (51%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWW 85
+GGGG GG GG G G W
Sbjct: 154 EGGGGYGGGEGGGYGGSGGGGGW 176
>At2g05580 unknown protein
Length = 302
Score = 56.6 bits (135), Expect = 2e-09
Identities = 30/57 (52%), Positives = 31/57 (53%), Gaps = 10/57 (17%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG+ GGG GG GGG R G R GGGSG GGGSG GGGGGG
Sbjct: 254 GQGGHKGGGGGGHVGGGGRGGGGGGR----------GGGGSGGGGGGGSGRGGGGGG 300
Score = 48.9 bits (115), Expect = 4e-07
Identities = 25/54 (46%), Positives = 26/54 (47%)
Query: 66 GGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GG GGG GG GGG G + Q GGG G GGG GGGGGG
Sbjct: 225 GGGGGGGQGGHKGGGGGGQGGGGHKGGGGGQGGHKGGGGGGHVGGGGRGGGGGG 278
Score = 48.5 bits (114), Expect = 5e-07
Identities = 26/55 (47%), Positives = 27/55 (48%), Gaps = 1/55 (1%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GG GGG GG GGG R G + Q GGG G GG GGGGGGG
Sbjct: 178 GGQKGGGGQGGQKGGGGRGQGGMKGGGGGG-QGGHKGGGGGGGQGGHKGGGGGGG 231
Score = 46.6 bits (109), Expect = 2e-06
Identities = 25/57 (43%), Positives = 25/57 (43%), Gaps = 14/57 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG GG GGG G GGG G GG GGGGG GG
Sbjct: 215 GGGGGQGGHKGGGGGGGQGGHKG--------------GGGGGQGGGGHKGGGGGQGG 257
Score = 46.6 bits (109), Expect = 2e-06
Identities = 25/57 (43%), Positives = 25/57 (43%), Gaps = 15/57 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG GG GGG Q GGG G GG GGGGGG G
Sbjct: 203 GGGGGQGGHKGGGGGGG---------------QGGHKGGGGGGGQGGHKGGGGGGQG 244
Score = 46.2 bits (108), Expect = 3e-06
Identities = 26/57 (45%), Positives = 28/57 (48%), Gaps = 1/57 (1%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG GG GG + G+ Q Q GGG G GG GGGGGG G
Sbjct: 154 GGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQKGGG-GRGQGGMKGGGGGGQG 209
Score = 45.8 bits (107), Expect = 3e-06
Identities = 24/56 (42%), Positives = 28/56 (49%), Gaps = 2/56 (3%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG GGG GG GGG + G++ R + GGG G G GGGGG GG
Sbjct: 169 GGQKGGGGQGGQKGGGGQG--GQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGGQGG 222
Score = 45.4 bits (106), Expect = 4e-06
Identities = 25/57 (43%), Positives = 27/57 (46%), Gaps = 2/57 (3%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG G GGG + G + Q GGG G GGG GGGGG G
Sbjct: 202 GGGGGGQGGHKGGGGGGGQG--GHKGGGGGGGQGGHKGGGGGGQGGGGHKGGGGGQG 256
Score = 43.9 bits (102), Expect = 1e-05
Identities = 27/65 (41%), Positives = 29/65 (44%), Gaps = 8/65 (12%)
Query: 64 GGGGNDGG---GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG-----G 115
G GG GG G GG GGG G++ GGG G GGG GG G
Sbjct: 83 GQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGQGGQKGGGGGQGGQKGGGGGGQGGQKG 142
Query: 116 GGGGG 120
GGGGG
Sbjct: 143 GGGGG 147
Score = 43.9 bits (102), Expect = 1e-05
Identities = 24/57 (42%), Positives = 27/57 (47%), Gaps = 2/57 (3%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G G GGG GG GGG G++ + Q GGG G GGG GG GGG
Sbjct: 139 GQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQ--KGGGGQGGQKGGGGQGGQKGGG 193
Score = 41.2 bits (95), Expect = 8e-05
Identities = 26/63 (41%), Positives = 28/63 (44%), Gaps = 15/63 (23%)
Query: 64 GGGGNDGGGSG-GSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG-----GGG 117
G GG GGG G G GG G++ GGG G GGG GG GGG
Sbjct: 105 GQGGQKGGGGGQGGQKGGGGGQGGQKG---------GGGGGQGGQKGGGGGGQGGQKGGG 155
Query: 118 GGG 120
GGG
Sbjct: 156 GGG 158
Score = 40.8 bits (94), Expect = 1e-04
Identities = 24/59 (40%), Positives = 26/59 (43%), Gaps = 18/59 (30%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG--GGGGGGG 120
G GG+ GGG GG GGG + GGG G GGG G GGGG G
Sbjct: 231 GQGGHKGGGGGGQGGGGHKG----------------GGGGQGGHKGGGGGGHVGGGGRG 273
Score = 40.8 bits (94), Expect = 1e-04
Identities = 25/59 (42%), Positives = 28/59 (47%), Gaps = 3/59 (5%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG--GGGG 120
G G GGG GG GGG G++ + GGG G GGG G GG GGGG
Sbjct: 128 GQKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGG-GQGGQKGGGG 185
Score = 38.9 bits (89), Expect = 4e-04
Identities = 25/60 (41%), Positives = 27/60 (44%), Gaps = 11/60 (18%)
Query: 64 GGGGNDGGGSG-GSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGG--GGG 120
G GG GGG G G GG G ++ GGG G GGG GG GG GGG
Sbjct: 115 GQGGQKGGGGGQGGQKGGGGGGQGGQKG--------GGGGGQGGQKGGGGGGQGGQKGGG 166
Score = 38.9 bits (89), Expect = 4e-04
Identities = 21/54 (38%), Positives = 23/54 (41%), Gaps = 13/54 (24%)
Query: 67 GNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G G G GG GGG G++ GG G GG GGGGG GG
Sbjct: 78 GGIGRGQGGQKGGGGGGQGGQK-------------GGGGGGQGGQKGGGGGQGG 118
Score = 37.7 bits (86), Expect = 0.001
Identities = 24/59 (40%), Positives = 24/59 (40%), Gaps = 14/59 (23%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG--GGGG 120
GG G GG G GGG G GGG G GGG G GG GGGG
Sbjct: 78 GGIGRGQGGQKGGGGGGQGGQKG------------GGGGGQGGQKGGGGGQGGQKGGGG 124
Score = 37.4 bits (85), Expect = 0.001
Identities = 26/63 (41%), Positives = 30/63 (47%), Gaps = 11/63 (17%)
Query: 65 GGGNDG--GGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG-----GGG 117
GG G GG GG GGG + G++ + Q GGG G GGG G GGG
Sbjct: 149 GGQKGGGGGGQGGQKGGGGQG--GQKGGGGQGGQ--KGGGGQGGQKGGGGRGQGGMKGGG 204
Query: 118 GGG 120
GGG
Sbjct: 205 GGG 207
>At5g59630 glycine-rich protein - like
Length = 253
Score = 56.2 bits (134), Expect = 2e-09
Identities = 28/57 (49%), Positives = 28/57 (49%), Gaps = 14/57 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 75 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 117
Score = 56.2 bits (134), Expect = 2e-09
Identities = 28/57 (49%), Positives = 28/57 (49%), Gaps = 14/57 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 72 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 114
Score = 56.2 bits (134), Expect = 2e-09
Identities = 28/57 (49%), Positives = 28/57 (49%), Gaps = 14/57 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 76 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 118
Score = 56.2 bits (134), Expect = 2e-09
Identities = 28/57 (49%), Positives = 28/57 (49%), Gaps = 14/57 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 78 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 120
Score = 52.4 bits (124), Expect = 4e-08
Identities = 26/58 (44%), Positives = 28/58 (47%), Gaps = 14/58 (24%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+ GG + GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 66 NSGGSSGGGGGGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 109
Score = 48.5 bits (114), Expect = 5e-07
Identities = 26/54 (48%), Positives = 26/54 (48%), Gaps = 14/54 (25%)
Query: 67 GNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G D SGGSSGGG G GGG G GGG GGGGGGGG
Sbjct: 61 GLDSSNSGGSSGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 100
Score = 45.8 bits (107), Expect = 3e-06
Identities = 24/58 (41%), Positives = 27/58 (46%), Gaps = 14/58 (24%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+G ++ GGS G GGG G GGG G GGG GGGGGGGG
Sbjct: 60 NGLDSSNSGGSSGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 103
Score = 36.6 bits (83), Expect = 0.002
Identities = 15/22 (68%), Positives = 16/22 (72%)
Query: 99 DDGGGSGSDDGGGSGGGGGGGG 120
DD G S+ GG SGGGGGGGG
Sbjct: 58 DDNGLDSSNSGGSSGGGGGGGG 79
Score = 33.1 bits (74), Expect = 0.023
Identities = 14/27 (51%), Positives = 15/27 (54%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQ 90
GGGG GGG GG GGG G + Q
Sbjct: 97 GGGGGGGGGGGGGGGGGGGGGGGGKLQ 123
>At4g36230 putative glycine-rich cell wall protein
Length = 221
Score = 56.2 bits (134), Expect = 2e-09
Identities = 29/57 (50%), Positives = 30/57 (51%), Gaps = 10/57 (17%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG SG G+ R R GGG G GGG GGGGGGGG
Sbjct: 131 GGGGGGGGGGGGGSGNGSG----------RGRGGGGGGGGGGGGGGGGGGGGGGGGG 177
Score = 52.8 bits (125), Expect = 3e-08
Identities = 29/66 (43%), Positives = 30/66 (44%), Gaps = 9/66 (13%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGS---------GG 114
GGGG GGG GS GGG G + R GGG G GGG GG
Sbjct: 95 GGGGGGGGGGQGSGGGGGEGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSGRGRGG 154
Query: 115 GGGGGG 120
GGGGGG
Sbjct: 155 GGGGGG 160
Score = 48.5 bits (114), Expect = 5e-07
Identities = 27/64 (42%), Positives = 31/64 (48%), Gaps = 18/64 (28%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDD--------GGGSGGGG 116
GGG GGG GG GGG + G + GG+G D+ GGG GGGG
Sbjct: 89 GGGGGGGGGGGGGGGGQGSGGGGG----------EGNGGNGKDNSHKRNKSSGGGGGGGG 138
Query: 117 GGGG 120
GGGG
Sbjct: 139 GGGG 142
Score = 47.4 bits (111), Expect = 1e-06
Identities = 26/57 (45%), Positives = 26/57 (45%), Gaps = 4/57 (7%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GGS G R G GGG G GGG GGGGG G
Sbjct: 132 GGGGGGGGGGGGSGNGSGRGRGGGGGGGGGG----GGGGGGGGGGGGGGGGGGGSDG 184
Score = 44.3 bits (103), Expect = 1e-05
Identities = 25/58 (43%), Positives = 26/58 (44%), Gaps = 22/58 (37%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+G G GGG GG GGG GGG G GGG GGGGGGGG
Sbjct: 146 NGSGRGRGGGGGGGGGGGG-------------------GGGGG---GGGGGGGGGGGG 181
Score = 42.7 bits (99), Expect = 3e-05
Identities = 26/58 (44%), Positives = 26/58 (44%), Gaps = 20/58 (34%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSD-DGGGSGGGGGGGG 120
GGGG GGG GG GGG GGG GSD GGG G G G GG
Sbjct: 159 GGGGGGGGGGGGGGGGGG-------------------GGGGGSDGKGGGWGFGFGWGG 197
Score = 40.8 bits (94), Expect = 1e-04
Identities = 23/57 (40%), Positives = 23/57 (40%), Gaps = 14/57 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GG G G G G G GG G
Sbjct: 157 GGGGGGGGGGGGGGGGGGGGGGG--------------GGSDGKGGGWGFGFGWGGDG 199
Score = 39.7 bits (91), Expect = 2e-04
Identities = 23/56 (41%), Positives = 23/56 (41%), Gaps = 15/56 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG GGG DG G G G G GG G GG
Sbjct: 162 GGGGGGGGGGGGGGGGGGGG---------------SDGKGGGWGFGFGWGGDGPGG 202
Score = 34.3 bits (77), Expect = 0.010
Identities = 13/23 (56%), Positives = 14/23 (60%)
Query: 98 CDDGGGSGSDDGGGSGGGGGGGG 120
C+ G G GGG GGGGGG G
Sbjct: 84 CESRAGGGGGGGGGGGGGGGGQG 106
>At2g41270 pseudogene
Length = 280
Score = 56.2 bits (134), Expect = 2e-09
Identities = 45/123 (36%), Positives = 49/123 (39%), Gaps = 29/123 (23%)
Query: 21 GGGDSGGGGWC-------WWRRWRRSVV--VVAVTVVVIVVEAVVVEAVVVDGGGGNDGG 71
GGG GG G C WWR R + VV VE VE GG GG
Sbjct: 123 GGGGGGGRGGCRWGCCGGWWRGRCRYCCRSQAEASEVVETVEPNDVEPQQ----GGRGGG 178
Query: 72 GSGGSSGGGARAWWGRRRQWWRWR-QWCDDGGGSGSD-------------DGGGSGGGGG 117
G GG GG R WG WWR R ++C S+ GG GGGGG
Sbjct: 179 GGGGGGRGGCR--WGCCGGWWRGRCRYCCRSQAEASEVVETVEPNDVEPQQGGRGGGGGG 236
Query: 118 GGG 120
GGG
Sbjct: 237 GGG 239
Score = 52.0 bits (123), Expect = 5e-08
Identities = 48/137 (35%), Positives = 53/137 (38%), Gaps = 30/137 (21%)
Query: 7 VAAAVVVAMLAVVEGGGDSGGGGWC-------WWR---RWRRSVVVVAVTVVVIVVEAVV 56
+A A V A VVE D GG G C WWR R+ A VV V V
Sbjct: 54 IAEAQVEANDPVVEPQQDWGGRGGCRWGCCGGWWRGRCRYCCRSQAEASEVVETVEPNDV 113
Query: 57 VEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWR-QWCDDGGGSGSD-------- 107
GGGG GGG GG WG WWR R ++C S+
Sbjct: 114 EPQQGGRGGGGGGGGGRGGCR-------WGCCGGWWRGRCRYCCRSQAEASEVVETVEPN 166
Query: 108 ----DGGGSGGGGGGGG 120
GG GGGGGGGG
Sbjct: 167 DVEPQQGGRGGGGGGGG 183
Score = 43.5 bits (101), Expect = 2e-05
Identities = 33/84 (39%), Positives = 34/84 (40%), Gaps = 15/84 (17%)
Query: 21 GGGDSGGGGWC-------WWRRWRRSVV--VVAVTVVVIVVEAVVVEAVVVDGGGGNDGG 71
GGG GG G C WWR R + VV VE VE GG GG
Sbjct: 178 GGGGGGGRGGCRWGCCGGWWRGRCRYCCRSQAEASEVVETVEPNDVEPQQ----GGRGGG 233
Query: 72 GSGGSSGGGARAWWGRRRQWWRWR 95
G GG GG R WG WWR R
Sbjct: 234 GGGGGGRGGCR--WGCCGGWWRGR 255
>At2g37830 hypothetical protein
Length = 106
Score = 55.5 bits (132), Expect = 4e-09
Identities = 31/85 (36%), Positives = 40/85 (46%)
Query: 36 WRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWR 95
W ++ +A V V++ A + A GGGG+ GGG GG GG +
Sbjct: 4 WIVIIIAIASFVGVLIFLAALTGAGGKGGGGGDGGGGGGGGEGGDGGGGEDGGGEDVEIG 63
Query: 96 QWCDDGGGSGSDDGGGSGGGGGGGG 120
+ GG G GGG GGGGGGGG
Sbjct: 64 DGANGGGFGGDGGGGGFGGGGGGGG 88
Score = 53.1 bits (126), Expect = 2e-08
Identities = 38/124 (30%), Positives = 47/124 (37%), Gaps = 47/124 (37%)
Query: 1 MVVAMMVAAAVVVAMLAVVEGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAV 60
+++A+ V++ + A+ GG GGGG
Sbjct: 7 IIIAIASFVGVLIFLAALTGAGGKGGGGG------------------------------- 35
Query: 61 VVDGGGGNDGG----GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG 116
DGGGG GG G GG GGG G +GGG G D GGG GGG
Sbjct: 36 --DGGGGGGGGEGGDGGGGEDGGGEDVEIGDGA----------NGGGFGGDGGGGGFGGG 83
Query: 117 GGGG 120
GGGG
Sbjct: 84 GGGG 87
Score = 51.6 bits (122), Expect = 6e-08
Identities = 30/71 (42%), Positives = 30/71 (42%), Gaps = 26/71 (36%)
Query: 63 DGGGGNDGGGS-------------GGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDG 109
DGGGG DGGG GG GGG G GGG G G
Sbjct: 48 DGGGGEDGGGEDVEIGDGANGGGFGGDGGGGGFGGGG-------------GGGGDGGGGG 94
Query: 110 GGSGGGGGGGG 120
GG GGGGGGGG
Sbjct: 95 GGGGGGGGGGG 105
>At2g36120 unknown protein
Length = 255
Score = 55.5 bits (132), Expect = 4e-09
Identities = 28/57 (49%), Positives = 31/57 (54%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG SG G A +G + GG G GGG+GGGGGGGG
Sbjct: 95 GGGGGPGGGYGGGSGEGGGAGYGG-----------GEAGGHGGGGGGGAGGGGGGGG 140
Score = 54.3 bits (129), Expect = 9e-09
Identities = 29/58 (50%), Positives = 32/58 (55%), Gaps = 2/58 (3%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGGG G G GG+ GGG A G + GGG G GGGSGGGGGGGG
Sbjct: 175 NGGGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAGGHGGGGGG--GGGSGGGGGGGG 230
Score = 53.5 bits (127), Expect = 2e-08
Identities = 27/57 (47%), Positives = 29/57 (50%), Gaps = 14/57 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG G GA +G GGG+G GGG GG GGGGG
Sbjct: 138 GGGGAHGGGYGGGQGAGAGGGYG--------------GGGAGGHGGGGGGGNGGGGG 180
Score = 52.4 bits (124), Expect = 4e-08
Identities = 28/61 (45%), Positives = 30/61 (48%), Gaps = 4/61 (6%)
Query: 64 GGGGNDGGGSGGSSGGGARAWW----GRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGGN GGG GGS GGA G + + GG G GG GGGGGGG
Sbjct: 171 GGGGNGGGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAGGHGGGGGGGGGSGGGGGGGG 230
Query: 120 G 120
G
Sbjct: 231 G 231
Score = 51.2 bits (121), Expect = 8e-08
Identities = 27/58 (46%), Positives = 29/58 (49%), Gaps = 2/58 (3%)
Query: 64 GGG--GNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG G GGG GG +GGG G + Q GGG G GG GGGGGGG
Sbjct: 117 GGGEAGGHGGGGGGGAGGGGGGGGGAHGGGYGGGQGAGAGGGYGGGGAGGHGGGGGGG 174
Score = 47.0 bits (110), Expect = 2e-06
Identities = 24/57 (42%), Positives = 26/57 (45%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG GG G A G + G G+G GGG GGGG GGG
Sbjct: 169 GGGGGGNGGGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAGGHGGGGGGGGGSGGG 225
Score = 45.1 bits (105), Expect = 6e-06
Identities = 27/59 (45%), Positives = 28/59 (46%), Gaps = 13/59 (22%)
Query: 64 GGGGNDGGGSG---GSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGGSG GS GG G GGG G GGG+GGGGGGG
Sbjct: 51 GIGGGPGGGSGYGGGSGEGGGAGGHGEGH----------IGGGGGGGHGGGAGGGGGGG 99
Score = 42.0 bits (97), Expect = 5e-05
Identities = 23/57 (40%), Positives = 24/57 (41%), Gaps = 18/57 (31%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG GG GGG A SG GGG+GGG G GG
Sbjct: 214 GGGGGGGGSGGGGGGGGGYA------------------AASGYGHGGGAGGGEGSGG 252
Score = 38.5 bits (88), Expect = 5e-04
Identities = 23/57 (40%), Positives = 23/57 (40%), Gaps = 16/57 (28%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G G GG SG G A G G G GGG GG GGG G
Sbjct: 53 GGGPGGGSGYGGGSGEGGGA----------------GGHGEGHIGGGGGGGHGGGAG 93
Score = 27.7 bits (60), Expect = 0.95
Identities = 12/19 (63%), Positives = 12/19 (63%)
Query: 101 GGGSGSDDGGGSGGGGGGG 119
G G G GGGSG GGG G
Sbjct: 49 GVGIGGGPGGGSGYGGGSG 67
Score = 25.8 bits (55), Expect = 3.6
Identities = 11/20 (55%), Positives = 11/20 (55%)
Query: 101 GGGSGSDDGGGSGGGGGGGG 120
G G G G G GGG G GG
Sbjct: 51 GIGGGPGGGSGYGGGSGEGG 70
Score = 25.4 bits (54), Expect = 4.7
Identities = 11/18 (61%), Positives = 11/18 (61%)
Query: 103 GSGSDDGGGSGGGGGGGG 120
G G GGG GGG G GG
Sbjct: 47 GLGVGIGGGPGGGSGYGG 64
>At4g22020 glycine-rich protein
Length = 396
Score = 54.7 bits (130), Expect = 7e-09
Identities = 28/57 (49%), Positives = 31/57 (54%), Gaps = 5/57 (8%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG + +G + G GSGS GGG GGGG GGG
Sbjct: 285 GGGGGGGGGGGGGGGGGNGSGYGSGSGYGSGM-----GKGSGSGGGGGGGGGGSGGG 336
Score = 52.4 bits (124), Expect = 4e-08
Identities = 27/56 (48%), Positives = 31/56 (55%), Gaps = 3/56 (5%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGGSGGS GG +G + + + GGG G GGG GGGGG G
Sbjct: 251 GGGGGGGGGSGGSKVGGG---YGHGSGFGGGVGFGNSGGGGGGGGGGGGGGGGGNG 303
Score = 50.4 bits (119), Expect = 1e-07
Identities = 27/57 (47%), Positives = 30/57 (52%), Gaps = 9/57 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG +GGGS G SG G+ + G GG SG GGG GGGG GGG
Sbjct: 143 GGGGGEGGGSSGGSGHGSGSGAGAG---------AGVGGSSGGAGGGGGGGGGEGGG 190
Score = 50.1 bits (118), Expect = 2e-07
Identities = 26/57 (45%), Positives = 31/57 (53%), Gaps = 14/57 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG +GGG+ G SG G+ + G GG SG+ GGG GGGGGG G
Sbjct: 219 GGGGGEGGGANGGSGHGSGSGAG--------------GGVSGAAGGGGGGGGGGGSG 261
Score = 50.1 bits (118), Expect = 2e-07
Identities = 26/57 (45%), Positives = 27/57 (46%), Gaps = 9/57 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG S GG+ G G G G GG GGGGGGGG
Sbjct: 139 GGGGGGGGGEGGGSSGGSGHGSG---------SGAGAGAGVGGSSGGAGGGGGGGGG 186
Score = 49.7 bits (117), Expect = 2e-07
Identities = 25/59 (42%), Positives = 29/59 (48%), Gaps = 7/59 (11%)
Query: 62 VDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
V G G GGG GG GG++ G + GGG G + GG GGGGGGGG
Sbjct: 244 VSGAAGGGGGGGGGGGSGGSKVGGG-------YGHGSGFGGGVGFGNSGGGGGGGGGGG 295
Score = 49.7 bits (117), Expect = 2e-07
Identities = 27/59 (45%), Positives = 30/59 (50%), Gaps = 12/59 (20%)
Query: 63 DGGGGNDGGGSGGSSGGGAR-AWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+ GGG GGG GG GGG + +G G GSG G GSGGGGGGGG
Sbjct: 283 NSGGGGGGGGGGGGGGGGGNGSGYG-----------SGSGYGSGMGKGSGSGGGGGGGG 330
Score = 49.3 bits (116), Expect = 3e-07
Identities = 26/57 (45%), Positives = 30/57 (52%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG +GGG+ G SG G+ A G GG +G GGG GGGG GGG
Sbjct: 182 GGGGGEGGGANGGSGHGSGAGAG-----------AGVGGAAGGVGGGGGGGGGEGGG 227
Score = 46.6 bits (109), Expect = 2e-06
Identities = 24/56 (42%), Positives = 27/56 (47%), Gaps = 11/56 (19%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG GGG GG GGGA G G G+G+ GG +GG GGGGG
Sbjct: 175 GGAGGGGGGGGGEGGGANGGSGH-----------GSGAGAGAGVGGAAGGVGGGGG 219
Score = 45.4 bits (106), Expect = 4e-06
Identities = 27/70 (38%), Positives = 33/70 (46%), Gaps = 24/70 (34%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG---------- 113
GGGG GGGSGG +G G+ + G GGG+G+ +GGG G
Sbjct: 324 GGGGGGGGGSGGGNGSGSGSGEGYGM-----------GGGAGTGNGGGGGVGFGMGIGFG 372
Query: 114 ---GGGGGGG 120
GGG GGG
Sbjct: 373 IGIGGGSGGG 382
Score = 44.3 bits (103), Expect = 1e-05
Identities = 28/63 (44%), Positives = 29/63 (45%), Gaps = 17/63 (26%)
Query: 62 VDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGG----SGGGGG 117
V G G GGG GG G G A G G GSGS GGG +GGGGG
Sbjct: 207 VGGAAGGVGGGGGGGGGEGGGANGG-------------SGHGSGSGAGGGVSGAAGGGGG 253
Query: 118 GGG 120
GGG
Sbjct: 254 GGG 256
Score = 44.3 bits (103), Expect = 1e-05
Identities = 25/57 (43%), Positives = 27/57 (46%), Gaps = 8/57 (14%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
+GGG + GGSG SG GA A G GGG G GGG GGG GG
Sbjct: 107 EGGGSSANGGSGHGSGSGAGAGVGGTTG--------GVGGGGGGGGGGGEGGGSSGG 155
Score = 44.3 bits (103), Expect = 1e-05
Identities = 25/56 (44%), Positives = 26/56 (45%), Gaps = 5/56 (8%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG GGG GG GGG+ A G G G G GG GGGGGGGG
Sbjct: 95 GGVGGGGGRGGGEGGGSSANGGSGHG-----SGSGAGAGVGGTTGGVGGGGGGGGG 145
Score = 43.5 bits (101), Expect = 2e-05
Identities = 27/64 (42%), Positives = 28/64 (43%), Gaps = 21/64 (32%)
Query: 62 VDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDG-----GGSGGGG 116
V GGGG GG GGSS G G GSG+ G GG GGGG
Sbjct: 97 VGGGGGRGGGEGGGSSANGGSG----------------HGSGSGAGAGVGGTTGGVGGGG 140
Query: 117 GGGG 120
GGGG
Sbjct: 141 GGGG 144
Score = 43.1 bits (100), Expect = 2e-05
Identities = 24/57 (42%), Positives = 27/57 (47%), Gaps = 12/57 (21%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GG +G + G G+G GG GGGGG GG
Sbjct: 61 GGGGGGGGGGGGGEGGDG---YGHGEGY---------GAGAGMSGYGGVGGGGGRGG 105
Score = 41.2 bits (95), Expect = 8e-05
Identities = 24/59 (40%), Positives = 26/59 (43%), Gaps = 5/59 (8%)
Query: 62 VDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
V G G GGG GG GGG G G+G+ GG SGG GGGGG
Sbjct: 129 VGGTTGGVGGGGGGGGGGGEGGGSSGGS-----GHGSGSGAGAGAGVGGSSGGAGGGGG 182
Score = 41.2 bits (95), Expect = 8e-05
Identities = 23/55 (41%), Positives = 25/55 (44%), Gaps = 3/55 (5%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG GGG GG GG +G + GG G GGG GGG GGG
Sbjct: 59 GGGGGGGGGGGGGGGEGGDGYGHGEGYGAGAGMSGYGGVGG---GGGRGGGEGGG 110
Score = 39.3 bits (90), Expect = 3e-04
Identities = 23/59 (38%), Positives = 26/59 (43%)
Query: 62 VDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+ G GG GGG G GG + G GG +G GGG GGGGGG G
Sbjct: 91 MSGYGGVGGGGGRGGGEGGGSSANGGSGHGSGSGAGAGVGGTTGGVGGGGGGGGGGGEG 149
Score = 33.1 bits (74), Expect = 0.023
Identities = 18/53 (33%), Positives = 24/53 (44%), Gaps = 8/53 (15%)
Query: 68 NDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
ND S G +++G+ + + GGG G GGG GGG GG G
Sbjct: 34 NDDDDCSSRSWRGCGSFYGKGAKRY--------GGGGGGGGGGGGGGGEGGDG 78
Score = 29.3 bits (64), Expect = 0.33
Identities = 19/57 (33%), Positives = 21/57 (36%), Gaps = 19/57 (33%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
+G G G G+G GGG G G G G G GGG GGG
Sbjct: 345 EGYGMGGGAGTGNGGGGGV-------------------GFGMGIGFGIGIGGGSGGG 382
Score = 25.8 bits (55), Expect = 3.6
Identities = 19/58 (32%), Positives = 20/58 (33%), Gaps = 20/58 (34%)
Query: 58 EAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGG 115
E + GG G GG GG G G G G GGGSGGG
Sbjct: 345 EGYGMGGGAGTGNGGGGGVGFG--------------------MGIGFGIGIGGGSGGG 382
>At1g05135 unknown protein
Length = 384
Score = 54.7 bits (130), Expect = 7e-09
Identities = 28/57 (49%), Positives = 30/57 (52%), Gaps = 7/57 (12%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG GGGA G + GGG G GGG+GGGGGGGG
Sbjct: 244 GGGAGGGGGGGGGGGGGANGGSGHGSGFGA-------GGGVGGGVGGGAGGGGGGGG 293
Score = 53.5 bits (127), Expect = 2e-08
Identities = 28/57 (49%), Positives = 30/57 (52%), Gaps = 7/57 (12%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG GG GGGA G + GGG G GGG+GGGGGGGG
Sbjct: 206 GVGGGGGGGGGGGGGGGANGGSGHGSGFGA-------GGGVGGGVGGGAGGGGGGGG 255
Score = 52.4 bits (124), Expect = 4e-08
Identities = 28/57 (49%), Positives = 31/57 (54%), Gaps = 7/57 (12%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG+ G + GGG+G GGG GGGGGGGG
Sbjct: 170 GGGGGAGGGGGGGVGGGSGHGSG-------FGAGGGIGGGAGGGVGGGGGGGGGGGG 219
Score = 50.4 bits (119), Expect = 1e-07
Identities = 27/57 (47%), Positives = 30/57 (52%), Gaps = 7/57 (12%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG+ G + GGG+G GGG G GGGGGG
Sbjct: 95 GGGGGAGGGGGGGIGGGSGHGGG-------FGAGGGVGGGAGGGIGGGGGAGGGGGG 144
Score = 50.4 bits (119), Expect = 1e-07
Identities = 27/57 (47%), Positives = 30/57 (52%), Gaps = 7/57 (12%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG+ G + GGG+G GGG GGGGGGG
Sbjct: 133 GGGGGAGGGGGGGVGGGSGHGGG-------FGAGGGVGGGAGGIGGGGGAGGGGGGG 182
Score = 50.1 bits (118), Expect = 2e-07
Identities = 28/57 (49%), Positives = 28/57 (49%), Gaps = 13/57 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGGSG SG GA G G G G GGG GGGGGGGG
Sbjct: 178 GGGGGVGGGSGHGSGFGAGGGIG-------------GGAGGGVGGGGGGGGGGGGGG 221
Score = 50.1 bits (118), Expect = 2e-07
Identities = 27/59 (45%), Positives = 28/59 (46%)
Query: 62 VDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
V GG G GG GG GGG G + GGG G GGG GGGGGGGG
Sbjct: 239 VGGGVGGGAGGGGGGGGGGGGGANGGSGHGSGFGAGGGVGGGVGGGAGGGGGGGGGGGG 297
Score = 50.1 bits (118), Expect = 2e-07
Identities = 27/57 (47%), Positives = 30/57 (52%), Gaps = 7/57 (12%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG+ G + GGG+G GGG G GGGGGG
Sbjct: 286 GGGGGGGGGGGGGVGGGSGHGGG-------FGAGGGLGGGAGGGLGGGGGAGGGGGG 335
Score = 49.7 bits (117), Expect = 2e-07
Identities = 28/60 (46%), Positives = 31/60 (51%), Gaps = 14/60 (23%)
Query: 62 VDGGGGNDGGGSGGSSGG-GARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+ GGGG GGG GG GG G + +G GGG G GGG GGGGGGGG
Sbjct: 169 IGGGGGAGGGGGGGVGGGSGHGSGFGA-------------GGGIGGGAGGGVGGGGGGGG 215
Score = 46.2 bits (108), Expect = 3e-06
Identities = 28/59 (47%), Positives = 30/59 (50%), Gaps = 9/59 (15%)
Query: 64 GGGGNDGG--GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG GSGG GGG G + GGG+G GGG G GGGGGG
Sbjct: 55 GGGGGFGGGRGSGGGIGGGGGQGGG-------FGAGGGVGGGAGGGLGGGGGAGGGGGG 106
Score = 45.4 bits (106), Expect = 4e-06
Identities = 26/60 (43%), Positives = 30/60 (49%), Gaps = 14/60 (23%)
Query: 62 VDGGGGNDGG-GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+ GG G+ GG G+GG GGGA G GGG+G GGG GGG G GG
Sbjct: 108 IGGGSGHGGGFGAGGGVGGGAGGGIG-------------GGGGAGGGGGGGVGGGSGHGG 154
Score = 45.1 bits (105), Expect = 6e-06
Identities = 27/60 (45%), Positives = 30/60 (50%), Gaps = 14/60 (23%)
Query: 62 VDGGGGNDGG-GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
V GG G+ GG G+GG GGGA G GGG+G GGG G GGG GG
Sbjct: 299 VGGGSGHGGGFGAGGGLGGGAGGGLG-------------GGGGAGGGGGGGLGHGGGVGG 345
Score = 45.1 bits (105), Expect = 6e-06
Identities = 25/56 (44%), Positives = 26/56 (45%), Gaps = 15/56 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGG+GG GGG A GGG G GGG GGG GGG
Sbjct: 310 GAGGGLGGGAGGGLGGGGGA---------------GGGGGGGLGHGGGVGGGHGGG 350
Score = 44.7 bits (104), Expect = 8e-06
Identities = 24/57 (42%), Positives = 26/57 (45%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG+GG GGG G + GG G G G GGGGG GG
Sbjct: 129 GGGIGGGGGAGGGGGGGVGGGSGHGGGFGAGGGVGGGAGGIGGGGGAGGGGGGGVGG 185
Score = 44.7 bits (104), Expect = 8e-06
Identities = 26/57 (45%), Positives = 28/57 (48%), Gaps = 12/57 (21%)
Query: 64 GGGGNDGG-GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GG G+ GG G GG GGG A G G G G GGG+GGGGGGG
Sbjct: 62 GGRGSGGGIGGGGGQGGGFGAGGG-----------VGGGAGGGLGGGGGAGGGGGGG 107
Score = 44.7 bits (104), Expect = 8e-06
Identities = 26/60 (43%), Positives = 28/60 (46%), Gaps = 20/60 (33%)
Query: 62 VDGGGGNDGG-GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+ GGGG GG G+GG GGGA GGG G G G GGGGG GG
Sbjct: 70 IGGGGGQGGGFGAGGGVGGGA-------------------GGGLGGGGGAGGGGGGGIGG 110
Score = 44.3 bits (103), Expect = 1e-05
Identities = 26/57 (45%), Positives = 26/57 (45%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGGSG G GA G GGG G G G GGGGG GG
Sbjct: 103 GGGGGIGGGSGHGGGFGAGGGVGGGA-----------GGGIGGGGGAGGGGGGGVGG 148
Score = 43.9 bits (102), Expect = 1e-05
Identities = 29/59 (49%), Positives = 31/59 (52%), Gaps = 9/59 (15%)
Query: 64 GGGGNDGG-GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG-GGGGGGG 120
GGGG GG GSGG GGG G + GGG+G GGG G GGGGGGG
Sbjct: 56 GGGGFGGGRGSGGGIGGGGGQGGG-------FGAGGGVGGGAGGGLGGGGGAGGGGGGG 107
Score = 43.9 bits (102), Expect = 1e-05
Identities = 26/56 (46%), Positives = 26/56 (46%), Gaps = 7/56 (12%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG GGG GG GGG G GGGSG G GSG GGGGG
Sbjct: 334 GGGLGHGGGVGGGHGGGVGIGIGIGIG-------VGVGGGSGQGSGSGSGSGGGGG 382
Score = 33.5 bits (75), Expect = 0.017
Identities = 19/36 (52%), Positives = 19/36 (52%), Gaps = 3/36 (8%)
Query: 85 WGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
WGRR R GGG G G GSGGG GGGG
Sbjct: 42 WGRR---CAGRGRFGRGGGGGFGGGRGSGGGIGGGG 74
Score = 32.0 bits (71), Expect = 0.050
Identities = 14/28 (50%), Positives = 15/28 (53%)
Query: 93 RWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
RW + C G G GGG GGG G GG
Sbjct: 41 RWGRRCAGRGRFGRGGGGGFGGGRGSGG 68
Score = 28.1 bits (61), Expect = 0.73
Identities = 13/20 (65%), Positives = 13/20 (65%), Gaps = 1/20 (5%)
Query: 64 GGGGNDGGGSG-GSSGGGAR 82
GGG G GSG GS GGG R
Sbjct: 364 GGGSGQGSGSGSGSGGGGGR 383
>At3g17110 hypothetical protein
Length = 175
Score = 53.5 bits (127), Expect = 2e-08
Identities = 27/57 (47%), Positives = 31/57 (54%), Gaps = 13/57 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG G G+ + +G G GSGS GGG+ GGGGGGG
Sbjct: 54 GGGGGGGGGGGGGDGSGSGSGYG-------------SGYGSGSGYGGGNNGGGGGGG 97
Score = 50.8 bits (120), Expect = 1e-07
Identities = 26/60 (43%), Positives = 29/60 (48%), Gaps = 4/60 (6%)
Query: 65 GGGNDGGGSGGSS----GGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGN+GGG GG GGG G + + G G GGG GGGGGGGG
Sbjct: 86 GGGNNGGGGGGGGRGGGGGGGNGGSGNGSGYGSGSGYGSGAGRGGGGGGGGGGGGGGGGG 145
Score = 50.1 bits (118), Expect = 2e-07
Identities = 25/57 (43%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG+ G G + +G + GGG G GGG GGGGG G
Sbjct: 95 GGGGRGGGGGGGNGGSGNGSGYGSGSGYG---SGAGRGGGGGGGGGGGGGGGGGSRG 148
Score = 49.7 bits (117), Expect = 2e-07
Identities = 26/58 (44%), Positives = 29/58 (49%), Gaps = 4/58 (6%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+ GGG GGG GG GGG G + G G+G GGG GGGGGGGG
Sbjct: 89 NNGGGGGGGGRGGGGGGGN----GGSGNGSGYGSGSGYGSGAGRGGGGGGGGGGGGGG 142
Score = 45.4 bits (106), Expect = 4e-06
Identities = 27/77 (35%), Positives = 32/77 (41%), Gaps = 19/77 (24%)
Query: 43 VAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGG 102
VA V+V+ + + G G G G G SGGG +GG
Sbjct: 9 VAFFVLVVFDVCLAARSPKALGRGSGSGSGYGSGSGGG-------------------NGG 49
Query: 103 GSGSDDGGGSGGGGGGG 119
G G GGG GGGGGGG
Sbjct: 50 GGGGGGGGGGGGGGGGG 66
Score = 43.9 bits (102), Expect = 1e-05
Identities = 24/56 (42%), Positives = 26/56 (45%), Gaps = 16/56 (28%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG GGG+R +G G GS G G G G GGG
Sbjct: 129 GGGGGGGGGGGGGGGGGSRG----------------NGSGYGSGYGEGYGSGYGGG 168
Score = 43.1 bits (100), Expect = 2e-05
Identities = 23/57 (40%), Positives = 24/57 (41%), Gaps = 20/57 (35%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G G G GSGG +GGG GGG G GGG GGGG G G
Sbjct: 34 GSGSGYGSGSGGGNGGG--------------------GGGGGGGGGGGGGGGGDGSG 70
Score = 42.7 bits (99), Expect = 3e-05
Identities = 29/82 (35%), Positives = 34/82 (41%), Gaps = 21/82 (25%)
Query: 39 SVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWC 98
S+ + V V+VV V + A G G GSG SG G
Sbjct: 3 SLKYLGVAFFVLVVFDVCLAARSPKALGRGSGSGSGYGSGSG------------------ 44
Query: 99 DDGGGSGSDDGGGSGGGGGGGG 120
GG+G GGG GGGGGGGG
Sbjct: 45 ---GGNGGGGGGGGGGGGGGGG 63
Score = 41.2 bits (95), Expect = 8e-05
Identities = 25/63 (39%), Positives = 27/63 (42%), Gaps = 19/63 (30%)
Query: 64 GGGGNDGGGSGG------SSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGG 117
GGGG DG GSG SG G + + GGG G GGG GGG G
Sbjct: 62 GGGGGDGSGSGSGYGSGYGSGSG-------------YGGGNNGGGGGGGGRGGGGGGGNG 108
Query: 118 GGG 120
G G
Sbjct: 109 GSG 111
Score = 40.0 bits (92), Expect = 2e-04
Identities = 23/54 (42%), Positives = 24/54 (43%), Gaps = 8/54 (14%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGG 117
G G GGG GG GGG G R G GSG +G GSG GGG
Sbjct: 123 GSGAGRGGGGGGGGGGGGGGGGGSRGN--------GSGYGSGYGEGYGSGYGGG 168
>At2g30560 putative glycine-rich protein
Length = 171
Score = 53.5 bits (127), Expect = 2e-08
Identities = 30/61 (49%), Positives = 31/61 (50%), Gaps = 15/61 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWC----DDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG SGGGA G C GGG G +GGG GGGGGG
Sbjct: 78 GGGGKGGGGGGGISGGGAGGKSG-----------CGGGKSGGGGGGGKNGGGCGGGGGGK 126
Query: 120 G 120
G
Sbjct: 127 G 127
Score = 48.1 bits (113), Expect = 7e-07
Identities = 27/57 (47%), Positives = 28/57 (48%), Gaps = 21/57 (36%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGGA+ GG G GG SGGGGGGGG
Sbjct: 16 GGGGGSGGGRGGGGGGGAK-------------------GGCGG--GGKSGGGGGGGG 51
Score = 47.4 bits (111), Expect = 1e-06
Identities = 25/57 (43%), Positives = 26/57 (44%), Gaps = 8/57 (14%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG G G G GG GGG + GR GG G GGG GGGGGGG
Sbjct: 2 GGKGGSGSGGGGKGGGGGGSGGGRGGG--------GGGGAKGGCGGGGKSGGGGGGG 50
>At3g23450 hypothetical protein
Length = 452
Score = 53.1 bits (126), Expect = 2e-08
Identities = 31/68 (45%), Positives = 34/68 (49%), Gaps = 16/68 (23%)
Query: 58 EAVVVDGG-----GGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGS 112
E +V GG GG GGG+GG GGGA +G GGG G GGG
Sbjct: 36 EKTLVGGGKGGGFGGGFGGGAGGGVGGGAGGGFGGGA-----------GGGFGGGGGGGG 84
Query: 113 GGGGGGGG 120
GGGGGGGG
Sbjct: 85 GGGGGGGG 92
Score = 52.0 bits (123), Expect = 5e-08
Identities = 27/57 (47%), Positives = 29/57 (50%), Gaps = 12/57 (21%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG+GG GGGA +G GGG G GGG GG GGGGG
Sbjct: 55 GAGGGVGGGAGGGFGGGAGGGFG------------GGGGGGGGGGGGGGGGFGGGGG 99
Score = 47.4 bits (111), Expect = 1e-06
Identities = 26/56 (46%), Positives = 26/56 (46%), Gaps = 13/56 (23%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG GGG G GGG G GGG GGG GGG
Sbjct: 81 GGGGGGGGGGGGGFGGGGGFGGGH-------------GGGVGGGVGGGHGGGVGGG 123
Score = 47.4 bits (111), Expect = 1e-06
Identities = 26/56 (46%), Positives = 26/56 (46%), Gaps = 13/56 (23%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG GGG G GGG G GGG GGG GGG
Sbjct: 77 GGGGGGGGGGGGGGGGGFGGGGGF-------------GGGHGGGVGGGVGGGHGGG 119
Score = 45.4 bits (106), Expect = 4e-06
Identities = 25/57 (43%), Positives = 26/57 (44%), Gaps = 12/57 (21%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG+GG GGG G GGG G GGG GGG GGG
Sbjct: 63 GAGGGFGGGAGGGFGGGGGGGGG------------GGGGGGGGFGGGGGFGGGHGGG 107
Score = 44.3 bits (103), Expect = 1e-05
Identities = 25/57 (43%), Positives = 26/57 (44%), Gaps = 9/57 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG G GGG G + G G G GGG GGGGGGGG
Sbjct: 401 GKGGGFGGGGGFGKGGGIGGGGGFGK---------GGGFGGGGFGGGGGGGGGGGGG 448
Score = 43.5 bits (101), Expect = 2e-05
Identities = 25/57 (43%), Positives = 25/57 (43%), Gaps = 19/57 (33%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG GGG G GGG G GGG GG
Sbjct: 95 GGGGGFGGGHGGGVGGGV-------------------GGGHGGGVGGGFGKGGGIGG 132
Score = 42.4 bits (98), Expect = 4e-05
Identities = 24/56 (42%), Positives = 25/56 (43%), Gaps = 14/56 (25%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG GGG GG G G +G GGG G GGG GGGGG G
Sbjct: 409 GGGFGKGGGIGGGGGFGKGGGFG--------------GGGFGGGGGGGGGGGGGIG 450
Score = 42.4 bits (98), Expect = 4e-05
Identities = 26/70 (37%), Positives = 30/70 (42%), Gaps = 11/70 (15%)
Query: 62 VDGGGGNDGGGSGGSSGG-----------GARAWWGRRRQWWRWRQWCDDGGGSGSDDGG 110
+ GGGG GGG G GG G +G+ + GG G GG
Sbjct: 378 IGGGGGFGGGGGFGKGGGIGGGIGKGGGFGGGGGFGKGGGIGGGGGFGKGGGFGGGGFGG 437
Query: 111 GSGGGGGGGG 120
G GGGGGGGG
Sbjct: 438 GGGGGGGGGG 447
Score = 41.6 bits (96), Expect = 6e-05
Identities = 25/58 (43%), Positives = 27/58 (46%), Gaps = 15/58 (25%)
Query: 64 GGGGNDGGGSGGS--SGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG GGG GG GGG +G+ GGG G GGG G GGGGG
Sbjct: 345 GGGFGKGGGIGGGIGKGGGIGGGFGK-------------GGGIGGGIGGGGGFGGGGG 389
Score = 40.8 bits (94), Expect = 1e-04
Identities = 24/59 (40%), Positives = 26/59 (43%)
Query: 62 VDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+ GG G GG GG GGG G + GGG G G G GGG GGGG
Sbjct: 364 IGGGFGKGGGIGGGIGGGGGFGGGGGFGKGGGIGGGIGKGGGFGGGGGFGKGGGIGGGG 422
Score = 40.0 bits (92), Expect = 2e-04
Identities = 24/57 (42%), Positives = 25/57 (43%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG G GGG G+ G G G GGG GGG GGGG
Sbjct: 337 GKGGGIGGGGGFGKGGGIGGGIGK-----------GGGIGGGFGKGGGIGGGIGGGG 382
Score = 39.3 bits (90), Expect = 3e-04
Identities = 24/57 (42%), Positives = 25/57 (43%), Gaps = 9/57 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG G GGG G+ GGG G GGG GGG G GG
Sbjct: 315 GKGGGIGGGGGFGKGGGIGGGIGKGG---------GIGGGGGFGKGGGIGGGIGKGG 362
Score = 37.4 bits (85), Expect = 0.001
Identities = 24/59 (40%), Positives = 26/59 (43%), Gaps = 13/59 (22%)
Query: 64 GGGGNDGGGSGGS--SGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG GGG +G+ G G G GGG GGG G GG
Sbjct: 211 GGGFGKGGGVGGGIGKGGGVGGGFGK-----------GGGVGGGIGKGGGIGGGIGKGG 258
Score = 37.4 bits (85), Expect = 0.001
Identities = 24/59 (40%), Positives = 26/59 (43%), Gaps = 13/59 (22%)
Query: 64 GGGGNDGGGSGGS--SGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG GGG +G+ G G G GGG GGG G GG
Sbjct: 191 GGGIGKGGGIGGGIGKGGGVGGGFGK-----------GGGVGGGIGKGGGVGGGFGKGG 238
Score = 36.6 bits (83), Expect = 0.002
Identities = 23/56 (41%), Positives = 24/56 (42%), Gaps = 11/56 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG GGG GG G G G + GGG G G G G GGGGG
Sbjct: 281 GGGIGKGGGIGGGIGKGGGIGGGIGK-----------GGGIGGGIGKGGGIGGGGG 325
Score = 35.8 bits (81), Expect = 0.003
Identities = 23/57 (40%), Positives = 24/57 (41%), Gaps = 9/57 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG G G G + G G G GGG GGG G GG
Sbjct: 201 GGGIGKGGGVGGGFGKGGGVGGGIGK---------GGGVGGGFGKGGGVGGGIGKGG 248
Score = 35.4 bits (80), Expect = 0.005
Identities = 23/57 (40%), Positives = 23/57 (40%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G G GG GGG G G G G GGG G GGG GG
Sbjct: 187 GGGIGGGIGKGGGIGGGIGKGGG-----------VGGGFGKGGGVGGGIGKGGGVGG 232
Score = 35.4 bits (80), Expect = 0.005
Identities = 23/57 (40%), Positives = 23/57 (40%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G G GG GGG G G G G GGG G GGG GG
Sbjct: 227 GGGVGGGFGKGGGVGGGIGKGGG-----------IGGGIGKGGGIGGGIGKGGGIGG 272
Score = 35.4 bits (80), Expect = 0.005
Identities = 23/57 (40%), Positives = 24/57 (41%), Gaps = 9/57 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG G G G + G G G GGG GGG G GG
Sbjct: 221 GGGIGKGGGVGGGFGKGGGVGGGIGK---------GGGIGGGIGKGGGIGGGIGKGG 268
Score = 35.4 bits (80), Expect = 0.005
Identities = 23/57 (40%), Positives = 24/57 (41%), Gaps = 9/57 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG G G G + G G G GGG GGG G GG
Sbjct: 131 GGGIGKGGGVGGGIGKGGGIGGGIGK---------GGGVGGGIGKGGGIGGGIGKGG 178
Score = 35.0 bits (79), Expect = 0.006
Identities = 23/57 (40%), Positives = 23/57 (40%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G G GG GGG G G G G GGG G GGG GG
Sbjct: 237 GGGVGGGIGKGGGIGGGIGKGGG-----------IGGGIGKGGGIGGGIGKGGGIGG 282
Score = 35.0 bits (79), Expect = 0.006
Identities = 23/57 (40%), Positives = 23/57 (40%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G G GG GGG G G G G GGG G GGG GG
Sbjct: 137 GGGVGGGIGKGGGIGGGIGKGGG-----------VGGGIGKGGGIGGGIGKGGGIGG 182
Score = 35.0 bits (79), Expect = 0.006
Identities = 23/57 (40%), Positives = 23/57 (40%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G G GG GGG G G G G GGG G GGG GG
Sbjct: 157 GGGVGGGIGKGGGIGGGIGKGGG-----------IGGGIGKGGGIGGGIGKGGGIGG 202
Score = 35.0 bits (79), Expect = 0.006
Identities = 23/57 (40%), Positives = 23/57 (40%), Gaps = 13/57 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G G GG GGG G GGG G G G GGG G GG
Sbjct: 287 GGGIGGGIGKGGGIGGGIGKGGGI-------------GGGIGKGGGIGGGGGFGKGG 330
Score = 34.7 bits (78), Expect = 0.008
Identities = 24/60 (40%), Positives = 26/60 (43%), Gaps = 10/60 (16%)
Query: 62 VDGGGGNDGGGSGG-SSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+ GG G GG GG GGG G + GGG G GGG GGG G GG
Sbjct: 354 IGGGIGKGGGIGGGFGKGGGIGGGIGGGGGF---------GGGGGFGKGGGIGGGIGKGG 404
Score = 34.7 bits (78), Expect = 0.008
Identities = 23/57 (40%), Positives = 23/57 (40%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G G GG GGG G G G G GGG G GGG GG
Sbjct: 257 GGGIGGGIGKGGGIGGGIGKGGG-----------IGGGIGKGGGIGGGIGKGGGIGG 302
Score = 33.9 bits (76), Expect = 0.013
Identities = 24/60 (40%), Positives = 25/60 (41%), Gaps = 12/60 (20%)
Query: 62 VDGGGGNDGG-GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
V GG G GG G G GGG G+ G G G GGG GGG G GG
Sbjct: 120 VGGGFGKGGGIGGGIGKGGGVGGGIGK-----------GGGIGGGIGKGGGVGGGIGKGG 168
Score = 33.1 bits (74), Expect = 0.023
Identities = 24/60 (40%), Positives = 25/60 (41%), Gaps = 12/60 (20%)
Query: 62 VDGGGGNDGGGSGG-SSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
V GG G GG GG GGG G+ G G G GGG GGG G GG
Sbjct: 160 VGGGIGKGGGIGGGIGKGGGIGGGIGK-----------GGGIGGGIGKGGGIGGGIGKGG 208
Score = 32.7 bits (73), Expect = 0.030
Identities = 23/60 (38%), Positives = 25/60 (41%), Gaps = 12/60 (20%)
Query: 62 VDGGGGNDGGGSGG-SSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+ GG G GG GG GGG G+ G G G GGG GGG G GG
Sbjct: 250 IGGGIGKGGGIGGGIGKGGGIGGGIGK-----------GGGIGGGIGKGGGIGGGIGKGG 298
Score = 32.7 bits (73), Expect = 0.030
Identities = 23/60 (38%), Positives = 25/60 (41%), Gaps = 12/60 (20%)
Query: 62 VDGGGGNDGGGSGG-SSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+ GG G GG GG GGG G+ G G G GGG GGG G GG
Sbjct: 270 IGGGIGKGGGIGGGIGKGGGIGGGIGK-----------GGGIGGGIGKGGGIGGGIGKGG 318
Score = 32.0 bits (71), Expect = 0.050
Identities = 24/62 (38%), Positives = 24/62 (38%), Gaps = 14/62 (22%)
Query: 62 VDGGGG---NDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
V GG G G G GG GGG G G G G GGG G GGG
Sbjct: 112 VGGGHGGGVGGGFGKGGGIGGGIGKGGG-----------VGGGIGKGGGIGGGIGKGGGV 160
Query: 119 GG 120
GG
Sbjct: 161 GG 162
>At5g35660 unknown protein
Length = 343
Score = 52.8 bits (125), Expect = 3e-08
Identities = 38/109 (34%), Positives = 40/109 (35%), Gaps = 40/109 (36%)
Query: 20 EGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGG--SGGSS 77
+GGG GGGW GGGG GGG GG
Sbjct: 266 QGGGGGHGGGW--------------------------------QGGGGGHGGGWQGGGGR 293
Query: 78 GGGARAWWGRRRQWW----RWRQWCDDGGGSGSDDGGGSGG--GGGGGG 120
GGG + G W R W GGG G GGG GG GGGGGG
Sbjct: 294 GGGWKGGGGHGGGWQGGGGRGGGWKGGGGGGGWRGGGGGGGWRGGGGGG 342
Score = 49.7 bits (117), Expect = 2e-07
Identities = 28/64 (43%), Positives = 29/64 (44%), Gaps = 9/64 (14%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQ-------WCDDGGGSGSDDGGGSGGGG 116
GGGG GG GG GG W G Q W+ W GG G GGG GGG
Sbjct: 176 GGGGQGGGWKGGGGQGGG--WKGGGGQGGGWKGGGGQGGGWKGGGGQGGGWKGGGGRGGG 233
Query: 117 GGGG 120
GGGG
Sbjct: 234 GGGG 237
Score = 48.9 bits (115), Expect = 4e-07
Identities = 36/109 (33%), Positives = 43/109 (39%), Gaps = 10/109 (9%)
Query: 22 GGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVV----DGGGGNDGGGSGG-- 75
GG GGGW R + + +++ E V GGG +GGG GG
Sbjct: 115 GGGGHGGGWKGGGGGRGGGGGGGAELETVEIDSASDEDVQNYRGGHGGGWKEGGGQGGGW 174
Query: 76 -SSGGGARAWWGRRRQWWRWRQWCDDGG---GSGSDDGGGSGGGGGGGG 120
GG W G Q W+ GG G G GG GGGG GGG
Sbjct: 175 KGGGGQGGGWKGGGGQGGGWKGGGGQGGGWKGGGGQGGGWKGGGGQGGG 223
Score = 47.4 bits (111), Expect = 1e-06
Identities = 26/57 (45%), Positives = 28/57 (48%), Gaps = 5/57 (8%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG+ GG GG GG W G W+ GG G GGG G GGGGGG
Sbjct: 85 GGGGHGGGWKGGGGHGGG--WKGGGGHGGGWK---GGGGHGGGWKGGGGGRGGGGGG 136
Score = 46.6 bits (109), Expect = 2e-06
Identities = 27/57 (47%), Positives = 27/57 (47%), Gaps = 3/57 (5%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG GGGA QW GGG G GG GGGGG GG
Sbjct: 221 GGGWKGGGGRGGGGGGGAELENEEINGASEEIQWQGGGGGHG---GGWQGGGGGHGG 274
Score = 45.4 bits (106), Expect = 4e-06
Identities = 30/76 (39%), Positives = 33/76 (42%), Gaps = 13/76 (17%)
Query: 58 EAVVVDGGGGNDGGG--------SGGSSGGG---ARAWWGRRRQWWRWRQWCDDGGG--S 104
E + GGGG GGG GG GGG W G + W+ GGG
Sbjct: 250 EEIQWQGGGGGHGGGWQGGGGGHGGGWQGGGGGHGGGWQGGGGRGGGWKGGGGHGGGWQG 309
Query: 105 GSDDGGGSGGGGGGGG 120
G GGG GGGGGGG
Sbjct: 310 GGGRGGGWKGGGGGGG 325
Score = 31.6 bits (70), Expect = 0.066
Identities = 22/58 (37%), Positives = 22/58 (37%), Gaps = 21/58 (36%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG--GGGGG 119
GGG GGG GG GG G G G GGG GG GGGG
Sbjct: 80 GGGWKGGGGHGGGWKGGG-------------------GHGGGWKGGGGHGGGWKGGGG 118
Score = 28.5 bits (62), Expect = 0.56
Identities = 14/26 (53%), Positives = 14/26 (53%), Gaps = 2/26 (7%)
Query: 97 WCDDGGGSGSDDGGGSGGGG--GGGG 120
W GG G GGG GGG GGGG
Sbjct: 83 WKGGGGHGGGWKGGGGHGGGWKGGGG 108
Score = 28.1 bits (61), Expect = 0.73
Identities = 27/89 (30%), Positives = 34/89 (37%), Gaps = 15/89 (16%)
Query: 41 VVVAVTVVVIVVEAVV---------VEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQW 91
++VA VV + E V VE V+G ND S G A + +
Sbjct: 15 LLVAAVVVAVAAETSVDDKKQGEKQVEIPTVEGK--NDAKDSLNKGDVGVEATVEKEEE- 71
Query: 92 WRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+ D G G GGG GGG GGG
Sbjct: 72 ---NNYEIDQHGGGWKGGGGHGGGWKGGG 97
Score = 26.2 bits (56), Expect = 2.8
Identities = 13/22 (59%), Positives = 13/22 (59%), Gaps = 2/22 (9%)
Query: 101 GGG--SGSDDGGGSGGGGGGGG 120
GGG G GGG GGGG GG
Sbjct: 80 GGGWKGGGGHGGGWKGGGGHGG 101
Score = 25.4 bits (54), Expect = 4.7
Identities = 11/22 (50%), Positives = 11/22 (50%)
Query: 99 DDGGGSGSDDGGGSGGGGGGGG 120
D GG GG GG GGGG
Sbjct: 77 DQHGGGWKGGGGHGGGWKGGGG 98
>At3g17050 unknown protein
Length = 349
Score = 50.8 bits (120), Expect = 1e-07
Identities = 35/82 (42%), Positives = 40/82 (48%), Gaps = 16/82 (19%)
Query: 42 VVAVTVVVIVVEAVVVEAVVVDGGGGND-GGGSGGSSG--GGARAWWGRRRQWWRWRQWC 98
++ V VV+ VVE E + GGGG GGG GG G GG A G
Sbjct: 11 LMLVVVVIGVVECRRFEKETLGGGGGGGLGGGFGGGKGFGGGIGAGGGF----------- 59
Query: 99 DDGGGSGSDDGGGSGGGGGGGG 120
GGG+G GGG GGG GGGG
Sbjct: 60 --GGGAGGGAGGGLGGGAGGGG 79
Score = 50.4 bits (119), Expect = 1e-07
Identities = 26/56 (46%), Positives = 31/56 (54%), Gaps = 11/56 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGG+GG +GGGA +G GGG+G GGG+GGG GGG
Sbjct: 246 GTGGGFGGGAGGGAGGGAGGGFGGGA-----------GGGAGGGFGGGAGGGAGGG 290
Score = 50.1 bits (118), Expect = 2e-07
Identities = 26/57 (45%), Positives = 29/57 (50%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG GG GGG+ +G GGG+G GGG GGGGG GG
Sbjct: 298 GAGGGHGGGVGGGFGGGSGGGFGGGA-----------GGGAGGGAGGGFGGGGGAGG 343
Score = 48.9 bits (115), Expect = 4e-07
Identities = 26/56 (46%), Positives = 29/56 (51%), Gaps = 11/56 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG+GG +GGG G GGG G GGG+GGG GGG
Sbjct: 76 GGGGGIGGGAGGGAGGGLGGGAGGGL-----------GGGHGGGIGGGAGGGAGGG 120
Score = 48.9 bits (115), Expect = 4e-07
Identities = 26/57 (45%), Positives = 28/57 (48%), Gaps = 19/57 (33%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG+GG SGGG GGG G GGG+GGGGG GG
Sbjct: 182 GHGGGIGGGAGGGSGGGL-------------------GGGIGGGAGGGAGGGGGAGG 219
Score = 48.5 bits (114), Expect = 5e-07
Identities = 26/56 (46%), Positives = 30/56 (53%), Gaps = 7/56 (12%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGG GG +GGGA +G GGG+G GGG+GGG GGG
Sbjct: 258 GAGGGAGGGFGGGAGGGAGGGFGGGAGGGA-------GGGAGGGFGGGAGGGHGGG 306
Score = 48.5 bits (114), Expect = 5e-07
Identities = 27/56 (48%), Positives = 28/56 (49%), Gaps = 11/56 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG GGGA G GGG G GGG+GGG GGG
Sbjct: 154 GGGGGLGGGHGGGIGGGAGGGAGGGL-----------GGGHGGGIGGGAGGGSGGG 198
Score = 48.5 bits (114), Expect = 5e-07
Identities = 25/56 (44%), Positives = 28/56 (49%), Gaps = 19/56 (33%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGG GG +GGGA GGG+G GGG+GGG GGG
Sbjct: 242 GAGGGTGGGFGGGAGGGA-------------------GGGAGGGFGGGAGGGAGGG 278
Score = 48.1 bits (113), Expect = 7e-07
Identities = 26/57 (45%), Positives = 27/57 (46%), Gaps = 15/57 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG GG GGGA GGG G GGG+GGG GGGG
Sbjct: 116 GAGGGLGGGHGGGIGGGAGG---------------GSGGGLGGGIGGGAGGGAGGGG 157
Score = 48.1 bits (113), Expect = 7e-07
Identities = 26/56 (46%), Positives = 28/56 (49%), Gaps = 13/56 (23%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGG GG +GGGA G GGG G GGG+GGG GGG
Sbjct: 136 GSGGGLGGGIGGGAGGGAGGGGGL-------------GGGHGGGIGGGAGGGAGGG 178
Score = 47.8 bits (112), Expect = 9e-07
Identities = 25/56 (44%), Positives = 29/56 (51%), Gaps = 15/56 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGG+GG +GGGA +G GG+G GGG GGG GGG
Sbjct: 274 GAGGGFGGGAGGGAGGGAGGGFG---------------GGAGGGHGGGVGGGFGGG 314
Score = 47.8 bits (112), Expect = 9e-07
Identities = 26/56 (46%), Positives = 27/56 (47%), Gaps = 15/56 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG GGGA G GG+G GGG GGG GGG
Sbjct: 218 GGGGGLGGGHGGGFGGGAGGGLG---------------GGAGGGTGGGFGGGAGGG 258
Score = 47.4 bits (111), Expect = 1e-06
Identities = 26/57 (45%), Positives = 28/57 (48%), Gaps = 19/57 (33%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG+GG SGGG GGG G GGG+GGGGG GG
Sbjct: 124 GHGGGIGGGAGGGSGGGL-------------------GGGIGGGAGGGAGGGGGLGG 161
Score = 46.2 bits (108), Expect = 3e-06
Identities = 25/57 (43%), Positives = 27/57 (46%), Gaps = 19/57 (33%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG GG +GGGA GGG+G GGG G GGG GG
Sbjct: 310 GFGGGSGGGFGGGAGGGA-------------------GGGAGGGFGGGGGAGGGFGG 347
Score = 46.2 bits (108), Expect = 3e-06
Identities = 25/56 (44%), Positives = 27/56 (47%), Gaps = 19/56 (33%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGG GG +GGGA GGG G+ GGG GGG GGG
Sbjct: 194 GSGGGLGGGIGGGAGGGA-------------------GGGGGAGGGGGLGGGHGGG 230
Score = 45.4 bits (106), Expect = 4e-06
Identities = 26/56 (46%), Positives = 27/56 (47%), Gaps = 13/56 (23%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGGSGG GGGA G G G G GGG+GGG GGG
Sbjct: 306 GVGGGFGGGSGGGFGGGAGGGAG-------------GGAGGGFGGGGGAGGGFGGG 348
Score = 44.3 bits (103), Expect = 1e-05
Identities = 26/58 (44%), Positives = 27/58 (45%), Gaps = 12/58 (20%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGG-SGGGGGGGG 120
GGG GGG GG +GGGA G GGG G GGG GG GGG G
Sbjct: 72 GGGAGGGGGIGGGAGGGAGGGLGGGA-----------GGGLGGGHGGGIGGGAGGGAG 118
Score = 43.9 bits (102), Expect = 1e-05
Identities = 25/56 (44%), Positives = 26/56 (45%), Gaps = 19/56 (33%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGG GG GGGA GGG+G GGG GGG GGG
Sbjct: 96 GAGGGLGGGHGGGIGGGA-------------------GGGAGGGLGGGHGGGIGGG 132
Score = 43.9 bits (102), Expect = 1e-05
Identities = 26/63 (41%), Positives = 28/63 (44%), Gaps = 16/63 (25%)
Query: 62 VDGGGGND-----GGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG 116
+ GG G GGG GG +GGGA G GGG G GGG GGG
Sbjct: 187 IGGGAGGGSGGGLGGGIGGGAGGGAGGGGG-----------AGGGGGLGGGHGGGFGGGA 235
Query: 117 GGG 119
GGG
Sbjct: 236 GGG 238
Score = 43.5 bits (101), Expect = 2e-05
Identities = 23/57 (40%), Positives = 26/57 (45%), Gaps = 19/57 (33%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG+GG +GGG GGG+G G G G GGG GG
Sbjct: 54 GAGGGFGGGAGGGAGGGL-------------------GGGAGGGGGIGGGAGGGAGG 91
Score = 43.1 bits (100), Expect = 2e-05
Identities = 27/59 (45%), Positives = 29/59 (48%), Gaps = 15/59 (25%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG--GGGGGG 120
G GG GGG+GG GGGA G GGG+G GGG GG GGG GG
Sbjct: 58 GFGGGAGGGAGGGLGGGAGGGGGI-------------GGGAGGGAGGGLGGGAGGGLGG 103
Score = 41.6 bits (96), Expect = 6e-05
Identities = 24/58 (41%), Positives = 25/58 (42%)
Query: 62 VDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
+ GG G GG GG GG G GGG G GGGSGGG GGG
Sbjct: 145 IGGGAGGGAGGGGGLGGGHGGGIGGGAGGGAGGGLGGGHGGGIGGGAGGGSGGGLGGG 202
>At5g46730 unknown protein
Length = 268
Score = 50.1 bits (118), Expect = 2e-07
Identities = 26/62 (41%), Positives = 32/62 (50%), Gaps = 7/62 (11%)
Query: 66 GGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDD-------GGGSGGGGGG 118
GG+ GGG GGS GGG A+ + +GGG+G+ GGG GGGGG
Sbjct: 117 GGHAGGGGGGSGGGGGSAYGAGGEHASGYGNGAGEGGGAGASGYGGGAYGGGGGHGGGGG 176
Query: 119 GG 120
GG
Sbjct: 177 GG 178
Score = 48.9 bits (115), Expect = 4e-07
Identities = 26/56 (46%), Positives = 27/56 (47%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG S GGA G GG+G GGG GG GGGG
Sbjct: 166 GGGGGHGGGGGGGSAGGAHGGSGYGGGEGGGAGGGGSHGGAGGYGGGGGGGSGGGG 221
Score = 48.5 bits (114), Expect = 5e-07
Identities = 27/59 (45%), Positives = 30/59 (50%), Gaps = 11/59 (18%)
Query: 64 GGGGNDGGGSGGSSGG--GARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG+ GGG GGS+GG G + G GG GS G G GGGGGGG
Sbjct: 167 GGGGHGGGGGGGSAGGAHGGSGYGGGEG---------GGAGGGGSHGGAGGYGGGGGGG 216
Score = 47.8 bits (112), Expect = 9e-07
Identities = 27/59 (45%), Positives = 30/59 (50%), Gaps = 2/59 (3%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWR--WRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG GGS GGGA G + + GGG+ GGG GGGGG GG
Sbjct: 205 GAGGYGGGGGGGSGGGGAYGGGGAHGGGYGSGGGEGGGYGGGAAGGYGGGHGGGGGHGG 263
Score = 47.4 bits (111), Expect = 1e-06
Identities = 28/70 (40%), Positives = 33/70 (47%), Gaps = 13/70 (18%)
Query: 64 GGGGND-------GGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDD------GG 110
GGGG GGG GGS GGG A+ + +GGG+G+ GG
Sbjct: 108 GGGGYGGAAGGHAGGGGGGSGGGGGSAYGAGGEHASGYGNGAGEGGGAGASGYGGGAYGG 167
Query: 111 GSGGGGGGGG 120
G G GGGGGG
Sbjct: 168 GGGHGGGGGG 177
Score = 46.6 bits (109), Expect = 2e-06
Identities = 26/57 (45%), Positives = 27/57 (46%), Gaps = 15/57 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG GG SG GA +G G G GGGSG GGGGGG
Sbjct: 52 GYGGESGGGYGGGSGEGAGGGYG---------------GAEGYASGGGSGHGGGGGG 93
Score = 45.1 bits (105), Expect = 6e-06
Identities = 23/56 (41%), Positives = 25/56 (44%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG GGG G GGG A + + GGG GG GGGGGGG
Sbjct: 161 GGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGGEGGGAGGGGSHGGAGGYGGGGGGG 216
Score = 44.3 bits (103), Expect = 1e-05
Identities = 25/67 (37%), Positives = 29/67 (42%), Gaps = 9/67 (13%)
Query: 63 DGGGGNDG---------GGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG 113
+GGG G GG GG GG A+ G + +GGG G GG G
Sbjct: 193 EGGGAGGGGSHGGAGGYGGGGGGGSGGGGAYGGGGAHGGGYGSGGGEGGGYGGGAAGGYG 252
Query: 114 GGGGGGG 120
GG GGGG
Sbjct: 253 GGHGGGG 259
Score = 42.0 bits (97), Expect = 5e-05
Identities = 23/63 (36%), Positives = 27/63 (42%), Gaps = 13/63 (20%)
Query: 58 EAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGG 117
E GG G+ GGG G +S GG + G +GGG G G GGG
Sbjct: 77 EGYASGGGSGHGGGGGGAASSGGYASGAG-------------EGGGGGYGGAAGGHAGGG 123
Query: 118 GGG 120
GGG
Sbjct: 124 GGG 126
Score = 42.0 bits (97), Expect = 5e-05
Identities = 25/66 (37%), Positives = 31/66 (46%), Gaps = 8/66 (12%)
Query: 63 DGGGGNDGGGSGGSSGGGA--RAWWGRRRQWWRWRQWCDDGGG------SGSDDGGGSGG 114
+G GG GG G +SGGG+ G + +GGG +G GGG GG
Sbjct: 67 EGAGGGYGGAEGYASGGGSGHGGGGGGAASSGGYASGAGEGGGGGYGGAAGGHAGGGGGG 126
Query: 115 GGGGGG 120
GGGGG
Sbjct: 127 SGGGGG 132
Score = 40.8 bits (94), Expect = 1e-04
Identities = 26/64 (40%), Positives = 28/64 (43%), Gaps = 19/64 (29%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGS------GSDDGGGSGGGG 116
+GGG G GG+ GGG G GGGS GS GGG GGG
Sbjct: 151 EGGGAGASGYGGGAYGGGGGHGGG-------------GGGGSAGGAHGGSGYGGGEGGGA 197
Query: 117 GGGG 120
GGGG
Sbjct: 198 GGGG 201
Score = 26.9 bits (58), Expect = 1.6
Identities = 11/18 (61%), Positives = 11/18 (61%)
Query: 102 GGSGSDDGGGSGGGGGGG 119
GG G GGGSGG GG
Sbjct: 35 GGGGHGGGGGSGGVSSGG 52
Score = 26.2 bits (56), Expect = 2.8
Identities = 10/16 (62%), Positives = 10/16 (62%)
Query: 105 GSDDGGGSGGGGGGGG 120
G GGG GGGGG G
Sbjct: 31 GQASGGGGHGGGGGSG 46
Score = 25.4 bits (54), Expect = 4.7
Identities = 10/17 (58%), Positives = 10/17 (58%)
Query: 103 GSGSDDGGGSGGGGGGG 119
G S GG GGGG GG
Sbjct: 31 GQASGGGGHGGGGGSGG 47
>At4g39260 glycine-rich protein (clone AtGRP8)
Length = 169
Score = 49.7 bits (117), Expect = 2e-07
Identities = 28/65 (43%), Positives = 31/65 (47%), Gaps = 19/65 (29%)
Query: 64 GGGGNDGGGSGG----------SSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG 113
GGGG GGG GG GGG R + G R+ +GGG G DGG G
Sbjct: 113 GGGGYSGGGGGGYERRSGGYGSGGGGGGRGYGGGGRR---------EGGGYGGGDGGSYG 163
Query: 114 GGGGG 118
GGGGG
Sbjct: 164 GGGGG 168
Score = 45.4 bits (106), Expect = 4e-06
Identities = 26/57 (45%), Positives = 28/57 (48%), Gaps = 4/57 (7%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG SGGG + R + GGG G GG GGG GGG
Sbjct: 105 GGGGYSGGGGGGYSGGGGGGYERRSGGYGSG----GGGGGRGYGGGGRREGGGYGGG 157
Score = 43.1 bits (100), Expect = 2e-05
Identities = 25/56 (44%), Positives = 27/56 (47%), Gaps = 1/56 (1%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G G GGG GGS GGG R+ G + GGG GG G GGGGGG
Sbjct: 86 GSGGGGGGRGGS-GGGYRSGGGGGYSGGGGGGYSGGGGGGYERRSGGYGSGGGGGG 140
Score = 43.1 bits (100), Expect = 2e-05
Identities = 25/55 (45%), Positives = 27/55 (48%), Gaps = 5/55 (9%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG GG GG SGGG + G + R GG GS GGG G GGGG
Sbjct: 98 GGGYRSGGGGGYSGGGGGGYSGGGGGGYERR-----SGGYGSGGGGGGRGYGGGG 147
Score = 30.8 bits (68), Expect = 0.11
Identities = 12/19 (63%), Positives = 12/19 (63%)
Query: 66 GGNDGGGSGGSSGGGARAW 84
GG GGG GGS GGG W
Sbjct: 151 GGGYGGGDGGSYGGGGGGW 169
>At2g32690 unknown protein
Length = 201
Score = 49.7 bits (117), Expect = 2e-07
Identities = 27/56 (48%), Positives = 28/56 (49%), Gaps = 15/56 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG+GG GGGA G GG G GGG GGGGGGG
Sbjct: 132 GGGGGLGGGAGGGYGGGAGGGLG---------------GGGGIGGGGGFGGGGGGG 172
Score = 48.1 bits (113), Expect = 7e-07
Identities = 27/56 (48%), Positives = 27/56 (48%), Gaps = 19/56 (33%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGGSG GGG GGGSG GGG GGGGGGG
Sbjct: 94 GGGGGLGGGSGLGGGGGL-------------------GGGSGLGGGGGLGGGGGGG 130
Score = 48.1 bits (113), Expect = 7e-07
Identities = 26/57 (45%), Positives = 28/57 (48%), Gaps = 18/57 (31%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG GGG+G GGG+GGG GGGG
Sbjct: 119 GGGGLGGGGGGGLGGGGGL------------------GGGAGGGYGGGAGGGLGGGG 157
Score = 48.1 bits (113), Expect = 7e-07
Identities = 27/57 (47%), Positives = 27/57 (47%), Gaps = 13/57 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG GGGGG GG
Sbjct: 118 GGGGGLGGGGGGGLGGGGGLGGGA-------------GGGYGGGAGGGLGGGGGIGG 161
Score = 45.1 bits (105), Expect = 6e-06
Identities = 28/60 (46%), Positives = 28/60 (46%), Gaps = 16/60 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGG---GGGGG 120
GGGG GGG G GGG G GGGSG GGG GGG GGGGG
Sbjct: 76 GGGGGLGGGGGLGGGGGLGGGGGL-------------GGGSGLGGGGGLGGGSGLGGGGG 122
Score = 43.9 bits (102), Expect = 1e-05
Identities = 27/59 (45%), Positives = 27/59 (45%), Gaps = 21/59 (35%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGG--GGG 120
GGGG GGG GG GGGA GGG G GGG G GGG GGG
Sbjct: 160 GGGGGFGGGGGGGFGGGA-------------------GGGFGKGIGGGGGLGGGYVGGG 199
Score = 43.5 bits (101), Expect = 2e-05
Identities = 25/57 (43%), Positives = 25/57 (43%), Gaps = 19/57 (33%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG G GGG GGG G GGG GGGGG GG
Sbjct: 64 GGGGGLGGGGGLGGGGGL-------------------GGGGGLGGGGGLGGGGGLGG 101
Score = 29.6 bits (65), Expect = 0.25
Identities = 13/20 (65%), Positives = 13/20 (65%)
Query: 101 GGGSGSDDGGGSGGGGGGGG 120
G G G GGG GGGGG GG
Sbjct: 58 GFGKGLGGGGGLGGGGGLGG 77
Score = 26.9 bits (58), Expect = 1.6
Identities = 12/20 (60%), Positives = 12/20 (60%)
Query: 101 GGGSGSDDGGGSGGGGGGGG 120
G G G G G GGG GGGG
Sbjct: 60 GKGLGGGGGLGGGGGLGGGG 79
>At2g18115 putative protein
Length = 296
Score = 48.1 bits (113), Expect = 7e-07
Identities = 26/57 (45%), Positives = 26/57 (45%), Gaps = 18/57 (31%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GSGG GGG GGG GS GGG GGGGGGGG
Sbjct: 197 GIGGEPSPGSGGGGGGGG------------------GGGGGGSGPGGGGGGGGGGGG 235
Score = 48.1 bits (113), Expect = 7e-07
Identities = 25/56 (44%), Positives = 26/56 (45%), Gaps = 9/56 (16%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG GGG +G R W GGG G GGG G GG G
Sbjct: 224 GGGGGGGGGGGGGGGGVGDGYGYGRGW---------GGGYGGGGGGGWGWGGDSNG 270
Score = 47.4 bits (111), Expect = 1e-06
Identities = 26/56 (46%), Positives = 27/56 (47%), Gaps = 11/56 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG+ GG GG GGG G DG G G GGG GGGGGGG
Sbjct: 217 GGGGSGPGGGGGGGGGGGGGGGG-----------VGDGYGYGRGWGGGYGGGGGGG 261
Score = 45.1 bits (105), Expect = 6e-06
Identities = 26/58 (44%), Positives = 27/58 (45%), Gaps = 20/58 (34%)
Query: 64 GGGGNDG-GGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG + G GG GG GGG GGGSG GGG GGGGGGGG
Sbjct: 199 GGEPSPGSGGGGGGGGGGG-------------------GGGSGPGGGGGGGGGGGGGG 237
Score = 43.9 bits (102), Expect = 1e-05
Identities = 23/56 (41%), Positives = 24/56 (42%), Gaps = 17/56 (30%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G G GGG GG GGG+ GGG G GGG GGGG G G
Sbjct: 205 GSGGGGGGGGGGGGGGS-----------------GPGGGGGGGGGGGGGGGGVGDG 243
Score = 43.5 bits (101), Expect = 2e-05
Identities = 27/63 (42%), Positives = 27/63 (42%), Gaps = 20/63 (31%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDG------GGSGGGGG 117
GGGG GGGSG GGG G GGG G DG G G GGG
Sbjct: 212 GGGGGGGGGSGPGGGGGGGGGGG--------------GGGGGVGDGYGYGRGWGGGYGGG 257
Query: 118 GGG 120
GGG
Sbjct: 258 GGG 260
Score = 43.1 bits (100), Expect = 2e-05
Identities = 24/57 (42%), Positives = 24/57 (42%), Gaps = 19/57 (33%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG SG G GGG G GGG GG G G G
Sbjct: 208 GGGGGGGGGGGGGSGPGG-------------------GGGGGGGGGGGGGGVGDGYG 245
Score = 35.8 bits (81), Expect = 0.003
Identities = 26/80 (32%), Positives = 32/80 (39%), Gaps = 23/80 (28%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRW----RQWCDD------------------- 100
GG DGG GG SG R G+ + ++ C+D
Sbjct: 135 GGSSGDGGAGGGGSGRVGRRGNGKGDGKGQGDDGGKEDCNDHEKNKGDDRNNGDGVGIGI 194
Query: 101 GGGSGSDDGGGSGGGGGGGG 120
G G G + GSGGGGGGGG
Sbjct: 195 GIGIGGEPSPGSGGGGGGGG 214
Score = 35.4 bits (80), Expect = 0.005
Identities = 24/68 (35%), Positives = 24/68 (35%), Gaps = 28/68 (41%)
Query: 64 GGGGNDGGGS-----------GGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGS 112
GGGG GGG GG GGG WG W G D GG
Sbjct: 230 GGGGGGGGGGVGDGYGYGRGWGGGYGGGGGGGWG----W-------------GGDSNGGV 272
Query: 113 GGGGGGGG 120
G G GGG
Sbjct: 273 WGPGNGGG 280
Score = 29.6 bits (65), Expect = 0.25
Identities = 24/82 (29%), Positives = 27/82 (32%), Gaps = 29/82 (35%)
Query: 21 GGGDSGGG---GWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSS 77
GGG GGG G+ + R W GGGG G G GG S
Sbjct: 232 GGGGGGGGVGDGYGYGRGWGGGY-----------------------GGGGGGGWGWGGDS 268
Query: 78 GGGARAWWGRRRQWWRWRQWCD 99
GG WG W C+
Sbjct: 269 NGGV---WGPGNGGGCWFPGCE 287
Score = 28.1 bits (61), Expect = 0.73
Identities = 10/22 (45%), Positives = 12/22 (54%)
Query: 98 CDDGGGSGSDDGGGSGGGGGGG 119
C+ G+G G G GGGG G
Sbjct: 128 CEGCNGAGGSSGDGGAGGGGSG 149
Score = 28.1 bits (61), Expect = 0.73
Identities = 11/22 (50%), Positives = 12/22 (54%)
Query: 98 CDDGGGSGSDDGGGSGGGGGGG 119
C+ GGS D G G GG G G
Sbjct: 131 CNGAGGSSGDGGAGGGGSGRVG 152
Score = 27.7 bits (60), Expect = 0.95
Identities = 19/49 (38%), Positives = 20/49 (40%), Gaps = 6/49 (12%)
Query: 74 GGSSGGGARAWWGRRRQWWRWRQWCDDGGGS--GSDDGGGSGGGGGGGG 120
G GG GR R C G GS G + GGS G GG GG
Sbjct: 101 GDVEEGGCEICRGRERDCGG----CTGGEGSCEGCNGAGGSSGDGGAGG 145
Score = 27.3 bits (59), Expect = 1.2
Identities = 11/21 (52%), Positives = 12/21 (56%)
Query: 100 DGGGSGSDDGGGSGGGGGGGG 120
+G G S DGG GGG G G
Sbjct: 132 NGAGGSSGDGGAGGGGSGRVG 152
>At1g75550 hypothetical protein
Length = 167
Score = 48.1 bits (113), Expect = 7e-07
Identities = 25/56 (44%), Positives = 25/56 (44%), Gaps = 10/56 (17%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG GGG WG W GGG G G GGG G G
Sbjct: 72 GGGGGGGGGGGGGGGGGGGGGWG----------WGGGGGGGGWYKWGCGGGGKGKG 117
Score = 46.2 bits (108), Expect = 3e-06
Identities = 25/57 (43%), Positives = 25/57 (43%), Gaps = 14/57 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG W G GGG G G GGGG G G
Sbjct: 75 GGGGGGGGGGGGGGGGGGWGWGG--------------GGGGGGWYKWGCGGGGKGKG 117
Score = 45.4 bits (106), Expect = 4e-06
Identities = 21/47 (44%), Positives = 22/47 (46%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGG 110
GGGG GGG GG GGG WG W +W GGG G G
Sbjct: 74 GGGGGGGGGGGGGGGGGGGWGWGGGGGGGGWYKWGCGGGGKGKGREG 120
Score = 42.4 bits (98), Expect = 4e-05
Identities = 25/63 (39%), Positives = 27/63 (42%), Gaps = 23/63 (36%)
Query: 63 DGGGGN-----DGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGG 117
+GG G+ GGG GG GGG GGG G G G GGGGG
Sbjct: 59 NGGSGSYRWGWGGGGGGGGGGGGG------------------GGGGGGGGGGWGWGGGGG 100
Query: 118 GGG 120
GGG
Sbjct: 101 GGG 103
Score = 42.4 bits (98), Expect = 4e-05
Identities = 18/29 (62%), Positives = 19/29 (65%), Gaps = 1/29 (3%)
Query: 92 WRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+RW W GGG G GGG GGGGGGGG
Sbjct: 65 YRWG-WGGGGGGGGGGGGGGGGGGGGGGG 92
Score = 29.3 bits (64), Expect = 0.33
Identities = 18/62 (29%), Positives = 19/62 (30%), Gaps = 31/62 (50%)
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
GGG GGGGW W GGGG GG GGG
Sbjct: 84 GGGGGGGGGWGW-------------------------------GGGGGGGGWYKWGCGGG 112
Query: 81 AR 82
+
Sbjct: 113 GK 114
>At3g20470 glycine-rich protein, putative
Length = 174
Score = 47.4 bits (111), Expect = 1e-06
Identities = 26/57 (45%), Positives = 27/57 (46%), Gaps = 13/57 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G GSGG GGG A G GGG+G GGG GGG G GG
Sbjct: 90 GGGFGGGAGSGGGLGGGGGAGGGF-------------GGGAGGGSGGGFGGGAGAGG 133
Score = 47.0 bits (110), Expect = 2e-06
Identities = 29/57 (50%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGGSGG GGGA A G GGG GS GGG GGG G GG
Sbjct: 112 GFGGGAGGGSGGGFGGGAGAGGGLGGGGGAGGGG-GFGGGGGSGIGGGFGGGAGAGG 167
Score = 45.8 bits (107), Expect = 3e-06
Identities = 25/57 (43%), Positives = 28/57 (48%), Gaps = 13/57 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG +GGG G GGG+G GGG+G GGG GG
Sbjct: 62 GGGFGGGGGLGGGAGGGGGLGGGA-------------GGGAGGGFGGGAGSGGGLGG 105
Score = 43.1 bits (100), Expect = 2e-05
Identities = 26/62 (41%), Positives = 28/62 (44%), Gaps = 16/62 (25%)
Query: 62 VDGGGGNDGG---GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
+ GG G GG G GG GGGA G GGG+G GGG GGG G
Sbjct: 53 IGGGSGLGGGGGFGGGGGLGGGAGGGGGL-------------GGGAGGGAGGGFGGGAGS 99
Query: 119 GG 120
GG
Sbjct: 100 GG 101
Score = 40.4 bits (93), Expect = 1e-04
Identities = 24/57 (42%), Positives = 24/57 (42%), Gaps = 21/57 (36%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG SG G GG G GGG GGG GGGG
Sbjct: 44 GGGLGGGGGIGGGSGLG---------------------GGGGFGGGGGLGGGAGGGG 79
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.142 0.499
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,545,024
Number of Sequences: 26719
Number of extensions: 217901
Number of successful extensions: 16579
Number of sequences better than 10.0: 472
Number of HSP's better than 10.0 without gapping: 406
Number of HSP's successfully gapped in prelim test: 69
Number of HSP's that attempted gapping in prelim test: 1528
Number of HSP's gapped (non-prelim): 4179
length of query: 120
length of database: 11,318,596
effective HSP length: 96
effective length of query: 24
effective length of database: 8,753,572
effective search space: 210085728
effective search space used: 210085728
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0090b.2


