

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0086.14
(430 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
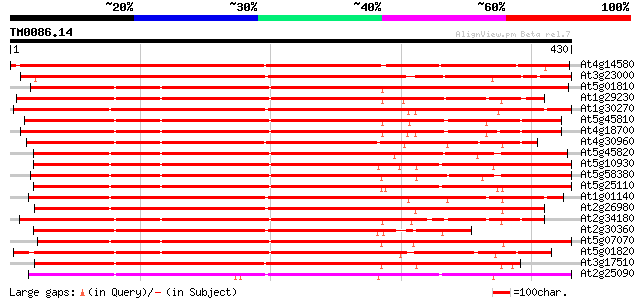
Score E
Sequences producing significant alignments: (bits) Value
At4g14580 CBL-interacting protein kinase 4 (CIPK4) 507 e-144
At3g23000 SNF1 related protein kinase (ATSRPK1) 488 e-138
At5g01810 serine/threonine protein kinase ATPK10 373 e-104
At1g29230 CBL-interacting protein kinase 18 (CIPK18) 357 9e-99
At1g30270 serine/threonine kinase, putative 351 5e-97
At5g45810 CBL-interacting protein kinase 19 (CIPK19) 350 1e-96
At4g18700 putative protein kinase 349 1e-96
At4g30960 CBL-interacting protein kinase 6 (CIPK6) 342 2e-94
At5g45820 CBL-interacting protein kinase 20 (CIPK20) 340 7e-94
At5g10930 serine/threonine protein kinase -like protein 340 1e-93
At5g58380 SOS2-like protein kinase PKS2 338 3e-93
At5g25110 serine/threonine protein kinase-like protein 336 1e-92
At1g01140 SOS2-like protein kinase PKS6/CBL-interacting protein ... 336 1e-92
At2g26980 putative protein kinase (At2g26980) 330 7e-91
At2g34180 putative protein kinase 327 6e-90
At2g30360 putative protein kinase 326 1e-89
At5g07070 serine/threonine protein kinase-like protein 325 2e-89
At5g01820 unknown protein 325 4e-89
At3g17510 CBL-interacting protein kinase 1 (CIPK1) 313 2e-85
At2g25090 CBL-interacting protein kinase 16 (CIPK16) 313 2e-85
>At4g14580 CBL-interacting protein kinase 4 (CIPK4)
Length = 426
Score = 507 bits (1306), Expect = e-144
Identities = 266/434 (61%), Positives = 330/434 (75%), Gaps = 13/434 (2%)
Query: 1 MEPPMRQPPPPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTV 60
ME P P P T +LGKY++ R LG GSFAKV+ ARS+ G +VA+KIIDK KT+
Sbjct: 1 MESPY--PKSPEKITGTVLLGKYELGRRLGSGSFAKVHVARSISTGELVAIKIIDKQKTI 58
Query: 61 DAAMEPRIVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLP 120
D+ MEPRI+REI+AMRRLH+HPN+L+IHEVMATK+KI+LVVE+AAGGELF+ + R G+L
Sbjct: 59 DSGMEPRIIREIEAMRRLHNHPNVLKIHEVMATKSKIYLVVEYAAGGELFTKLIRFGRLN 118
Query: 121 ENTARRYFQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEH-LQNGL 179
E+ ARRYFQQL SAL FCHR+G+AHRD+KPQNLLLD GNLKVSDFGLSALPEH NGL
Sbjct: 119 ESAARRYFQQLASALSFCHRDGIAHRDVKPQNLLLDKQGNLKVSDFGLSALPEHRSNNGL 178
Query: 180 LHTACGTPAYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISR 239
LHTACGTPAYTAPE++ GYDG+KADAWSCG+ LFVLLAGY+PFDD+NI AMYRKI +
Sbjct: 179 LHTACGTPAYTAPEVI-AQRGYDGAKADAWSCGVFLFVLLAGYVPFDDANIVAMYRKIHK 237
Query: 240 RDYQFPEWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTLNLD 299
RDY+FP WI+KPAR +I +LLDPNPETRMS+E + G WF+KSL++ SE +SS LD
Sbjct: 238 RDYRFPSWISKPARSIIYKLLDPNPETRMSIEAVMGTVWFQKSLEI---SEFQSSVFELD 294
Query: 300 SGYGNRVRGL-GVNAFDLISMSSGLDLSGLFQDEGKRKEKRFTSGAKLEVVEEKVKEIGG 358
+ + AFDLIS+SSGLDLSGLF + KRKEKRFT+ E V EK IG
Sbjct: 295 RFLEKEAKSSNAITAFDLISLSSGLDLSGLF-ERRKRKEKRFTARVSAERVVEKAGMIGE 353
Query: 359 VLGFKVEVGKDSTAIGLVKGKVALVVQVFEILPDELLLVAVKVVEGGLEFE---ENHWGD 415
LGF+VE +++ +GL KG+ A+VV+V E + L++ VKVV G E E E+HW +
Sbjct: 354 KLGFRVEKKEETKVVGLGKGRTAVVVEVVE-FAEGLVVADVKVVVEGEEEEEEVESHWSE 412
Query: 416 WKLGLQDLVLSWYN 429
+ L+++VLSW+N
Sbjct: 413 LIVELEEIVLSWHN 426
>At3g23000 SNF1 related protein kinase (ATSRPK1)
Length = 429
Score = 488 bits (1256), Expect = e-138
Identities = 259/432 (59%), Positives = 327/432 (74%), Gaps = 21/432 (4%)
Query: 9 PPPRSSAATT----ILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAM 64
P +SS ATT +LGKY++ R LG GSFAKV+ ARS+ +VAVKII+K KT+++ M
Sbjct: 7 PQNQSSPATTPAKILLGKYELGRRLGSGSFAKVHLARSIESDELVAVKIIEKKKTIESGM 66
Query: 65 EPRIVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTA 124
EPRI+REIDAMRRL HHPNIL+IHEVMATK+KI+LV+E A+GGELFS + RRG+LPE+TA
Sbjct: 67 EPRIIREIDAMRRLRHHPNILKIHEVMATKSKIYLVMELASGGELFSKVLRRGRLPESTA 126
Query: 125 RRYFQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTAC 184
RRYFQQL SALRF H++GVAHRD+KPQNLLLD GNLKVSDFGLSALPEHLQNGLLHTAC
Sbjct: 127 RRYFQQLASALRFSHQDGVAHRDVKPQNLLLDEQGNLKVSDFGLSALPEHLQNGLLHTAC 186
Query: 185 GTPAYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQF 244
GTPAYTAPE++ GYDG+KADAWSCG++LFVLL G +PFDDSNIAAMYRKI RRDY+F
Sbjct: 187 GTPAYTAPEVIS-RRGYDGAKADAWSCGVILFVLLVGDVPFDDSNIAAMYRKIHRRDYRF 245
Query: 245 PEWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKV-EAESETESSTLNLDSGYG 303
P WI+K A+ +I ++LDPNP TRMS+E + +WFKKSL+ E S + + S
Sbjct: 246 PSWISKQAKSIIYQMLDPNPVTRMSIETVMKTNWFKKSLETSEFHRNVFDSEVEMKSSVN 305
Query: 304 NRVRGLGVNAFDLISMSSGLDLSGLFQDEGKRKEKRFTSGAKLEVVEEKVKEIGGVLGFK 363
+ + AFDLIS+SSGLDLSGLF+ + K+KE+RFT+ VEEK K IG LG+
Sbjct: 306 S------ITAFDLISLSSGLDLSGLFEAK-KKKERRFTAKVSGVEVEEKAKMIGEKLGYV 358
Query: 364 VEVGK-----DSTAIGLVKGKVALVVQVFEILPDELLLVAVKVVEGGLEFEENHWGDWKL 418
V+ + +GL +G+ +VV+ E+ D +++V VKVVEG E +++ W D
Sbjct: 359 VKKKMMKKEGEVKVVGLGRGRTVIVVEAVELTVD-VVVVEVKVVEG--EEDDSRWSDLIT 415
Query: 419 GLQDLVLSWYNE 430
L+D+VLSW+N+
Sbjct: 416 ELEDIVLSWHND 427
>At5g01810 serine/threonine protein kinase ATPK10
Length = 421
Score = 373 bits (958), Expect = e-104
Identities = 198/419 (47%), Positives = 276/419 (65%), Gaps = 10/419 (2%)
Query: 17 TTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMR 76
+ ++ +Y+V + LG+G+FAKVY AR L G VA+K+IDK + + M +I REI AMR
Sbjct: 6 SVLMLRYEVGKFLGQGTFAKVYHARHLKTGDSVAIKVIDKERILKVGMTEQIKREISAMR 65
Query: 77 RLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALR 136
L H PNI+ +HEVMATK+KI+ V+E GGELF+ +S GKL E+ AR+YFQQLV A+
Sbjct: 66 LLRH-PNIVELHEVMATKSKIYFVMEHVKGGELFNKVST-GKLREDVARKYFQQLVRAVD 123
Query: 137 FCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPE-HLQNGLLHTACGTPAYTAPEIL 195
FCH GV HRDLKP+NLLLD GNLK+SDFGLSAL + Q+GLLHT CGTPAY APE++
Sbjct: 124 FCHSRGVCHRDLKPENLLLDEHGNLKISDFGLSALSDSRRQDGLLHTTCGTPAYVAPEVI 183
Query: 196 RLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFV 255
+G YDG KAD WSCG++LFVLLAGYLPF DSN+ +Y+KI + + +FP W+ A+ +
Sbjct: 184 SRNG-YDGFKADVWSCGVILFVLLAGYLPFRDSNLMELYKKIGKAEVKFPNWLAPGAKRL 242
Query: 256 IRRLLDPNPETRMSVEDLYGNSWFKKSLK------VEAESETESSTLNLDSGYGNRVRGL 309
++R+LDPNP TR+S E + +SWF+K L+ VE E+E ++ S + R +
Sbjct: 243 LKRILDPNPNTRVSTEKIMKSSWFRKGLQEEVKESVEEETEVDAEAEGNASAEKEKKRCI 302
Query: 310 GVNAFDLISMSSGLDLSGLFQDEGKRKEKRFTSGAKLEVVEEKVKEIGGVLGFKVEVGKD 369
+NAF++IS+S+G DLSGLF+ +++E RFTS + + EK+ EIG L KV +
Sbjct: 303 NLNAFEIISLSTGFDLSGLFEKGEEKEEMRFTSNREASEITEKLVEIGKDLKMKVRKKEH 362
Query: 370 STAIGLVKGKVALVVQVFEILPDELLLVAVKVVEGGLEFEENHWGDWKLGLQDLVLSWY 428
+ + + +VFEI P ++V K E++ + L D VL+W+
Sbjct: 363 EWRVKMSAEATVVEAEVFEIAPSYHMVVLKKSGGDTAEYKRVMKESIRPALIDFVLAWH 421
>At1g29230 CBL-interacting protein kinase 18 (CIPK18)
Length = 520
Score = 357 bits (915), Expect = 9e-99
Identities = 199/439 (45%), Positives = 278/439 (62%), Gaps = 43/439 (9%)
Query: 6 RQPPPPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAME 65
+ P PRS ++GKY++ +LLG G+FAKVY A+++ G VA+K+IDK K + + +
Sbjct: 57 QSPRSPRSPRNNILMGKYELGKLLGHGTFAKVYLAQNIKSGDKVAIKVIDKEKIMKSGLV 116
Query: 66 PRIVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTAR 125
I REI +RR+ H P I+ + EVMATK+KI+ V+E+ GGELF+ +++ G+LPE TAR
Sbjct: 117 AHIKREISILRRVRH-PYIVHLFEVMATKSKIYFVMEYVGGGELFNTVAK-GRLPEETAR 174
Query: 126 RYFQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHL-QNGLLHTAC 184
RYFQQL+S++ FCH GV HRDLKP+NLLLD GNLKVSDFGLSA+ E L Q+GL HT C
Sbjct: 175 RYFQQLISSVSFCHGRGVYHRDLKPENLLLDNKGNLKVSDFGLSAVAEQLRQDGLCHTFC 234
Query: 185 GTPAYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQF 244
GTPAY APE+L G YD +KAD WSCG++LFVL+AG++PF D NI MY+KI + +++
Sbjct: 235 GTPAYIAPEVLTRKG-YDAAKADVWSCGVILFVLMAGHIPFYDKNIMVMYKKIYKGEFRC 293
Query: 245 PEWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLK-------------------V 285
P W + ++ RLLD NP+TR+++ ++ N WFKK K
Sbjct: 294 PRWFSSDLVRLLTRLLDTNPDTRITIPEIMKNRWFKKGFKHVKFYIEDDKLCREDEDEEE 353
Query: 286 EAESETESSTLNLDS--------GYGNRVRGLGVNAFDLISMSSGLDLSGLFQDEGKRKE 337
EA S SST++ G G+ R +NAFD+IS SSG DLSGLF++EG +
Sbjct: 354 EASSSGRSSTVSESDAEFDVKRMGIGSMPRPSSLNAFDIISFSSGFDLSGLFEEEG-GEG 412
Query: 338 KRFTSGAKLEVVEEKVKEIGGVLGFKVEVGKDSTAIGL------VKGKVALVVQVFEILP 391
RF SGA + + K++EI ++ F V K ++ L KG + + ++FE+ P
Sbjct: 413 TRFVSGAPVSKIISKLEEIAKIVSFTVR--KKEWSLRLEGCREGAKGPLTIAAEIFELTP 470
Query: 392 DELLLVAVKVVEGGLEFEE 410
LV V+V + G + EE
Sbjct: 471 S---LVVVEVKKKGGDREE 486
>At1g30270 serine/threonine kinase, putative
Length = 482
Score = 351 bits (900), Expect = 5e-97
Identities = 197/449 (43%), Positives = 283/449 (62%), Gaps = 25/449 (5%)
Query: 4 PMRQPPPPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAA 63
P R P SS T +GKY++ R LG G+FAKV AR++ +G VA+K+IDK K +
Sbjct: 12 PSRSTPSGSSSGGRTRVGKYELGRTLGEGTFAKVKFARNVENGDNVAIKVIDKEKVLKNK 71
Query: 64 MEPRIVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENT 123
M +I REI M+ L HPN++R+ EVMA+KTKI+ V+EF GGELF IS G+L E+
Sbjct: 72 MIAQIKREISTMK-LIKHPNVIRMFEVMASKTKIYFVLEFVTGGELFDKISSNGRLKEDE 130
Query: 124 ARRYFQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHL-QNGLLHT 182
AR+YFQQL++A+ +CH GV HRDLKP+NLLLDA G LKVSDFGLSALP+ + ++GLLHT
Sbjct: 131 ARKYFQQLINAVDYCHSRGVYHRDLKPENLLLDANGALKVSDFGLSALPQQVREDGLLHT 190
Query: 183 ACGTPAYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDY 242
CGTP Y APE++ + GYDG+KAD WSCG++LFVL+AGYLPF+DSN+ ++Y+KI + ++
Sbjct: 191 TCGTPNYVAPEVIN-NKGYDGAKADLWSCGVILFVLMAGYLPFEDSNLTSLYKKIFKAEF 249
Query: 243 QFPEWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTLNLDSGY 302
P W + A+ +I+R+LDPNP TR++ ++ N WFKK K + S ++D+ +
Sbjct: 250 TCPPWFSASAKKLIKRILDPNPATRITFAEVIENEWFKKGYKAPKFENADVSLDDVDAIF 309
Query: 303 GN-----------RVRGL----GVNAFDLISMSSGLDLSGLFQDE-GKRKEK-RFTSGAK 345
+ R GL +NAF+LIS S GL+L LF+ + G K K RFTS +
Sbjct: 310 DDSGESKNLVVERREEGLKTPVTMNAFELISTSQGLNLGSLFEKQMGLVKRKTRFTSKSS 369
Query: 346 LEVVEEKVKEIGGVLGFKVEVGKDSTAI----GLVKGKVALVVQVFEILPDELLLVAVKV 401
+ K++ +GF V+ + KG++A+ +VF++ P ++ K
Sbjct: 370 ANEIVTKIEAAAAPMGFDVKTNNYKMKLTGEKSGRKGQLAVATEVFQVAPSLYMVEMRKS 429
Query: 402 VEGGLEFEENHWGDWKLGLQDLVLSWYNE 430
LEF + + + GL+D+V +E
Sbjct: 430 GGDTLEFHK-FYKNLTTGLKDIVWKTIDE 457
>At5g45810 CBL-interacting protein kinase 19 (CIPK19)
Length = 483
Score = 350 bits (897), Expect = 1e-96
Identities = 201/450 (44%), Positives = 280/450 (61%), Gaps = 44/450 (9%)
Query: 12 RSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVRE 71
+S+ ILGKY++ RLLG G+FAKVY AR+ G VA+K+IDK K + + + I RE
Sbjct: 17 QSNHQALILGKYEMGRLLGHGTFAKVYLARNAQSGESVAIKVIDKEKVLKSGLIAHIKRE 76
Query: 72 IDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQL 131
I +RR+ H PNI+++ EVMATK+KI+ V+E+ GGELF+ +++ G+L E AR+YFQQL
Sbjct: 77 ISILRRVRH-PNIVQLFEVMATKSKIYFVMEYVKGGELFNKVAK-GRLKEEMARKYFQQL 134
Query: 132 VSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHL-QNGLLHTACGTPAYT 190
+SA+ FCH GV HRDLKP+NLLLD GNLKVSDFGLSA+ + + Q+GL HT CGTPAY
Sbjct: 135 ISAVSFCHFRGVYHRDLKPENLLLDENGNLKVSDFGLSAVSDQIRQDGLFHTFCGTPAYV 194
Query: 191 APEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITK 250
APE+L G YDG+K D WSCG++LFVL+AG+LPF D N+ AMY+KI R D++ P W
Sbjct: 195 APEVLARKG-YDGAKVDIWSCGVILFVLMAGFLPFHDRNVMAMYKKIYRGDFRCPRWFPV 253
Query: 251 PARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLK-------------------------- 284
++ R+L+ PE R ++ D+ SWFKK K
Sbjct: 254 EINRLLIRMLETKPERRFTMPDIMETSWFKKGFKHIKFYVEDDHQLCNVADDDEIESIES 313
Query: 285 VEAESETESSTLNLDSGYGNR-----VRGLGVNAFDLISMSSGLDLSGLFQDEGKRKEKR 339
V S T S + +S G R R +NAFDLIS S G DLSGLF+D+G + R
Sbjct: 314 VSGRSSTVSEPEDFESFDGRRRGGSMPRPASLNAFDLISFSPGFDLSGLFEDDG--EGSR 371
Query: 340 FTSGAKLEVVEEKVKEIGGVLGFKV-----EVGKDSTAIGLVKGKVALVVQVFEILPDEL 394
F SGA + + K++EI ++ F V +V + + G +KG +++ ++FE+ P L
Sbjct: 372 FVSGAPVGQIISKLEEIARIVSFTVRKKDCKVSLEGSREGSMKGPLSIAAEIFELTP-AL 430
Query: 395 LLVAVKVVEGG-LEFEENHWGDWKLGLQDL 423
++V VK G +E++E + K LQ+L
Sbjct: 431 VVVEVKKKGGDKMEYDEFCNKELKPKLQNL 460
>At4g18700 putative protein kinase
Length = 489
Score = 349 bits (896), Expect = 1e-96
Identities = 203/451 (45%), Positives = 278/451 (61%), Gaps = 43/451 (9%)
Query: 9 PPPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRI 68
P RSS ILG+Y++ +LLG G+FAKVY AR++ VA+K+IDK K + + I
Sbjct: 12 PKERSSPQALILGRYEMGKLLGHGTFAKVYLARNVKTNESVAIKVIDKEKVLKGGLIAHI 71
Query: 69 VREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYF 128
REI +RR+ H PNI+++ EVMATK KI+ V+E+ GGELF+ +++ G+L E AR+YF
Sbjct: 72 KREISILRRVRH-PNIVQLFEVMATKAKIYFVMEYVRGGELFNKVAK-GRLKEEVARKYF 129
Query: 129 QQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHL-QNGLLHTACGTP 187
QQL+SA+ FCH GV HRDLKP+NLLLD GNLKVSDFGLSA+ + + Q+GL HT CGTP
Sbjct: 130 QQLISAVTFCHARGVYHRDLKPENLLLDENGNLKVSDFGLSAVSDQIRQDGLFHTFCGTP 189
Query: 188 AYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEW 247
AY APE+L G YD +K D WSCG++LFVL+AGYLPF D N+ AMY+KI R +++ P W
Sbjct: 190 AYVAPEVLARKG-YDAAKVDIWSCGVILFVLMAGYLPFHDRNVMAMYKKIYRGEFRCPRW 248
Query: 248 ITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLK-------------VEAESETESS 294
+ ++ +LL+ NPE R + ++ NSWFKK K V + E ES
Sbjct: 249 FSTELTRLLSKLLETNPEKRFTFPEIMENSWFKKGFKHIKFYVEDDKLCNVVDDDELESD 308
Query: 295 TLNLDSGYG------------NRVRGL----GVNAFDLISMSSGLDLSGLFQDEGKRKEK 338
++ D RV GL +NAFD+IS S G DLSGLF D+G +
Sbjct: 309 SVESDRDSAASESEIEYLEPRRRVGGLPRPASLNAFDIISFSQGFDLSGLFDDDG--EGS 366
Query: 339 RFTSGAKLEVVEEKVKEIGGVLGFKV-----EVGKDSTAIGLVKGKVALVVQVFEILPDE 393
RF SGA + + K++EI V+ F V V + + G VKG + + ++FE+ P
Sbjct: 367 RFVSGAPVSKIISKLEEIAKVVSFTVRKKDCRVSLEGSRQG-VKGPLTIAAEIFELTP-S 424
Query: 394 LLLVAVKVVEGG-LEFEENHWGDWKLGLQDL 423
L++V VK G E+E+ + K LQ+L
Sbjct: 425 LVVVEVKKKGGDKTEYEDFCNNELKPKLQNL 455
>At4g30960 CBL-interacting protein kinase 6 (CIPK6)
Length = 441
Score = 342 bits (878), Expect = 2e-94
Identities = 187/406 (46%), Positives = 266/406 (65%), Gaps = 21/406 (5%)
Query: 14 SAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREID 73
S+ + G+Y++ RLLG G+FAKVY AR++ G VA+K++ K K V M +I REI
Sbjct: 15 SSTGLLHGRYELGRLLGHGTFAKVYHARNIQTGKSVAMKVVGKEKVVKVGMVDQIKREIS 74
Query: 74 AMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVS 133
MR + H PNI+ +HEVMA+K+KI+ +E GGELF+ +++ G+L E+ AR YFQQL+S
Sbjct: 75 VMRMVKH-PNIVELHEVMASKSKIYFAMELVRGGELFAKVAK-GRLREDVARVYFQQLIS 132
Query: 134 ALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHL-QNGLLHTACGTPAYTAP 192
A+ FCH GV HRDLKP+NLLLD GNLKV+DFGLSA EHL Q+GLLHT CGTPAY AP
Sbjct: 133 AVDFCHSRGVYHRDLKPENLLLDEEGNLKVTDFGLSAFTEHLKQDGLLHTTCGTPAYVAP 192
Query: 193 EILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPA 252
E++ L GYDG+KAD WSCG++LFVLLAGYLPF D N+ MYRKI R D++ P W++ A
Sbjct: 193 EVI-LKKGYDGAKADLWSCGVILFVLLAGYLPFQDDNLVNMYRKIYRGDFKCPGWLSSDA 251
Query: 253 RFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTLNLDSG------YGNRV 306
R ++ +LLDPNP TR+++E + + WFKK + +E ++T+ + ++
Sbjct: 252 RRLVTKLLDPNPNTRITIEKVMDSPWFKKQ-ATRSRNEPVAATITTTEEDVDFLVHKSKE 310
Query: 307 RGLGVNAFDLISMSSGLDLSGLFQDEGK--RKEKRFTSGAKLEVVEEKVKEIGGVLGFKV 364
+NAF +I++S G DLS LF+++ K ++E RF + V ++E V G K
Sbjct: 311 ETETLNAFHIIALSEGFDLSPLFEEKKKEEKREMRFATSRPASSVISSLEEAARV-GNKF 369
Query: 365 EVGKDSTAIGLV------KGKVALVVQVFEILPDELLLVAVKVVEG 404
+V K + + + KGK+A+ ++F + P ++V VK G
Sbjct: 370 DVRKSESRVRIEGKQNGRKGKLAVEAEIFAVAP-SFVVVEVKKDHG 414
>At5g45820 CBL-interacting protein kinase 20 (CIPK20)
Length = 439
Score = 340 bits (873), Expect = 7e-94
Identities = 189/425 (44%), Positives = 272/425 (63%), Gaps = 25/425 (5%)
Query: 19 ILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRL 78
++ KY++ RLLG+G+FAKVY AR++ G VA+K+IDK K + +I REI MR L
Sbjct: 8 LMRKYELGRLLGQGTFAKVYHARNIKTGESVAIKVIDKQKVAKVGLIDQIKREISVMR-L 66
Query: 79 HHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFC 138
HP+++ +HEVMA+KTKI+ +E+ GGELF +S+ GKL EN AR+YFQQL+ A+ +C
Sbjct: 67 VRHPHVVFLHEVMASKTKIYFAMEYVKGGELFDKVSK-GKLKENIARKYFQQLIGAIDYC 125
Query: 139 HRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQ-NGLLHTACGTPAYTAPEILRL 197
H GV HRDLKP+NLLLD G+LK+SDFGLSAL E Q +GLLHT CGTPAY APE++
Sbjct: 126 HSRGVYHRDLKPENLLLDENGDLKISDFGLSALRESKQQDGLLHTTCGTPAYVAPEVIGK 185
Query: 198 SGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIR 257
G YDG+KAD WSCG+VL+VLLAG+LPF + N+ MYRKI++ +++ P W + ++
Sbjct: 186 KG-YDGAKADVWSCGVVLYVLLAGFLPFHEQNLVEMYRKITKGEFKCPNWFPPEVKKLLS 244
Query: 258 RLLDPNPETRMSVEDLYGNSWFKKSL-KVEAESETES---STLNLDSGYGNRVRGLGVNA 313
R+LDPNP +R+ +E + NSWF+K K+E ES +L D V+ + NA
Sbjct: 245 RILDPNPNSRIKIEKIMENSWFQKGFKKIETPKSPESHQIDSLISDVHAAFSVKPMSYNA 304
Query: 314 FDLI-SMSSGLDLSGLFQDEGKRKEKRFTSGAKLEVVEEKVKEIG----------GVLGF 362
FDLI S+S G DLSGLF+ E +R E +FT+ + + K +EI +G
Sbjct: 305 FDLISSLSQGFDLSGLFEKE-ERSESKFTTKKDAKEIVSKFEEIATSSERFNLTKSDVGV 363
Query: 363 KVEVGKDSTAIGLVKGKVALVVQVFEILPDELLLVAVKVVEGGLEFEENHWGDWKLGLQD 422
K+E ++ KG +A+ V++FE+ ++ K +E+++ + + L+D
Sbjct: 364 KMEDKREGR-----KGHLAIDVEIFEVTNSFHMVEFKKSGGDTMEYKQFCDRELRPSLKD 418
Query: 423 LVLSW 427
+V W
Sbjct: 419 IVWKW 423
>At5g10930 serine/threonine protein kinase -like protein
Length = 445
Score = 340 bits (871), Expect = 1e-93
Identities = 200/434 (46%), Positives = 276/434 (63%), Gaps = 27/434 (6%)
Query: 19 ILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDA-AMEPRIVREIDAMRR 77
+ GKY++ RLLG+G+FAKVY + +I G VA+K+I+K + + M +I REI M+
Sbjct: 8 LFGKYEMGRLLGKGTFAKVYYGKEIIGGECVAIKVINKDQVMKRPGMMEQIKREISIMK- 66
Query: 78 LHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRF 137
L HPNI+ + EVMATKTKI V+EF GGELF IS+ GKL E+ ARRYFQQL+SA+ +
Sbjct: 67 LVRHPNIVELKEVMATKTKIFFVMEFVKGGELFCKISK-GKLHEDAARRYFQQLISAVDY 125
Query: 138 CHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEH-LQNGLLHTACGTPAYTAPEILR 196
CH GV+HRDLKP+NLLLD G+LK+SDFGLSALPE LQ+GLLHT CGTPAY APE+L+
Sbjct: 126 CHSRGVSHRDLKPENLLLDENGDLKISDFGLSALPEQILQDGLLHTQCGTPAYVAPEVLK 185
Query: 197 LSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVI 256
G YDG+KAD WSCG+VL+VLLAG LPF D N+ MYRKI R D++FP W + AR +I
Sbjct: 186 KKG-YDGAKADIWSCGVVLYVLLAGCLPFQDENLMNMYRKIFRADFEFPPWFSPEARRLI 244
Query: 257 RRLLDPNPETRMSVEDLYGNSWFKK------SLKVEAESETESSTLN----LDSGYGNRV 306
+LL +P+ R+S+ + W +K + K++ ++SS N D N+
Sbjct: 245 SKLLVVDPDRRISIPAIMRTPWLRKNFTPPLAFKIDEPICSQSSKNNEEEEEDGDCENQT 304
Query: 307 RGLG---VNAFDLI-SMSSGLDLSGLFQDEGKRK-EKRFTSGAKLEVVEEKVKEIGGVLG 361
+ NAF+ I SMSSG DLS LF E KRK + FTS + V EK++ + +
Sbjct: 305 EPISPKFFNAFEFISSMSSGFDLSSLF--ESKRKVQSVFTSRSSATEVMEKIETVTKEMN 362
Query: 362 FKVEVGKD-----STAIGLVKGKVALVVQVFEILPDELLLVAVKVVEGGLEFEENHWGDW 416
KV+ KD KG++++ +VFE+ P+ ++ K LE++ + +
Sbjct: 363 MKVKRTKDFKVKMEGKTEGRKGRLSMTAEVFEVAPEISVVEFCKSAGDTLEYDRLYEEEV 422
Query: 417 KLGLQDLVLSWYNE 430
+ L D+V SW+ +
Sbjct: 423 RPALNDIVWSWHGD 436
>At5g58380 SOS2-like protein kinase PKS2
Length = 479
Score = 338 bits (868), Expect = 3e-93
Identities = 194/454 (42%), Positives = 278/454 (60%), Gaps = 49/454 (10%)
Query: 17 TTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMR 76
+ + KY V RLLG+G+FAKVY RS++ VA+K+IDK K + + +I REI MR
Sbjct: 6 SVLTDKYDVGRLLGQGTFAKVYYGRSILTNQSVAIKMIDKEKVMKVGLIEQIKREISVMR 65
Query: 77 RLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALR 136
+ HPN++ ++EVMATKT+I+ V+E+ GGELF+ +++ GKL ++ A +YF QL++A+
Sbjct: 66 -IARHPNVVELYEVMATKTRIYFVMEYCKGGELFNKVAK-GKLRDDVAWKYFYQLINAVD 123
Query: 137 FCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPE-HLQNGLLHTACGTPAYTAPEIL 195
FCH V HRD+KP+NLLLD NLKVSDFGLSAL + Q+GLLHT CGTPAY APE++
Sbjct: 124 FCHSREVYHRDIKPENLLLDDNENLKVSDFGLSALADCKRQDGLLHTTCGTPAYVAPEVI 183
Query: 196 RLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFV 255
G YDG+KAD WSCG+VLFVLLAGYLPF DSN+ MYRKI + D++ P W R +
Sbjct: 184 NRKG-YDGTKADIWSCGVVLFVLLAGYLPFHDSNLMEMYRKIGKADFKAPSWFAPEVRRL 242
Query: 256 IRRLLDPNPETRMSVEDLYGNSWFKKSL---------KVEAESETESSTLNL-DSGYGNR 305
+ ++LDPNPETR+++ + +SWF+K L +V+ + E+ T ++G G
Sbjct: 243 LCKMLDPNPETRITIARIRESSWFRKGLHMKQKKMEKRVKEINSVEAGTAGTNENGAGPS 302
Query: 306 VRGLG-------------------VNAFDLISMSSGLDLSGLFQDEGKRKEKRFTSGAKL 346
G G +NAFDLI++S+G DL+GLF D+ KR E RFTS
Sbjct: 303 ENGAGPSENGDRVTEENHTDEPTNLNAFDLIALSAGFDLAGLFGDDNKR-ESRFTSQKPA 361
Query: 347 EVVEEKVKEIGGVLG----------FKVEVGKDSTAIGLVKGKVALVVQVFEILPDELLL 396
V+ K++E+ L FK+E K+ KG +++ ++F++ P+ L+
Sbjct: 362 SVIISKLEEVAQRLKLSIRKREAGLFKLERLKEGR-----KGILSMDAEIFQVTPNFHLV 416
Query: 397 VAVKVVEGGLEFEENHWGDWKLGLQDLVLSWYNE 430
K LE+++ D + L D+V W E
Sbjct: 417 EVKKSNGDTLEYQKLVAEDLRPALSDIVWVWQGE 450
>At5g25110 serine/threonine protein kinase-like protein
Length = 488
Score = 336 bits (862), Expect = 1e-92
Identities = 193/432 (44%), Positives = 271/432 (62%), Gaps = 25/432 (5%)
Query: 19 ILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRL 78
+ KY++ RLLG+G+F KVY + + G VA+KII+K + M +I REI MR L
Sbjct: 39 LFAKYEMGRLLGKGTFGKVYYGKEITTGESVAIKIINKDQVKREGMMEQIKREISIMR-L 97
Query: 79 HHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFC 138
HPNI+ + EVMATKTKI ++E+ GGELFS I + GKL E++AR+YFQQL+SA+ FC
Sbjct: 98 VRHPNIVELKEVMATKTKIFFIMEYVKGGELFSKIVK-GKLKEDSARKYFQQLISAVDFC 156
Query: 139 HRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEH-LQNGLLHTACGTPAYTAPEILRL 197
H GV+HRDLKP+NLL+D G+LKVSDFGLSALPE LQ+GLLHT CGTPAY APE+LR
Sbjct: 157 HSRGVSHRDLKPENLLVDENGDLKVSDFGLSALPEQILQDGLLHTQCGTPAYVAPEVLRK 216
Query: 198 SGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIR 257
G YDG+K D WSCG++L+VLLAG+LPF D N+ MYRKI + ++++P W + ++ +I
Sbjct: 217 KG-YDGAKGDIWSCGIILYVLLAGFLPFQDENLMKMYRKIFKSEFEYPPWFSPESKRLIS 275
Query: 258 RLLDPNPETRMSVEDLYGNSWFKKSL------KVE------AESETESSTLNLDSGYGNR 305
+LL +P R+S+ + WF+K++ K++ E ET ++T +
Sbjct: 276 KLLVVDPNKRISIPAIMRTPWFRKNINSPIEFKIDELEIQNVEDETPTTTATTATTTTTP 335
Query: 306 VRGLGVNAFDLI-SMSSGLDLSGLFQDEGKRK-EKRFTSGAKLEVVEEKVKEIGGVLGFK 363
V NAF+ I SMSSG DLS LF E KRK FTS + K++ IG + K
Sbjct: 336 VSPKFFNAFEFISSMSSGFDLSSLF--ESKRKLRSMFTSRWSASEIMGKLEGIGKEMNMK 393
Query: 364 VEVGKDSTA--IGLV---KGKVALVVQVFEILPDELLLVAVKVVEGGLEFEENHWGDWKL 418
V+ KD G KG++A+ +VFE+ P+ ++ K LE+ + +
Sbjct: 394 VKRTKDFKVKLFGKTEGRKGQIAVTAEVFEVAPEVAVVELCKSAGDTLEYNRLYEEHVRP 453
Query: 419 GLQDLVLSWYNE 430
L+++V SW+ +
Sbjct: 454 ALEEIVWSWHGD 465
>At1g01140 SOS2-like protein kinase PKS6/CBL-interacting protein
kinase 9 (CIPK9)
Length = 447
Score = 336 bits (862), Expect = 1e-92
Identities = 186/430 (43%), Positives = 278/430 (64%), Gaps = 25/430 (5%)
Query: 15 AATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDA 74
A+ T +G Y++ R LG GSFAKV A++ + G A+KI+D+ K M ++ REI
Sbjct: 11 ASRTRVGNYEMGRTLGEGSFAKVKYAKNTVTGDQAAIKILDREKVFRHKMVEQLKREIST 70
Query: 75 MRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSA 134
M+ L HPN++ I EVMA+KTKI++V+E GGELF I+++G+L E+ ARRYFQQL++A
Sbjct: 71 MK-LIKHPNVVEIIEVMASKTKIYIVLELVNGGELFDKIAQQGRLKEDEARRYFQQLINA 129
Query: 135 LRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHL-QNGLLHTACGTPAYTAPE 193
+ +CH GV HRDLKP+NL+LDA G LKVSDFGLSA + ++GLLHTACGTP Y APE
Sbjct: 130 VDYCHSRGVYHRDLKPENLILDANGVLKVSDFGLSAFSRQVREDGLLHTACGTPNYVAPE 189
Query: 194 ILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPAR 253
+L GYDG+ AD WSCG++LFVL+AGYLPFD+ N+ +Y++I + ++ P W ++ A+
Sbjct: 190 VLS-DKGYDGAAADVWSCGVILFVLMAGYLPFDEPNLMTLYKRICKAEFSCPPWFSQGAK 248
Query: 254 FVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVEA-ESETESSTL-NLDSGYGN------- 304
VI+R+L+PNP TR+S+ +L + WFKK K + + + E T+ ++D+ + N
Sbjct: 249 RVIKRILEPNPITRISIAELLEDEWFKKGYKPPSFDQDDEDITIDDVDAAFSNSKECLVT 308
Query: 305 --RVRGLGVNAFDLISMSSGLDLSGLFQDEGK--RKEKRFTSGAKLEVVEEKVKEIGGVL 360
+ + + +NAF+LIS SS L LF+ + + +KE RFTS + K++E L
Sbjct: 309 EKKEKPVSMNAFELISSSSEFSLENLFEKQAQLVKKETRFTSQRSASEIMSKMEETAKPL 368
Query: 361 GFKVEVGKDSTAIGLV------KGKVALVVQVFEILPDELLLVAVKVVEGGLEFEENHWG 414
GF V KD+ I + KG++++ +VFE+ P ++ K LEF + +
Sbjct: 369 GFNVR--KDNYKIKMKGDKSGRKGQLSVATEVFEVAPSLHVVELRKTGGDTLEFHK-FYK 425
Query: 415 DWKLGLQDLV 424
++ GL+D+V
Sbjct: 426 NFSSGLKDVV 435
>At2g26980 putative protein kinase (At2g26980)
Length = 441
Score = 330 bits (847), Expect = 7e-91
Identities = 180/408 (44%), Positives = 262/408 (64%), Gaps = 19/408 (4%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+GKY+V R +G G+FAKV AR+ G VA+KI+DK K + M +I REI M+ L
Sbjct: 11 VGKYEVGRTIGEGTFAKVKFARNSETGEPVALKILDKEKVLKHKMAEQIRREIATMK-LI 69
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
HPN+++++EVMA+KTKI +++E+ GGELF I G++ E+ ARRYFQQL+ A+ +CH
Sbjct: 70 KHPNVVQLYEVMASKTKIFIILEYVTGGELFDKIVNDGRMKEDEARRYFQQLIHAVDYCH 129
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQ-NGLLHTACGTPAYTAPEILRLS 198
GV HRDLKP+NLLLD+ GNLK+SDFGLSAL + ++ +GLLHT+CGTP Y APE+L
Sbjct: 130 SRGVYHRDLKPENLLLDSYGNLKISDFGLSALSQQVRDDGLLHTSCGTPNYVAPEVLN-D 188
Query: 199 GGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRR 258
GYDG+ AD WSCG+VL+VLLAGYLPFDDSN+ +Y+KIS ++ P W++ A +I R
Sbjct: 189 RGYDGATADMWSCGVVLYVLLAGYLPFDDSNLMNLYKKISSGEFNCPPWLSLGAMKLITR 248
Query: 259 LLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTL-NLDSGYGNRVRGL-------- 309
+LDPNP TR++ ++++ + WFKK K E + S + ++D+ + + L
Sbjct: 249 ILDPNPMTRVTPQEVFEDEWFKKDYKPPVFEERDDSNMDDIDAVFKDSEEHLVTEKREEQ 308
Query: 310 --GVNAFDLISMSSGLDLSGLFQDEGK-RKEKRFTSGAKLEVVEEKVKEIGGVLGFKVEV 366
+NAF++ISMS GL+L LF E + ++E R T + EK++E LGF V+
Sbjct: 309 PAAINAFEIISMSRGLNLENLFDPEQEFKRETRITLRGGANEIIEKIEEAAKPLGFDVQK 368
Query: 367 GKDSTAIGLV----KGKVALVVQVFEILPDELLLVAVKVVEGGLEFEE 410
+ V KG + + ++F++ P ++ K LEF +
Sbjct: 369 KNYKMRLENVKAGRKGNLNVATEIFQVAPSLHMVQVSKSKGDTLEFHK 416
>At2g34180 putative protein kinase
Length = 502
Score = 327 bits (839), Expect = 6e-90
Identities = 189/439 (43%), Positives = 270/439 (61%), Gaps = 47/439 (10%)
Query: 8 PPPPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPR 67
P PR+ + ++ KY++ +LLG GSFAKVY AR++ G VA+K+IDK K V + +
Sbjct: 42 PRSPRTPQGSILMDKYEIGKLLGHGSFAKVYLARNIHSGEDVAIKVIDKEKIVKSGLAGH 101
Query: 68 IVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRY 127
I REI +RR+ H P I+ + EVMATKTKI++V+E+ GGEL++ ++R G+L E TARRY
Sbjct: 102 IKREISILRRVRH-PYIVHLLEVMATKTKIYIVMEYVRGGELYNTVAR-GRLREGTARRY 159
Query: 128 FQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHL-QNGLLHTACGT 186
FQQL+S++ FCH GV HRDLK +NLLLD GN+KVSDFGLS + E L Q G+ T CGT
Sbjct: 160 FQQLISSVAFCHSRGVYHRDLKLENLLLDDKGNVKVSDFGLSVVSEQLKQEGICQTFCGT 219
Query: 187 PAYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPE 246
PAY APE+L G Y+G+KAD WSCG++LFVL+AGYLPFDD NI MY KI + ++ P+
Sbjct: 220 PAYLAPEVLTRKG-YEGAKADIWSCGVILFVLMAGYLPFDDKNILVMYTKIYKGQFKCPK 278
Query: 247 WITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLK------------VEAESETESS 294
W + ++ R+LD NP+TR+++ ++ + WFKK K E + +
Sbjct: 279 WFSPELARLVTRMLDTNPDTRITIPEIMKHRWFKKGFKHVKFYIENDKLCREDDDNDDDD 338
Query: 295 TLNLDSGYGNRV----------------RGLGVNAFDLISMSSGLDLSGLFQDEGKRKEK 338
+ +L SG + R +NAFD++S S DLSGLF++ G +
Sbjct: 339 SSSLSSGRSSTASEGDAEFDIKRVDSMPRPASLNAFDILSFS---DLSGLFEEGG--QGA 393
Query: 339 RFTSGAKLEVVEEKVKEIGGVLGFKVEVGKDSTAIGL------VKGKVALVVQVFEILPD 392
RF S A + + K++EI + F V K ++ L KG + + V++FE+ P
Sbjct: 394 RFVSAAPMTKIISKLEEIAKEVKFMVR--KKDWSVRLEGCREGAKGPLTIRVEIFELTP- 450
Query: 393 ELLLVAVKVVEGGL-EFEE 410
L++V VK G + E+EE
Sbjct: 451 SLVVVEVKKKGGNIEEYEE 469
>At2g30360 putative protein kinase
Length = 435
Score = 326 bits (836), Expect = 1e-89
Identities = 179/347 (51%), Positives = 240/347 (68%), Gaps = 22/347 (6%)
Query: 19 ILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTV-DAAMEPRIVREIDAMRR 77
+ GKY++ +LLG G+FAKV+ AR G VAVKI++K K + + A+ I REI MRR
Sbjct: 17 LFGKYELGKLLGCGAFAKVFHARDRRTGQSVAVKILNKKKLLTNPALANNIKREISIMRR 76
Query: 78 LHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRF 137
L H PNI+++HEVMATK+KI +EF GGELF+ IS+ G+L E+ +RRYFQQL+SA+ +
Sbjct: 77 LSH-PNIVKLHEVMATKSKIFFAMEFVKGGELFNKISKHGRLSEDLSRRYFQQLISAVGY 135
Query: 138 CHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQ-NGLLHTACGTPAYTAPEILR 196
CH GV HRDLKP+NLL+D GNLKVSDFGLSAL + ++ +GLLHT CGTPAY APEIL
Sbjct: 136 CHARGVYHRDLKPENLLIDENGNLKVSDFGLSALTDQIRPDGLLHTLCGTPAYVAPEILS 195
Query: 197 LSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVI 256
GY+G+K D WSCG+VLFVL+AGYLPF+D N+ MY+KI + +Y+FP W++ + +
Sbjct: 196 -KKGYEGAKVDVWSCGIVLFVLVAGYLPFNDPNVMNMYKKIYKGEYRFPRWMSPDLKRFV 254
Query: 257 RRLLDPNPETRMSVEDLYGNSWFK----KSLKV---EAESETESSTLNLDSGYGNRVRGL 309
RLLD NPETR++++++ + WF K +K E E + S+L V+ L
Sbjct: 255 SRLLDINPETRITIDEILKDPWFVRGGFKQIKFHDDEIEDQKVESSL-------EAVKSL 307
Query: 310 GVNAFDLISMSSGLDLSGLFQ--DEGKRKEKRFTSGAKLEVVEEKVK 354
NAFDLIS SSGLDLSGLF + +RF S E++ E+V+
Sbjct: 308 --NAFDLISYSSGLDLSGLFAGCSNSSGESERFLSEKSPEMLAEEVE 352
>At5g07070 serine/threonine protein kinase-like protein
Length = 456
Score = 325 bits (834), Expect = 2e-89
Identities = 182/431 (42%), Positives = 265/431 (61%), Gaps = 25/431 (5%)
Query: 22 KYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLHHH 81
+Y+V RLLG+G+FAKVY RS VA+K+IDK K + + +I REI MR + H
Sbjct: 11 RYEVGRLLGQGTFAKVYFGRSNHTNESVAIKMIDKDKVMRVGLSQQIKREISVMR-IAKH 69
Query: 82 PNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCHRN 141
PN++ ++EVMATK++I+ V+E+ GGELF+ +++ GKL E+ A +YF QL+SA+ FCH
Sbjct: 70 PNVVELYEVMATKSRIYFVIEYCKGGELFNKVAK-GKLKEDVAWKYFYQLISAVDFCHSR 128
Query: 142 GVAHRDLKPQNLLLDAAGNLKVSDFGLSALPE-HLQNGLLHTACGTPAYTAPEILRLSGG 200
GV HRD+KP+NLLLD NLKVSDFGLSAL + Q+GLLHT CGTPAY APE++ G
Sbjct: 129 GVYHRDIKPENLLLDDNDNLKVSDFGLSALADCKRQDGLLHTTCGTPAYVAPEVINRKG- 187
Query: 201 YDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRRLL 260
Y+G+KAD WSCG+VLFVLLAGYLPF D+N+ MYRKI + D++ P W + ++ ++L
Sbjct: 188 YEGTKADIWSCGVVLFVLLAGYLPFHDTNLMEMYRKIGKADFKCPSWFAPEVKRLLCKML 247
Query: 261 DPNPETRMSVEDLYGNSWFKKSL--------KVEAESETESSTLNLDSGYGNRVRG---- 308
DPN ETR+++ + +SWF+K L K+E + E++ G G G
Sbjct: 248 DPNHETRITIAKIKESSWFRKGLHLKQKKMEKMEKQQVREATNPMEAGGSGQNENGENHE 307
Query: 309 ----LGVNAFDLISMSSGLDLSGLFQDEGKRKEKRFTSGAKLEVVEEKVKEIGGVLGFKV 364
+NAFD+I++S+G L+GLF D ++E RF S + K+ E+ L K+
Sbjct: 308 PPRLATLNAFDIIALSTGFGLAGLFGDVYDKRESRFASQKPASEIISKLVEVAKCLKLKI 367
Query: 365 -EVGKDSTAIGLVK----GKVALVVQVFEILPDELLLVAVKVVEGGLEFEENHWGDWKLG 419
+ G + VK G + + ++F++ P L+ K +E+++ D +
Sbjct: 368 RKQGAGLFKLERVKEGKNGILTMDAEIFQVTPTFHLVEVKKCNGDTMEYQKLVEEDLRPA 427
Query: 420 LQDLVLSWYNE 430
L D+V W E
Sbjct: 428 LADIVWVWQGE 438
>At5g01820 unknown protein
Length = 442
Score = 325 bits (832), Expect = 4e-89
Identities = 190/423 (44%), Positives = 268/423 (62%), Gaps = 24/423 (5%)
Query: 4 PMRQPPPPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAA 63
P+ PP R + GKY+V +L+G G+FAKVY RS G VA+K++ K +
Sbjct: 6 PVEFPPENRRGQ---LFGKYEVGKLVGCGAFAKVYHGRSTATGQSVAIKVVSKQRLQKGG 62
Query: 64 MEPRIVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENT 123
+ I REI M RL H P+I+R+ EV+ATK+KI V+EFA GGELF+ +S+ G+ E+
Sbjct: 63 LNGNIQREIAIMHRLRH-PSIVRLFEVLATKSKIFFVMEFAKGGELFAKVSK-GRFCEDL 120
Query: 124 ARRYFQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQ-NGLLHT 182
+RRYFQQL+SA+ +CH G+ HRDLKP+NLLLD +LK+SDFGLSAL + ++ +GLLHT
Sbjct: 121 SRRYFQQLISAVGYCHSRGIFHRDLKPENLLLDEKLDLKISDFGLSALTDQIRPDGLLHT 180
Query: 183 ACGTPAYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDY 242
CGTPAY APE+L G YDG+K D WSCG++LFVL AGYLPF+D N+ MYRKI + ++
Sbjct: 181 LCGTPAYVAPEVLAKKG-YDGAKIDIWSCGIILFVLNAGYLPFNDHNLMVMYRKIYKGEF 239
Query: 243 QFPEWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVE-AESETESSTLNL--- 298
+ P+W + R ++ RLLD NP+TR+++E++ + WFK+ ++ E S + L
Sbjct: 240 RIPKWTSPDLRRLLTRLLDTNPQTRITIEEIIHDPWFKQGYDDRMSKFHLEDSDMKLPAD 299
Query: 299 --DSGYGNRVRGLGVNAFDLISMSSGLDLSGLFQDEGK-RKEKRFTSGAKLEVVEEKVKE 355
DS G R +NAFD+IS S G +LSGLF D K + +RF S E V E+++E
Sbjct: 300 ETDSEMGAR----RMNAFDIISGSPGFNLSGLFGDARKYDRVERFVSAWTAERVVERLEE 355
Query: 356 IGGVLGFKVEVGKDST---AIGLVKGKVALVVQVFEILPDELLLVAVKVVEGGLEFEENH 412
I V + V K T I KG A+VV++ + L DEL+++ V+ + +
Sbjct: 356 I--VSAENLTVAKKETWGMKIEGQKGNFAMVVEINQ-LTDELVMIEVRKRQRAAASGRDL 412
Query: 413 WGD 415
W D
Sbjct: 413 WTD 415
>At3g17510 CBL-interacting protein kinase 1 (CIPK1)
Length = 444
Score = 313 bits (801), Expect = 2e-85
Identities = 173/391 (44%), Positives = 244/391 (62%), Gaps = 21/391 (5%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
LGKY++ R LG G+F KV A+ + G AVKIIDKS+ D +I REI ++ L
Sbjct: 17 LGKYELGRTLGEGNFGKVKFAKDTVSGHSFAVKIIDKSRIADLNFSLQIKREIRTLKMLK 76
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
H P+I+R+HEV+A+KTKI++V+E GGELF I GKL E R+ FQQL+ + +CH
Sbjct: 77 H-PHIVRLHEVLASKTKINMVMELVTGGELFDRIVSNGKLTETDGRKMFQQLIDGISYCH 135
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQ-NGLLHTACGTPAYTAPEILRLS 198
GV HRDLK +N+LLDA G++K++DFGLSALP+H + +GLLHT CG+P Y APE+L +
Sbjct: 136 SKGVFHRDLKLENVLLDAKGHIKITDFGLSALPQHFRDDGLLHTTCGSPNYVAPEVL-AN 194
Query: 199 GGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRR 258
GYDG+ +D WSCG++L+V+L G LPFDD N+A +Y+KI + D P W++ AR +I+R
Sbjct: 195 RGYDGAASDIWSCGVILYVILTGCLPFDDRNLAVLYQKICKGDPPIPRWLSPGARTMIKR 254
Query: 259 LLDPNPETRMSVEDLYGNSWFKKSL-------KVEAESETESSTLNLDSGYGNRVRGLG- 310
+LDPNP TR++V + + WFK E E +T+ ++ +G
Sbjct: 255 MLDPNPVTRITVVGIKASEWFKLEYIPSIPDDDDEEEVDTDDDAFSIQELGSEEGKGSDS 314
Query: 311 ---VNAFDLISMSSGLDLSGLFQDEGKRKEK-RFTSGAKLEVVEEKVKEIGGVLGFKVEV 366
+NAF LI MSS LDLSG F+ E + + RFTS + + + EK++ +GF V+
Sbjct: 315 PTIINAFQLIGMSSFLDLSGFFEQENVSERRIRFTSNSSAKDLLEKIETAVTEMGFSVQK 374
Query: 367 GKDSTAIGL----VKGKVALVV--QVFEILP 391
+ KG+V L V +VFEI P
Sbjct: 375 KHAKLRVKQEERNQKGQVGLSVTAEVFEIKP 405
>At2g25090 CBL-interacting protein kinase 16 (CIPK16)
Length = 469
Score = 313 bits (801), Expect = 2e-85
Identities = 189/444 (42%), Positives = 262/444 (58%), Gaps = 32/444 (7%)
Query: 15 AATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDA-AMEPRIVREID 73
++T + KY + RLLG G+FAKVY + G VA+K+I K M +I REI
Sbjct: 7 SSTVLFDKYNIGRLLGTGNFAKVYHGTEISTGDDVAIKVIKKDHVFKRRGMMEQIEREIA 66
Query: 74 AMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVS 133
MR L H PN++ + EVMATK KI V+E+ GGELF I R GKLPE+ AR+YFQQL+S
Sbjct: 67 VMRLLRH-PNVVELREVMATKKKIFFVMEYVNGGELFEMIDRDGKLPEDLARKYFQQLIS 125
Query: 134 ALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSAL--PEHL------QNGLLHTACG 185
A+ FCH GV HRD+KP+NLLLD G+LKV+DFGLSAL PE L + LLHT CG
Sbjct: 126 AVDFCHSRGVFHRDIKPENLLLDGEGDLKVTDFGLSALMMPEGLGGRRGSSDDLLHTRCG 185
Query: 186 TPAYTAPEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFP 245
TPAY APE+LR + GYDG+ AD WSCG+VL+ LLAG+LPF D N+ +Y KI + + +FP
Sbjct: 186 TPAYVAPEVLR-NKGYDGAMADIWSCGIVLYALLAGFLPFIDENVMTLYTKIFKAECEFP 244
Query: 246 EWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKK------------SLKVEAESETES 293
W + ++ ++ RLL P+PE R+S+ ++ WF+K ++ E T+
Sbjct: 245 PWFSLESKELLSRLLVPDPEQRISMSEIKMIPWFRKNFTPSVAFSIDETIPSPPEPPTKK 304
Query: 294 STLNLDSGYGNRVRGLGVNAFDLI-SMSSGLDLSGLFQDEGKRKEKR-FTSGAKLEVVEE 351
+L+ + NAF I SMSSG DLS LF E KRK KR FTS + V+E
Sbjct: 305 KKKDLNEKEDDGASPRSFNAFQFITSMSSGFDLSNLF--EIKRKPKRMFTSKFPAKSVKE 362
Query: 352 KVKEIGGVLGFKVEVGKD-----STAIGLVKGKVALVVQVFEILPDELLLVAVKVVEGGL 406
+++ + +V+ KD KG++++ +VFE+ P+ ++ K L
Sbjct: 363 RLETAAREMDMRVKHVKDCKMKLQRRTEGRKGRLSVTAEVFEVAPEVSVVEFCKTSGDTL 422
Query: 407 EFEENHWGDWKLGLQDLVLSWYNE 430
E+ D + L+D+V SW +
Sbjct: 423 EYYLFCEDDVRPALKDIVWSWQGD 446
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,699,338
Number of Sequences: 26719
Number of extensions: 420208
Number of successful extensions: 3602
Number of sequences better than 10.0: 986
Number of HSP's better than 10.0 without gapping: 868
Number of HSP's successfully gapped in prelim test: 118
Number of HSP's that attempted gapping in prelim test: 1428
Number of HSP's gapped (non-prelim): 1136
length of query: 430
length of database: 11,318,596
effective HSP length: 102
effective length of query: 328
effective length of database: 8,593,258
effective search space: 2818588624
effective search space used: 2818588624
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0086.14


