

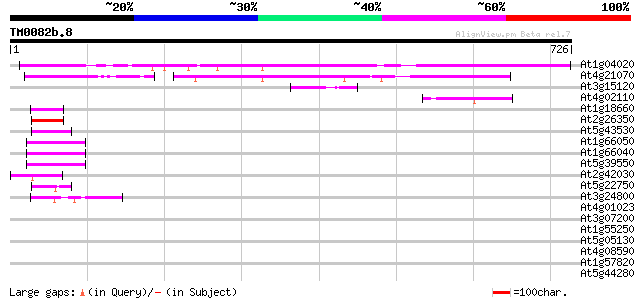
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0082b.8
(726 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g04020 unknown protein 542 e-154
At4g21070 putative protein (fragment) 281 7e-76
At3g15120 chaperone-like ATPase 62 1e-09
At4g02110 predicted protein of unknown function 48 2e-05
At1g18660 unknown protein 48 2e-05
At2g26350 zinc-binding peroxisomal integral membrane protein (PE... 47 4e-05
At5g43530 DNA repair protein-like 47 5e-05
At1g66050 hypothetical protein 45 2e-04
At1g66040 hypothetical protein 45 2e-04
At5g39550 zinc finger -like protein 44 2e-04
At2g42030 putative RING zinc finger protein 43 5e-04
At5g22750 putative protein 43 7e-04
At3g24800 PRT1 43 7e-04
At4g01023 unknown protein 42 0.001
At3g07200 putative RING zinc finger protein 42 0.001
At1g55250 unknown protein 42 0.002
At5g05130 helicase-like transcription factor-like protein 41 0.002
At4g08590 unknown protein with zinc finger motive 40 0.005
At1g57820 putative transcription factor protein (At1g57820) 40 0.005
At5g44280 putative protein 39 0.008
>At1g04020 unknown protein
Length = 714
Score = 542 bits (1396), Expect = e-154
Identities = 313/756 (41%), Positives = 428/756 (56%), Gaps = 93/756 (12%)
Query: 13 LMNPWMLHFQKLALELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSECAVCKTKYA 72
LMNPW+LH QKL LELKCPLCL L +PVLLPCDH+FCDSC+ SS S C VCK+K+
Sbjct: 8 LMNPWVLHLQKLELELKCPLCLKLLNRPVLLPCDHVFCDSCVHKSSQVESGCPVCKSKHP 67
Query: 73 QTDIRNVPFVENMVAIYRSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNF 132
+ R++ F+E++++IY+SL+A+ L Q Q D +Y N
Sbjct: 68 KKARRDLRFMESVISIYKSLNAAVSVH------------LPQLQIPNDCNYKN------- 108
Query: 133 SQSSPNSNGFGVGENRKSMITMHVKPEELEMSSGGRAGFRNDVKPYPMQRS---RVEIGD 189
+ NSN GE+ S +T +++ SGG D P P R + D
Sbjct: 109 -DALNNSNSPKHGESEDSEMT----DKDVSKRSGGTDSSSRDGSPLPTSEESDPRPKHQD 163
Query: 190 YVEMDVNQVT---------QAAVYSPPFCDTKGSDNDCSELDSDHPFNP---EILENSSL 237
+ E ++ AA ++P + + N S+ P N I +S +
Sbjct: 164 WTEKQLSDHLLLYEFESEYDAANHTPESYTEQAAKNVRDITASEQPSNAARKRICGDSFI 223
Query: 238 KRASTGKGNLKERKSQFRSESSASETDKPT-----------------RDLKRKKYLTKGD 280
+ +S N K + + +D PT +D KRK+ +T D
Sbjct: 224 QESSP---NPKTQDPTLLRLMESLRSDDPTDYVKAQNHQQLPKSHTEQDSKRKRDITASD 280
Query: 281 DHIQH--VSTHHSKLVDSHCGLDL--KSGKEPGELLPANIPIDLNPSTS---ICSFCQSS 333
H V + L+ +D K + L I L ++S IC FCQS+
Sbjct: 281 AMENHLKVPKRENNLMQKSADIDCNGKCSANSDDQLSEKISKALEQTSSNITICGFCQSA 340
Query: 334 ETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRCCVDWAPQVYFVDETCKNLKAEVARGA 393
SEATG MLHY+ G+ V GD + NVIHVH C++WAPQVY+ +T KNLKAE+ARG
Sbjct: 341 RVSEATGEMLHYSRGRPVDGDDIFRSNVIHVHSACIEWAPQVYYEGDTVKNLKAELARGM 400
Query: 394 KLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVSTCRWDQEKYLLLCPVHSNAKFPHEKS 453
K+KC+ C LKGAALGC+VKSC+R+YHVPCA ++S CRWD E +LLLCP HS+ KFP+EKS
Sbjct: 401 KIKCTKCSLKGAALGCFVKSCRRSYHVPCAREISRCRWDYEDFLLLCPAHSSVKFPNEKS 460
Query: 454 RPKKQATQEHPA--SAHLSSQLSNPLGAFHDDGKKVVFCGSALSDEEKGSIENLRVMAVV 511
+ + P A L S P AF K++V CGSALS +K +E+L V
Sbjct: 461 GHRVSRAEPLPKINPAELCSLEQTP--AF---TKELVLCGSALSKSDKKLMESLAV---- 511
Query: 512 VSGRGEVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILNGRWVLKI 571
+ AT++++W VTHVIA+TD GAC+RTLKVLM ILNG+W++
Sbjct: 512 --------------RFNATISRYWNPSVTHVIASTDEKGACTRTLKVLMGILNGKWIINA 557
Query: 572 DWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKLFSGLKFYFSGDYVSS 631
W+KA +K V+EEP+EI +D QGC GPK RL A N+PKLF GLKFYF GD+
Sbjct: 558 AWMKASLKASQPVDEEPFEIQIDTQGCQDGPKTARLRAETNKPKLFEGLKFYFFGDFYKG 617
Query: 632 YKEDLEDLVEVGGGAVLTSKDKL--EAKRHEFEVTSNLLIVYNLDPPQGSKLGDEVSTLW 689
YKEDL++LV+V GG +L ++D+L E+ + + S+ ++VYN+DPP G LG+EV+ +W
Sbjct: 618 YKEDLQNLVKVAGGTILNTEDELGAESSNNVNDQRSSSIVVYNIDPPHGCALGEEVTIIW 677
Query: 690 QRLNEAEDLAANTGSQVIGHTWILESIAACKLQPFI 725
QR N+AE LA+ TGS+++GHTW+LESIA KL P I
Sbjct: 678 QRANDAEALASQTGSRLVGHTWVLESIAGYKLHPVI 713
>At4g21070 putative protein (fragment)
Length = 1495
Score = 281 bits (720), Expect = 7e-76
Identities = 175/492 (35%), Positives = 233/492 (46%), Gaps = 74/492 (15%)
Query: 212 KGSDNDCSELDSDHPFNPEILENSSLK---RASTGKGNLKERKSQFRSESSASETDKPTR 268
KG + HP E SLK R S +LK+ + + ++S + +
Sbjct: 1011 KGDQDQAHGPSDTHPEKRSPTEKPSLKKRGRKSNASSSLKDLSGKTQKKTSEKKLKLDSH 1070
Query: 269 DLKRKKYLTKGDDHIQHVSTHHSKLVDSHCGLDLKSGKEPGELLPANIPIDLNPSTS--- 325
+ K G+ + DS GK+ + +N S+S
Sbjct: 1071 MISSKATQPHGNGILTAGLNQGGDKQDSRNNRKSTVGKDDHTMQVIEKCSTINKSSSGGS 1130
Query: 326 --------------ICSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRCCVDW 371
C+FCQ SE +EA+G M HY G+ V D VIHVH+ C +W
Sbjct: 1131 AHLRRCNGSLTKKFTCAFCQCSEDTEASGEMTHYYRGEPVSADFNGGSKVIHVHKNCAEW 1190
Query: 372 APQVYFVDETCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVSTCRW 431
AP VYF D T NL E+ R ++ CS CGLKGAALGCY KSCK ++HV CA + CRW
Sbjct: 1191 APNVYFNDLTIVNLDVELTRSRRISCSCCGLKGAALGCYNKSCKNSFHVTCAKLIPECRW 1250
Query: 432 D-------------------------QEKYLLLCPVHSNAKFPHEKSRPKKQATQEHPAS 466
D Q K+++LCP+ ++ K P E++ K + + P
Sbjct: 1251 DNVSVAYIVEALYSPLHSSFNTFSYQQVKFVMLCPLDASIKLPCEEANSKDRKCKRTPKE 1310
Query: 467 AHLSSQLSNPLGA----------FHDDGKKVVFCGSALSDEEKGSIENLRVMAVVVSGRG 516
L SQ G FH KK+V S L+ EEK
Sbjct: 1311 P-LHSQPKQVSGKANIRELHIKQFHGFSKKLVLSCSGLTVEEK----------------- 1352
Query: 517 EVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILNGRWVLKIDWIKA 576
++ FA G T++K W S VTHVIA+ + NGAC RTLK +MAIL G+W+L IDWIKA
Sbjct: 1353 -TVIAEFAELSGVTISKNWDSTVTHVIASINENGACKRTLKFMMAILEGKWILTIDWIKA 1411
Query: 577 CMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKLFSGLKFYFSGDYVSSYKEDL 636
CMK V EEPYEI++D G GP GR AL +PKLF+GLKFY GD+ +YK L
Sbjct: 1412 CMKNTKYVSEEPYEITMDVHGIREGPYLGRQRALKKKPKLFTGLKFYIMGDFELAYKGYL 1471
Query: 637 EDLVEVGGGAVL 648
+DL+ GG +L
Sbjct: 1472 QDLIVAAGGTIL 1483
Score = 84.3 bits (207), Expect = 2e-16
Identities = 52/169 (30%), Positives = 91/169 (53%), Gaps = 18/169 (10%)
Query: 20 HFQKLALELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNV 79
H +++ ELKCP+CLSL+ V L C+H+FC++C+V S + C VCK Y + +IR
Sbjct: 588 HLERMGRELKCPICLSLYNSAVSLSCNHVFCNACIVKSMKMDATCPVCKIPYHRREIRGA 647
Query: 80 PFVENMVAIYRSL-DASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQSSPN 138
P ++++V+IY+++ DAS + Q + S +Q RD+S ++KA +
Sbjct: 648 PHMDSLVSIYKNMEDASGIKLFVSQNNPSPSDKEKQ---VRDAS---VEKAS-------D 694
Query: 139 SNGFGVGENRKSMITMHVKPEELEMSSGGRAGFRNDVKPYPMQRSRVEI 187
N G + R S + K +E+++ + G +KP + RV++
Sbjct: 695 KNRQGSRKGRASKRNEYGKTKEIDVDAPGPI----VMKPSSQTKKRVQL 739
>At3g15120 chaperone-like ATPase
Length = 1954
Score = 62.0 bits (149), Expect = 1e-09
Identities = 33/88 (37%), Positives = 45/88 (50%), Gaps = 12/88 (13%)
Query: 364 VHRCCVDWAPQVYFVDETC-KNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPC 422
VH+ C W+P+VYF C KN++A + RG LKC+ C GA GC PC
Sbjct: 560 VHQNCAVWSPEVYFAGVGCLKNIRAALFRGRSLKCTRCDRPGATTGCR----------PC 609
Query: 423 AMDVSTCRWDQEKYLLLCPVHSNAKFPH 450
A + C +D K+L+ C H + PH
Sbjct: 610 AR-ANGCIFDHRKFLIACTDHRHHFQPH 636
>At4g02110 predicted protein of unknown function
Length = 1293
Score = 48.1 bits (113), Expect = 2e-05
Identities = 34/121 (28%), Positives = 49/121 (40%), Gaps = 10/121 (8%)
Query: 535 WTSDVTHVIAATDANGACSRTLKVLMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLD 594
W+ TH IA RT K A +G W+LK D++ + L++EEPYE
Sbjct: 1094 WSYQATHFIAPE-----IRRTEKFFAAAASGSWILKTDYVADSKEAGKLLQEEPYEWHSS 1148
Query: 595 NQGCNG-----GPKAGRLSALANEPKLFSGLKFYFSGDYVSSYKEDLEDLVEVGGGAVLT 649
+G PK RL GL+ GD + L+ V+ G G +L
Sbjct: 1149 GLSADGAINLESPKKWRLVREKTGHGALYGLRIVVYGDCTIPCLDTLKRAVKAGDGTILA 1208
Query: 650 S 650
+
Sbjct: 1209 T 1209
>At1g18660 unknown protein
Length = 486
Score = 48.1 bits (113), Expect = 2e-05
Identities = 18/43 (41%), Positives = 25/43 (57%)
Query: 27 ELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSECAVCKT 69
+ C +CL L +P PC H FC SCL S G++C +C+T
Sbjct: 193 DFDCTVCLKLLYEPATTPCGHTFCRSCLFQSMDRGNKCPLCRT 235
>At2g26350 zinc-binding peroxisomal integral membrane protein
(PEX10)
Length = 381
Score = 47.0 bits (110), Expect = 4e-05
Identities = 18/41 (43%), Positives = 25/41 (60%)
Query: 29 KCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSECAVCKT 69
KC LCLS + P PC H+FC SC+++ EC +C+T
Sbjct: 326 KCTLCLSTRQHPTATPCGHVFCWSCIMEWCNEKQECPLCRT 366
>At5g43530 DNA repair protein-like
Length = 1277
Score = 46.6 bits (109), Expect = 5e-05
Identities = 21/54 (38%), Positives = 31/54 (56%), Gaps = 2/54 (3%)
Query: 29 KCPLCLSLFEKPVLLPCDHLFCDSCLVDS--SLSGSECAVCKTKYAQTDIRNVP 80
+CP+CL + PVL PC H C CL+ S S S C +C+T +T++ + P
Sbjct: 1039 ECPICLESADDPVLTPCAHRMCRECLLTSWRSPSCGLCPICRTILKRTELISCP 1092
>At1g66050 hypothetical protein
Length = 598
Score = 44.7 bits (104), Expect = 2e-04
Identities = 22/79 (27%), Positives = 40/79 (49%), Gaps = 2/79 (2%)
Query: 22 QKLALELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSE--CAVCKTKYAQTDIRNV 79
+K LE+ C +C+ L E+PV PC H FC C ++ + C +C++K + +N
Sbjct: 121 EKKKLEIFCSICIQLPERPVTTPCGHNFCLKCFEKWAVGQGKLTCMICRSKIPRHVAKNP 180
Query: 80 PFVENMVAIYRSLDASFCA 98
+V+ R + + C+
Sbjct: 181 RINLALVSAIRLANVTKCS 199
Score = 36.6 bits (83), Expect = 0.051
Identities = 18/49 (36%), Positives = 24/49 (48%), Gaps = 2/49 (4%)
Query: 6 KPNNKSKLMNPWMLHFQKLALELKCPLCLSLFEKPVLLPCDHLFCDSCL 54
K N ++K N M +L E C +C + PV PC H FC +CL
Sbjct: 451 KKNKRAKKGNNAMK--ARLLKEFSCQICRKVLSLPVTTPCAHNFCKACL 497
>At1g66040 hypothetical protein
Length = 622
Score = 44.7 bits (104), Expect = 2e-04
Identities = 22/79 (27%), Positives = 40/79 (49%), Gaps = 2/79 (2%)
Query: 22 QKLALELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSE--CAVCKTKYAQTDIRNV 79
+K LE+ C +C+ L E+PV PC H FC C ++ + C +C++K + +N
Sbjct: 121 EKKKLEIFCSICIQLPERPVTTPCGHNFCLKCFEKWAVGQGKLTCMICRSKIPRHVAKNP 180
Query: 80 PFVENMVAIYRSLDASFCA 98
+V+ R + + C+
Sbjct: 181 RINLALVSAIRLANVTKCS 199
Score = 36.6 bits (83), Expect = 0.051
Identities = 18/49 (36%), Positives = 24/49 (48%), Gaps = 2/49 (4%)
Query: 6 KPNNKSKLMNPWMLHFQKLALELKCPLCLSLFEKPVLLPCDHLFCDSCL 54
K N ++K N M +L E C +C + PV PC H FC +CL
Sbjct: 476 KKNKRAKKGNNAMK--ARLLKEFSCQICRKVLSLPVTTPCAHNFCKACL 522
>At5g39550 zinc finger -like protein
Length = 617
Score = 44.3 bits (103), Expect = 2e-04
Identities = 21/79 (26%), Positives = 40/79 (50%), Gaps = 2/79 (2%)
Query: 22 QKLALELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSE--CAVCKTKYAQTDIRNV 79
+K LE+ C +C+ L E+P+ PC H FC C ++ + C +C++K + +N
Sbjct: 121 EKKKLEIFCSICIQLPERPITTPCGHNFCLKCFEKWAVGQGKLTCMICRSKIPRHVAKNP 180
Query: 80 PFVENMVAIYRSLDASFCA 98
+V+ R + + C+
Sbjct: 181 RINLALVSAIRLANVTKCS 199
Score = 35.4 bits (80), Expect = 0.11
Identities = 13/32 (40%), Positives = 17/32 (52%)
Query: 23 KLALELKCPLCLSLFEKPVLLPCDHLFCDSCL 54
+L E C +C + PV PC H FC +CL
Sbjct: 488 RLLKEFSCQICREVLSLPVTTPCAHNFCKACL 519
>At2g42030 putative RING zinc finger protein
Length = 425
Score = 43.1 bits (100), Expect = 5e-04
Identities = 28/79 (35%), Positives = 37/79 (46%), Gaps = 11/79 (13%)
Query: 1 MEDSGKPNNKSKLMNPWMLHFQKLALE---------LKCPLCLSLFEKPVLLPCDHLFCD 51
+EDS K N SK+M + +K +E C +CL L + PV+ C HL+C
Sbjct: 103 IEDSKKCENGSKVMAEDNVTEEKRDVEKSVGSDGSFFDCYICLDLSKDPVVTNCGHLYCW 162
Query: 52 SCLVD--SSLSGSECAVCK 68
SCL EC VCK
Sbjct: 163 SCLYQWLQVSEAKECPVCK 181
>At5g22750 putative protein
Length = 1029
Score = 42.7 bits (99), Expect = 7e-04
Identities = 21/55 (38%), Positives = 29/55 (52%), Gaps = 4/55 (7%)
Query: 29 KCPLCLSLFEKPVLLPCDHLFCDSCLVDS---SLSGSECAVCKTKYAQTDIRNVP 80
+CP+CL E VL PC H C CL+ S S SG C VC+ ++ ++ P
Sbjct: 793 ECPICLEALEDAVLTPCAHRLCRECLLASWRNSTSGL-CPVCRNTVSKQELITAP 846
>At3g24800 PRT1
Length = 410
Score = 42.7 bits (99), Expect = 7e-04
Identities = 39/134 (29%), Positives = 55/134 (40%), Gaps = 27/134 (20%)
Query: 27 ELKCPLCLSLFEKPVLLPCDHLFCDSCLVD-----SSLSGSECAVCKTKYAQTDIRNVPF 81
+L C C L +PV+L C H++C+ C+VD + EC VC D R P
Sbjct: 189 DLLCSACKELLVRPVVLNCGHVYCEGCVVDMAEESEKIKCQECNVC-------DPRGFPK 241
Query: 82 V---------ENMVAIYRSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNF 132
V EN Y S +S +Q+ +S+ Q S SN D+
Sbjct: 242 VCLILEQLLEENFPEEYNSR-----SSKVQKTLAHNSKGNIQSYLKEGPSLSNDNNNDDP 296
Query: 133 SQSSPNSN-GFGVG 145
++P SN FG G
Sbjct: 297 WLANPGSNVHFGAG 310
Score = 40.0 bits (92), Expect = 0.005
Identities = 19/45 (42%), Positives = 26/45 (57%), Gaps = 4/45 (8%)
Query: 30 CPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSG---SECAVCKTKY 71
C +CL L KP++L C HL C C V S++G S C +C+ Y
Sbjct: 26 CCVCLELLYKPIVLSCGHLSCFWC-VHKSMNGFRESHCPICRDPY 69
>At4g01023 unknown protein
Length = 255
Score = 42.4 bits (98), Expect = 0.001
Identities = 17/43 (39%), Positives = 22/43 (50%)
Query: 25 ALELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSECAVC 67
AL L C +C + F PV+ C+H FCD C + C VC
Sbjct: 100 ALPLACSICQNPFLDPVVTNCNHYFCDKCALKHHTENDTCFVC 142
>At3g07200 putative RING zinc finger protein
Length = 182
Score = 42.4 bits (98), Expect = 0.001
Identities = 16/53 (30%), Positives = 27/53 (50%)
Query: 27 ELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNV 79
+ CP+CL F + V C H+FC C+ ++ ++C C+ K D+ V
Sbjct: 124 KFSCPICLCPFTQEVSTKCGHIFCKKCIKNALSLQAKCPTCRKKITVKDLIRV 176
>At1g55250 unknown protein
Length = 899
Score = 41.6 bits (96), Expect = 0.002
Identities = 19/53 (35%), Positives = 29/53 (53%), Gaps = 1/53 (1%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSCLVDS-SLSGSECAVCKTKYAQTDIRNV 79
LKC +C ++ V++ C HLFC C+ S + +C C T + Q D+R V
Sbjct: 845 LKCGVCFDRPKEVVIVKCYHLFCQQCIQRSLEIRHRKCPGCGTAFGQNDVRLV 897
>At5g05130 helicase-like transcription factor-like protein
Length = 862
Score = 41.2 bits (95), Expect = 0.002
Identities = 16/55 (29%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query: 27 ELKCPLCLSLFEKPVLLPCDHLFCDSCLVDS-SLSGSECAVCKTKYAQTDIRNVP 80
+ CP+C+S ++ C H+FC +C++ + S C +C+ Q+D+ N P
Sbjct: 613 DFDCPICISPPTNIIITRCAHIFCRACILQTLQRSKPLCPLCRGSLTQSDLYNAP 667
>At4g08590 unknown protein with zinc finger motive
Length = 465
Score = 40.0 bits (92), Expect = 0.005
Identities = 22/90 (24%), Positives = 42/90 (46%), Gaps = 1/90 (1%)
Query: 2 EDSGKPNNKSKLMNPWMLHFQKLALELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSG 61
+++ K + + +N + F + +L C LC L ++PV + C H FC C G
Sbjct: 81 DENNKSDGEIASLNDGVDAFTAICEDLNCSLCNQLPDRPVTILCGHNFCLKCFDKWIDQG 140
Query: 62 SE-CAVCKTKYAQTDIRNVPFVENMVAIYR 90
++ CA C++ N ++V++ R
Sbjct: 141 NQICATCRSTIPDKMAANPRVNSSLVSVIR 170
>At1g57820 putative transcription factor protein (At1g57820)
Length = 645
Score = 40.0 bits (92), Expect = 0.005
Identities = 27/99 (27%), Positives = 39/99 (39%), Gaps = 1/99 (1%)
Query: 2 EDSGKPNNKSKLMNPWMLHFQKLALELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSG 61
++ K K K NP + L L C C+ L E+PV PC H C C G
Sbjct: 118 DEEEKRKKKGKGKNPNLDVLSALGDNLMCSFCMQLPERPVTKPCGHNACLKCFEKWMGQG 177
Query: 62 SE-CAVCKTKYAQTDIRNVPFVENMVAIYRSLDASFCAS 99
C C++ + +N ++VA R S A+
Sbjct: 178 KRTCGKCRSIIPEKMAKNPRINSSLVAAIRLAKVSKSAA 216
Score = 37.0 bits (84), Expect = 0.039
Identities = 14/32 (43%), Positives = 18/32 (55%)
Query: 23 KLALELKCPLCLSLFEKPVLLPCDHLFCDSCL 54
+L E KC +C + PV PC H FC +CL
Sbjct: 511 RLLKEFKCQICQQVLTLPVTTPCAHNFCKACL 542
>At5g44280 putative protein
Length = 486
Score = 39.3 bits (90), Expect = 0.008
Identities = 20/72 (27%), Positives = 42/72 (57%), Gaps = 4/72 (5%)
Query: 27 ELKCPLCLSLFEKP-VLLPCDHLFCDSCLVDS-SLSGSECAVCKTKYA-QTDIRNVPFVE 83
+++CP+CL + +K ++ C H FC C+ S L +EC C+ A + +R+ P +
Sbjct: 133 DVQCPICLGIIKKTRTVMECLHRFCRECIDKSMRLGNNECPACRKHCASRRSLRDDPKFD 192
Query: 84 NMV-AIYRSLDA 94
++ A++ ++D+
Sbjct: 193 ALIAALFTNIDS 204
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.132 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,159,656
Number of Sequences: 26719
Number of extensions: 761394
Number of successful extensions: 2579
Number of sequences better than 10.0: 155
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 95
Number of HSP's that attempted gapping in prelim test: 2455
Number of HSP's gapped (non-prelim): 197
length of query: 726
length of database: 11,318,596
effective HSP length: 107
effective length of query: 619
effective length of database: 8,459,663
effective search space: 5236531397
effective search space used: 5236531397
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0082b.8


