

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0082b.2
(153 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
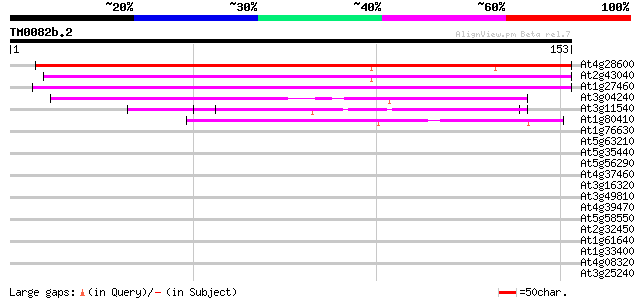
Score E
Sequences producing significant alignments: (bits) Value
At4g28600 unknown protein 164 2e-41
At2g43040 pollen-specific calmodulin-binding protein (NPG) 137 3e-33
At1g27460 unknown protein 115 9e-27
At3g04240 O-linked GlcNAc transferase like protein 50 4e-07
At3g11540 spindly (gibberellin signal transduction protein) 44 3e-05
At1g80410 putative N-terminal acetyltransferase (At1g80410) 40 6e-04
At1g76630 hypothetical protein, 3' partial 35 0.016
At5g63210 putative protein 32 0.13
At5g35440 putative protein 31 0.23
At5g56290 peroxisomal targeting signal type 1 receptor 31 0.30
At4g37460 unknown protein 31 0.30
At3g16320 hypothetical protein 30 0.39
At3g49810 unknown protein 29 1.1
At4g39470 unknown protein 28 1.5
At5g58550 putative protein 28 1.9
At2g32450 putative O-GlcNAc transferase 28 1.9
At1g61640 unknown protein 28 1.9
At1g33400 unknown protein 28 1.9
At4g08320 unknown protein 28 2.5
At3g25240 hypothetical protein 27 3.3
>At4g28600 unknown protein
Length = 739
Score = 164 bits (414), Expect = 2e-41
Identities = 87/150 (58%), Positives = 107/150 (71%), Gaps = 4/150 (2%)
Query: 8 NLEEEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKA 67
+LE WHDLA+ YI+LSQW DAE CLS+S+ Y++ R H+ G ++ +G +EA++A
Sbjct: 589 SLELGTWHDLAHIYINLSQWRDAESCLSRSRLIAPYSSVRYHIEGVLYNRRGQLEEAMEA 648
Query: 68 FRDALNIDPRHVPSLISTAVALRRWSNQSD-PVVRSFLMDALRYDRLNASAWYNLGILHK 126
F AL+IDP HVPSL S A L N+S VVRSFLM+ALR DRLN SAWYNLG + K
Sbjct: 649 FTTALDIDPMHVPSLTSKAEILLEVGNRSGIAVVRSFLMEALRIDRLNHSAWYNLGKMFK 708
Query: 127 AEGRV---LEAVECFQAANSLEETAPIEPF 153
AEG V EAVECFQAA +LEET P+EPF
Sbjct: 709 AEGSVSSMQEAVECFQAAVTLEETMPVEPF 738
>At2g43040 pollen-specific calmodulin-binding protein (NPG)
Length = 704
Score = 137 bits (344), Expect = 3e-33
Identities = 69/147 (46%), Positives = 89/147 (59%), Gaps = 3/147 (2%)
Query: 10 EEEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFR 69
E E+WH LAY Y SLS W+D EVCL K+ +QY+AS H G M E + +K A+ AF
Sbjct: 554 EFEVWHGLAYLYSSLSHWNDVEVCLKKAGELKQYSASMLHTEGRMWEGRKEFKPALAAFL 613
Query: 70 DALNIDPRHVPSLISTAVALRRWSNQSD---PVVRSFLMDALRYDRLNASAWYNLGILHK 126
D L +D VP ++ L PV RS L DALR D N AWY LG++HK
Sbjct: 614 DGLLLDGSSVPCKVAVGALLSERGKDHQPTLPVARSLLSDALRIDPTNRKAWYYLGMVHK 673
Query: 127 AEGRVLEAVECFQAANSLEETAPIEPF 153
++GR+ +A +CFQAA+ LEE+ PIE F
Sbjct: 674 SDGRIADATDCFQAASMLEESDPIESF 700
>At1g27460 unknown protein
Length = 694
Score = 115 bits (288), Expect = 9e-27
Identities = 60/147 (40%), Positives = 88/147 (59%)
Query: 7 RNLEEEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVK 66
+ E E W DLA Y L W DAE CL K+++ Y+ + G EAK L++EA+
Sbjct: 547 QKFETEAWQDLASVYGKLGSWSDAETCLEKARSMCYYSPRGWNETGLCLEAKSLHEEALI 606
Query: 67 AFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHK 126
+F +L+I+P HVPS++S A + + ++S P +SFLM+ALR D N AW LG + K
Sbjct: 607 SFFLSLSIEPDHVPSIVSIAEVMMKSGDESLPTAKSFLMNALRLDPRNHDAWMKLGHVAK 666
Query: 127 AEGRVLEAVECFQAANSLEETAPIEPF 153
+G +A E +QAA LE +AP++ F
Sbjct: 667 KQGLSQQAAEFYQAAYELELSAPVQSF 693
>At3g04240 O-linked GlcNAc transferase like protein
Length = 977
Score = 50.4 bits (119), Expect = 4e-07
Identities = 36/138 (26%), Positives = 64/138 (46%), Gaps = 18/138 (13%)
Query: 12 EIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDA 71
+ W +LA AY+ + +A C ++ + +G + +A+GL EA + +A
Sbjct: 156 DAWSNLASAYMRKGRLSEATQCCQQALSLNPLLVDAHSNLGNLMKAQGLIHEAYSCYLEA 215
Query: 72 LNIDPRHVPSLISTAVALRRWSNQSDPVVRS--------FLMDALRYDRLNASAWYNLGI 123
+ I P + A+A WSN + + S + +A++ A+ NLG
Sbjct: 216 VRIQP-------TFAIA---WSNLAGLFMESGDLNRALQYYKEAVKLKPAFPDAYLNLGN 265
Query: 124 LHKAEGRVLEAVECFQAA 141
++KA GR EA+ C+Q A
Sbjct: 266 VYKALGRPTEAIMCYQHA 283
Score = 37.7 bits (86), Expect = 0.002
Identities = 29/99 (29%), Positives = 47/99 (47%), Gaps = 2/99 (2%)
Query: 51 IGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRY 110
+G +++A G EA+ ++ AL + P + + A ++ Q D +R + AL
Sbjct: 263 LGNVYKALGRPTEAIMCYQHALQMRPNSAMAFGNIA-SIYYEQGQLDLAIRHY-KQALSR 320
Query: 111 DRLNASAWYNLGILHKAEGRVLEAVECFQAANSLEETAP 149
D A+ NLG K GRV EAV C+ +L+ P
Sbjct: 321 DPRFLEAYNNLGNALKDIGRVDEAVRCYNQCLALQPNHP 359
Score = 35.8 bits (81), Expect = 0.009
Identities = 31/128 (24%), Positives = 54/128 (41%), Gaps = 5/128 (3%)
Query: 26 QWHDAEVCLSKSKAFRQYTASRCHVIGTMHEA---KGLYKEAVKAFRDALNIDPRHVPSL 82
Q + ++C+++++ + G M A KG A++ + A+ + P +
Sbjct: 99 QLQEYDMCIARNEEALRIQPQFAECYGNMANAWKEKGDTDRAIRYYLIAIELRPNFADAW 158
Query: 83 ISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAVECFQAAN 142
+ A A R S+ AL + L A NLG L KA+G + EA C+ A
Sbjct: 159 SNLASAYMRKGRLSEAT--QCCQQALSLNPLLVDAHSNLGNLMKAQGLIHEAYSCYLEAV 216
Query: 143 SLEETAPI 150
++ T I
Sbjct: 217 RIQPTFAI 224
Score = 35.8 bits (81), Expect = 0.009
Identities = 31/136 (22%), Positives = 57/136 (41%), Gaps = 2/136 (1%)
Query: 12 EIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDA 71
E +++L A + + +A C ++ A + +G ++ + A F+
Sbjct: 326 EAYNNLGNALKDIGRVDEAVRCYNQCLALQPNHPQAMANLGNIYMEWNMMGPASSLFKAT 385
Query: 72 LNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRV 131
L + + A+ ++ N SD + S + LR D L A A N G +K GRV
Sbjct: 386 LAVTTGLSAPFNNLAIIYKQQGNYSDAI--SCYNEVLRIDPLAADALVNRGNTYKEIGRV 443
Query: 132 LEAVECFQAANSLEET 147
EA++ + A + T
Sbjct: 444 TEAIQDYMHAINFRPT 459
Score = 35.8 bits (81), Expect = 0.009
Identities = 27/95 (28%), Positives = 44/95 (45%), Gaps = 2/95 (2%)
Query: 50 VIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALR 109
+IG ++ Y + +AL I P+ + A A + +D +R +L+ A+
Sbjct: 92 LIGAIYYQLQEYDMCIARNEEALRIQPQFAECYGNMANAWKE-KGDTDRAIRYYLI-AIE 149
Query: 110 YDRLNASAWYNLGILHKAEGRVLEAVECFQAANSL 144
A AW NL + +GR+ EA +C Q A SL
Sbjct: 150 LRPNFADAWSNLASAYMRKGRLSEATQCCQQALSL 184
>At3g11540 spindly (gibberellin signal transduction protein)
Length = 914
Score = 44.3 bits (103), Expect = 3e-05
Identities = 28/85 (32%), Positives = 46/85 (53%), Gaps = 2/85 (2%)
Query: 57 AKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNAS 116
A+ + +A+ + L D ++V + I + L+ N+ + F +A+R D NA
Sbjct: 55 ARNKFADALALYEAMLEKDSKNVEAHIGKGICLQT-QNKGNLAFDCF-SEAIRLDPHNAC 112
Query: 117 AWYNLGILHKAEGRVLEAVECFQAA 141
A + GILHK EGR++EA E +Q A
Sbjct: 113 ALTHCGILHKEEGRLVEAAESYQKA 137
Score = 41.2 bits (95), Expect = 2e-04
Identities = 27/116 (23%), Positives = 55/116 (47%), Gaps = 9/116 (7%)
Query: 33 CLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPS-------LIST 85
C S++ + A G +H+ +G EA ++++ AL D + P+ L
Sbjct: 99 CFSEAIRLDPHNACALTHCGILHKEEGRLVEAAESYQKALMADASYKPAAECLAIVLTDL 158
Query: 86 AVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAVECFQAA 141
+L+ N + + + + +AL+ D A A+YNLG+++ + A+ C++ A
Sbjct: 159 GTSLKLAGNTQEGIQKYY--EALKIDPHYAPAYYNLGVVYSEMMQYDNALSCYEKA 212
Score = 39.3 bits (90), Expect = 8e-04
Identities = 23/89 (25%), Positives = 45/89 (49%), Gaps = 2/89 (2%)
Query: 51 IGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRY 110
+GT + G +E ++ + +AL IDP + P+ + V Q D + + AL
Sbjct: 158 LGTSLKLAGNTQEGIQKYYEALKIDPHYAPAYYNLGVVYSEMM-QYDNALSCYEKAALER 216
Query: 111 DRLNASAWYNLGILHKAEGRVLEAVECFQ 139
+ A A+ N+G+++K G + A+ C++
Sbjct: 217 P-MYAEAYCNMGVIYKNRGDLEMAITCYE 244
Score = 34.3 bits (77), Expect = 0.027
Identities = 31/118 (26%), Positives = 54/118 (45%), Gaps = 5/118 (4%)
Query: 31 EVCLSKSKAF---RQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAV 87
E CL+ S F + A +GT + +G + V ++ AL + + ++ + V
Sbjct: 244 ERCLAVSPNFEIAKNNMAIALTDLGTKVKLEGDVTQGVAYYKKALYYNWHYADAMYNLGV 303
Query: 88 ALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAVECFQAANSLE 145
A +V F A ++ A A NLG+L+K + +AVEC+Q A S++
Sbjct: 304 AYGEMLKFDMAIV--FYELAFHFNPHCAEACNNLGVLYKDRDNLDKAVECYQMALSIK 359
>At1g80410 putative N-terminal acetyltransferase (At1g80410)
Length = 714
Score = 39.7 bits (91), Expect = 6e-04
Identities = 29/105 (27%), Positives = 50/105 (47%), Gaps = 5/105 (4%)
Query: 49 HVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPV-VRSFLMDA 107
HV+G ++ + Y+EA+K +R+AL IDP ++ L ++ + + S V R L+
Sbjct: 81 HVLGLLYRSDREYREAIKCYRNALRIDPDNLEILRDLSLLQAQMRDLSGFVETRQQLLTL 140
Query: 108 LRYDRLNASAWYNLGILHKAEGRVLEAVECFQA-ANSLEETAPIE 151
R+N W + +AVE +A +LE+ P E
Sbjct: 141 KPNHRMN---WIGFAVSQHLNANASKAVEILEAYEGTLEDDYPPE 182
Score = 29.6 bits (65), Expect = 0.67
Identities = 18/81 (22%), Positives = 39/81 (47%), Gaps = 2/81 (2%)
Query: 61 YKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYN 120
YK+ +KA L P H +L + L +++ + ++ D + W+
Sbjct: 25 YKKGLKAADAILKKFPDHGETLSMKGLTLNCMDRKTEAY--ELVRLGVKNDIKSHVCWHV 82
Query: 121 LGILHKAEGRVLEAVECFQAA 141
LG+L++++ EA++C++ A
Sbjct: 83 LGLLYRSDREYREAIKCYRNA 103
>At1g76630 hypothetical protein, 3' partial
Length = 971
Score = 35.0 bits (79), Expect = 0.016
Identities = 37/153 (24%), Positives = 61/153 (39%), Gaps = 14/153 (9%)
Query: 3 RDYARNLEEEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYT--ASRCHVIGTMHEAKGL 60
RD + + W YI L Q +E S A R Y + +G ++ G+
Sbjct: 111 RDASEKSPKAFWAFCRLGYIQLHQKKWSEAVQSLQHAIRGYPTMSDLWEALGLAYQRLGM 170
Query: 61 YKEAVKAFRDALNIDPRHVPSLISTA-VALRRWSNQSDPVVRSFLMDALRYDRLNASAWY 119
+ A+KA+ A+ +D + +L+ +A + L S + AL+ N S Y
Sbjct: 171 FTAAIKAYGRAIELDETKIFALVESANIFLMLGSYRKVTFGVELFEQALKISPQNISVLY 230
Query: 120 NLGILHKAEGRVLEAVECFQ------AANSLEE 146
L A G + + EC AA+ LE+
Sbjct: 231 GL-----ASGLLSWSKECINLGAFGWAASLLED 258
>At5g63210 putative protein
Length = 262
Score = 32.0 bits (71), Expect = 0.13
Identities = 33/139 (23%), Positives = 59/139 (41%), Gaps = 5/139 (3%)
Query: 14 WHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALN 73
WH L + Q+ ++ L + + + +G + + EA + ++ AL
Sbjct: 99 WHQLGLHSLCSQQYKLSQKYLKAAVGRSRECSYAWSNLGISLQLSDEHSEAEEVYKRALT 158
Query: 74 IDPR-HVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVL 132
+ +++S L R Q + V ++ AL A A+ NLG++ AE R
Sbjct: 159 VSKEDQAHAILSNLGNLYRQKKQYE-VSKAMFSKALELKPGYAPAYNNLGLVFVAERRWE 217
Query: 133 EAVECFQ---AANSLEETA 148
EA CF+ A+SL + A
Sbjct: 218 EAKSCFEKSLEADSLLDAA 236
>At5g35440 putative protein
Length = 597
Score = 31.2 bits (69), Expect = 0.23
Identities = 13/32 (40%), Positives = 20/32 (61%)
Query: 119 YNLGILHKAEGRVLEAVECFQAANSLEETAPI 150
YN G+L+ A G+ L A +CFQ A+ + P+
Sbjct: 226 YNCGLLYLASGKPLLAAQCFQKASHVFRRQPL 257
>At5g56290 peroxisomal targeting signal type 1 receptor
Length = 728
Score = 30.8 bits (68), Expect = 0.30
Identities = 24/99 (24%), Positives = 45/99 (45%), Gaps = 2/99 (2%)
Query: 50 VIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALR 109
V+G ++ + A+ +F+ AL + P SL + A + S QS + ++ AL
Sbjct: 595 VLGVLYNLSREFDRAITSFQTALQLKPNDY-SLWNKLGATQANSVQSADAISAY-QQALD 652
Query: 110 YDRLNASAWYNLGILHKAEGRVLEAVECFQAANSLEETA 148
AW N+GI + +G E++ + A ++ A
Sbjct: 653 LKPNYVRAWANMGISYANQGMYKESIPYYVRALAMNPKA 691
>At4g37460 unknown protein
Length = 869
Score = 30.8 bits (68), Expect = 0.30
Identities = 26/94 (27%), Positives = 42/94 (44%), Gaps = 6/94 (6%)
Query: 58 KGLYKEAVKAFRDALNIDPRHVPSLIS--TAVALRRWSNQSDPVVRSFLMDALRYDRLNA 115
+G Y +A+ F L +P + +LI TA A +R + + + F A++ + +
Sbjct: 310 EGNYTKAISIFDKVLKEEPTYPEALIGRGTAYAFQR---ELESAIADFTK-AIQSNPAAS 365
Query: 116 SAWYNLGILHKAEGRVLEAVECFQAANSLEETAP 149
AW G A G +EAVE A E +P
Sbjct: 366 EAWKRRGQARAALGEYVEAVEDLTKALVFEPNSP 399
>At3g16320 hypothetical protein
Length = 727
Score = 30.4 bits (67), Expect = 0.39
Identities = 31/133 (23%), Positives = 58/133 (43%), Gaps = 9/133 (6%)
Query: 19 YAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRH 78
+ + +L ++ DAE C K+ + + +G + + ++ A F+ AL I+PR
Sbjct: 541 HEFAALEEFEDAERCYRKALGIDTRHYNAWYGLGMTYLRQEKFEFAQHQFQLALQINPRS 600
Query: 79 VPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWY-------NLGILHKAEGRV 131
+ +AL S ++D + + A+ D N Y +LG HKA+ +
Sbjct: 601 SVIMCYYGIALHE-SKRNDEAL-MMMEKAVLTDAKNPLPKYYKAHILTSLGDYHKAQKVL 658
Query: 132 LEAVECFQAANSL 144
E EC +S+
Sbjct: 659 EELKECAPQESSV 671
>At3g49810 unknown protein
Length = 448
Score = 28.9 bits (63), Expect = 1.1
Identities = 19/66 (28%), Positives = 30/66 (44%), Gaps = 6/66 (9%)
Query: 59 GLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAW 118
G+ E + + +D + +PS+ ++L DPV L Y+RLN W
Sbjct: 44 GVVDEKLDLKKMIKELDLQDIPSVFICPISLEP---MQDPVT---LCTGQTYERLNIHKW 97
Query: 119 YNLGIL 124
+NLG L
Sbjct: 98 FNLGHL 103
>At4g39470 unknown protein
Length = 341
Score = 28.5 bits (62), Expect = 1.5
Identities = 11/28 (39%), Positives = 19/28 (67%)
Query: 50 VIGTMHEAKGLYKEAVKAFRDALNIDPR 77
+IG MH +G ++E +K F+ +N +PR
Sbjct: 245 LIGQMHIVEGQFEEGLKIFQQMVNDNPR 272
>At5g58550 putative protein
Length = 925
Score = 28.1 bits (61), Expect = 1.9
Identities = 18/49 (36%), Positives = 24/49 (48%), Gaps = 1/49 (2%)
Query: 34 LSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSL 82
LSK+ AFR + H+ HEA G A + AL +DP H +L
Sbjct: 857 LSKAIAFRPELQT-LHLRAAFHEATGNLSLATQDCEAALCLDPNHTETL 904
>At2g32450 putative O-GlcNAc transferase
Length = 802
Score = 28.1 bits (61), Expect = 1.9
Identities = 33/123 (26%), Positives = 50/123 (39%), Gaps = 21/123 (17%)
Query: 30 AEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTA--- 86
A+ S+ +AF + A IG + L+KEA+ +F+ A + P V
Sbjct: 219 ADGARSREEAFDGHMA-----IGKVLYEHQLFKEALVSFKRACELQPTDVRPHFKAGNCL 273
Query: 87 VALRRWSNQSDPVVRSFLMDALRYDRLNASAW--------YNLGILHKAEGRVLEAVECF 138
L ++ D FL+ AL + W NLGI + EG VL A E +
Sbjct: 274 YVLGKYKESKD----EFLL-ALEAAESGGNQWAYLLPQIYVNLGISLEGEGMVLSACEYY 328
Query: 139 QAA 141
+ A
Sbjct: 329 REA 331
>At1g61640 unknown protein
Length = 621
Score = 28.1 bits (61), Expect = 1.9
Identities = 14/51 (27%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Query: 82 LISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVL 132
LIS ++ +SNQ++ + + ++ R+ ++ ++ LG+LH +GR L
Sbjct: 8 LISRVISRTLFSNQNNRTAINVSVKLPQF-RIQSNGYHTLGLLHNVKGRFL 57
>At1g33400 unknown protein
Length = 798
Score = 28.1 bits (61), Expect = 1.9
Identities = 25/94 (26%), Positives = 41/94 (43%), Gaps = 7/94 (7%)
Query: 61 YKEAVKAFRDALNIDPRHV----PSLISTAVALRRWSNQSDPVVRSFLMD---ALRYDRL 113
+ EA++ + AL + P SL+++ R + +++ L D ALR D
Sbjct: 79 FDEALRLYSKALRVAPLDAIDGDKSLLASLFLNRANVLHNLGLLKESLRDCHRALRIDPY 138
Query: 114 NASAWYNLGILHKAEGRVLEAVECFQAANSLEET 147
A AWY G L+ G +A + SLE +
Sbjct: 139 YAKAWYRRGKLNTLLGNYKDAFRDITVSMSLESS 172
>At4g08320 unknown protein
Length = 266
Score = 27.7 bits (60), Expect = 2.5
Identities = 18/77 (23%), Positives = 39/77 (50%), Gaps = 2/77 (2%)
Query: 60 LYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWY 119
LY EAV+ + A+ + ++ + A A + + S+ + + ++ D + A+
Sbjct: 30 LYLEAVELYSFAIALTDKNAVFYCNRAAAYTQINMCSEAI--KDCLKSIEIDPNYSKAYS 87
Query: 120 NLGILHKAEGRVLEAVE 136
LG+ + A+G+ EA+E
Sbjct: 88 RLGLAYYAQGKYAEAIE 104
>At3g25240 hypothetical protein
Length = 281
Score = 27.3 bits (59), Expect = 3.3
Identities = 14/43 (32%), Positives = 22/43 (50%)
Query: 28 HDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
HDA VC+SK + + A H I +H+ ++AV+ D
Sbjct: 128 HDAAVCVSKWTSSSKLIAGSYHFIDVVHKPSDNDQKAVRYLVD 170
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.133 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,213,495
Number of Sequences: 26719
Number of extensions: 112689
Number of successful extensions: 350
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 303
Number of HSP's gapped (non-prelim): 55
length of query: 153
length of database: 11,318,596
effective HSP length: 90
effective length of query: 63
effective length of database: 8,913,886
effective search space: 561574818
effective search space used: 561574818
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0082b.2


