

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0082a.8
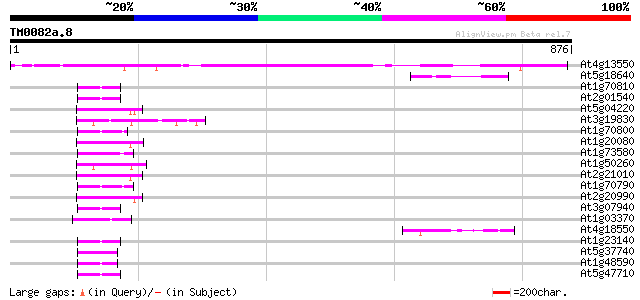
(876 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g13550 putative protein 720 0.0
At5g18640 triacylglycerol lipase-like protein 58 3e-08
At1g70810 unknown protein 53 9e-07
At2g01540 unknown protein (At2g01540) 52 1e-06
At5g04220 calcium lipid binding protein - like 51 3e-06
At3g19830 unknown protein 50 6e-06
At1g70800 unknown protein 50 6e-06
At1g20080 unknown protein 50 6e-06
At1g73580 hypothetical protein 50 7e-06
At1g50260 hypothetical protein 50 7e-06
At2g21010 hypothetical protein 49 1e-05
At1g70790 unknown protein 49 1e-05
At2g20990 unknown protein 49 2e-05
At3g07940 putative GTPase activating protein 47 4e-05
At1g03370 unknown protein 47 6e-05
At4g18550 lipase-like protein 46 8e-05
At1g23140 unknown protein 46 8e-05
At5g37740 unknown protein 45 1e-04
At1g48590 unknown protein 44 5e-04
At5g47710 unknown protein 43 7e-04
>At4g13550 putative protein
Length = 805
Score = 720 bits (1858), Expect = 0.0
Identities = 415/914 (45%), Positives = 539/914 (58%), Gaps = 178/914 (19%)
Query: 1 MASLQLRYLLSPPSLNFPSQHSSKPRFSRSFPPHFPGNLRAFSLAWRKQRRKRVFSTCCS 60
M SLQL + S H +P+ + P F N R F+ + R R
Sbjct: 1 MTSLQLHF----------SVHFLRPKHRLRYSPIF--NTRTFNFPGKITFRLRAKPNSLR 48
Query: 61 SKTGSQVERISVQEDDERAERPPFDINLAVILAGFAFEAYTTPPGTSDPYVVFQMDSQTV 120
S+ + +S+ E DER+ PFDINLAVILAGFAFE+Y +PPGTSDPYVV +D Q
Sbjct: 49 CFAQSETKEVSLPEKDERS---PFDINLAVILAGFAFESYASPPGTSDPYVVMDLDGQVA 105
Query: 121 KSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRMGNAGVDLEWLCD--- 177
KS KWGTKEP WNE+F FNIKLPP K +++AAWDANLV PHKRMGN+ ++LE +CD
Sbjct: 106 KSKTKWGTKEPKWNEDFVFNIKLPPAKKIEIAAWDANLVTPHKRMGNSEINLESVCDAVL 165
Query: 178 ------GDVHEILVELEGMGGGGKVQLEVKYKTFDEIDDEKKWWKMPFVSDFLKIN---- 227
G++H++LVEL+G+GGGGKVQLE+KYK F E+++EKKWW+ PFVS+FL+ N
Sbjct: 166 FCFASEGNLHKVLVELDGIGGGGKVQLEIKYKGFGEVEEEKKWWRFPFVSEFLQRNEIKS 225
Query: 228 ---------GFDSALKKVTGSNTVQTRQFVEYAFGQLKSFNKAYLQKGQMSDTDNDEYDT 278
+S LK + S V RQFVEYAFGQLKS N A L+ ++ +N D+
Sbjct: 226 VLKNFVDSEAVESVLKNLVDSEAVPARQFVEYAFGQLKSLNDAPLKNTEL--LNNTAEDS 283
Query: 279 EGSGELNESPFVLEMPPREAGSSEASNEACSEQRNTEEFHSHESDTE-NGHTSE-PSTQT 336
EG A S ++S++ S ++ S + D + +GH +E
Sbjct: 284 EG-----------------ASSEDSSDQHRSTNLSSSGKLSKDKDGDGDGHGNELEDDNE 326
Query: 337 SGEELSNQLFWRDFTNVINVNVVQKLGLTVPGKFKWDGLEFLDKIGSQSQNIAEATYIQS 396
SG S FW + +++ N+VQKLGL P K KW+G E L+ G QS+ AEA YI+S
Sbjct: 327 SGSIQSESNFWDNIPDIVGQNIVQKLGLPSPEKLKWNGTELLENFGLQSRKTAEAGYIES 386
Query: 397 GLAMPRGIDDTEDKTSGKPAIAAIQSALPEVKKATESLMRQTDSILGGLMLLAATVSKMK 456
GLA + ++K G+ AI A +S+L ++K AT+ L++Q D++ G LM+L A V +
Sbjct: 387 GLATADTREADDEKEDGQVAINASKSSLADMKNATQELLKQADNVFGALMVLKAVVPHLS 446
Query: 457 DEGSSSEERKIEEDSAKVGGNDI---QCLPKIPSSENGSVLDDKKAEEMRELFSTAETAM 513
+ S E+ IE++ + +D+ KI N D+K AEEM+ LFS+AE+AM
Sbjct: 447 KD-SVGSEKVIEKNGSSSVTDDVSGSSKTEKISGLVNVDGADEKNAEEMKTLFSSAESAM 505
Query: 514 EAWAMLATSLGHASFIKSEFEKICFLDNAPTDTQVAIWRDSMRRRLVVAFRGTEQSQWKD 573
EAWAMLAT+LGH SFIKSEFEK+CFL+N TDTQVAIWRD+ R+R+V+AFRGTEQ
Sbjct: 506 EAWAMLATALGHPSFIKSEFEKLCFLENDITDTQVAIWRDARRKRVVIAFRGTEQ----- 560
Query: 574 LITDLMIVPAGLASSSFSSAQASILDPKGVFVGVRYPIARLNPERIGGDFKQEVQVHSGF 633
S F SA S+
Sbjct: 561 ------------VHSGFLSAYDSV------------------------------------ 572
Query: 634 LSAYDSVRTRIISLIRLAIGYVDDHFEPLHKWHIYVTGHSLGGALATLLALELSSNQLTN 693
R RIISL+++ IGY+DD E KWH+YVTGHSLGGALATLLALELSS+QL
Sbjct: 573 -------RIRIISLLKMTIGYIDDVTEREDKWHVYVTGHSLGGALATLLALELSSSQLAK 625
Query: 694 MVDYVEEPKTHHIRQIMIANYGRISAMALSCPIFDNNAHLRGAISITMYNFGSPRVGNKR 753
RGAI++TMYNFGSPRVGNK+
Sbjct: 626 ----------------------------------------RGAITVTMYNFGSPRVGNKQ 645
Query: 754 FAEVYNEKIKDSWRVVNHRDIIPTVPRLMGYCHVNQPVFLAAG----------------V 797
FAE+YN+K+KDSWRVVNHRDIIPTVPRLMGYCHV PV+L+AG V
Sbjct: 646 FAEIYNQKVKDSWRVVNHRDIIPTVPRLMGYCHVAHPVYLSAGDVYELCKLYNMIFQYKV 705
Query: 798 LRNSLENKDILGDGYEGDVLGESTPDVIVSEFMKGEKELIEKLLQTEINIFRSIRDGTAF 857
++ E+ + DGY +V+GE+TPD++VS FMKGEKEL+EK+LQTEI IF ++RDG+A
Sbjct: 706 IKMLQEDIEFQKDGYHAEVIGEATPDILVSRFMKGEKELVEKILQTEIKIFNALRDGSAL 765
Query: 858 MQHMEDFYYVTLLE 871
MQHMEDFYY+TLLE
Sbjct: 766 MQHMEDFYYITLLE 779
>At5g18640 triacylglycerol lipase-like protein
Length = 369
Score = 57.8 bits (138), Expect = 3e-08
Identities = 41/156 (26%), Positives = 67/156 (42%), Gaps = 52/156 (33%)
Query: 626 EVQVHSGFLSAYDS--VRTRIISLIRLAIGYVDDHFEPLHKWHIYVTGHSLGGALATLLA 683
+ VH GF SAY + VR ++ ++ A + +I VTGHS+GGA+A+ A
Sbjct: 145 DAMVHHGFYSAYHNTTVRPAVLDAVKRAKESYGANL------NIMVTGHSMGGAMASFCA 198
Query: 684 LELSSNQLTNMVDYVEEPKTHHIRQIMIANYGRISAMALSCPIFDNNAHLRGAISITMYN 743
L+L N+ G ++ +
Sbjct: 199 LDLVVNE--------------------------------------------GEENVQVMT 214
Query: 744 FGSPRVGNKRFAEVYNEKIKDSWRVVNHRDIIPTVP 779
FG PRVGN FA +N + +++R+++ RDI+P +P
Sbjct: 215 FGQPRVGNAAFASYFNLLVPNTFRIIHDRDIVPHLP 250
>At1g70810 unknown protein
Length = 165
Score = 52.8 bits (125), Expect = 9e-07
Identities = 23/68 (33%), Positives = 41/68 (59%), Gaps = 1/68 (1%)
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
+SDP+VV M SQ +K+ + P WNEE T ++ P++P+ + +D + H +M
Sbjct: 25 SSDPFVVITMGSQKLKTRVVENNCNPEWNEELTLALR-HPDEPVNLIVYDKDTFTSHDKM 83
Query: 166 GNAGVDLE 173
G+A +D++
Sbjct: 84 GDAKIDIK 91
>At2g01540 unknown protein (At2g01540)
Length = 180
Score = 52.4 bits (124), Expect = 1e-06
Identities = 24/68 (35%), Positives = 40/68 (58%), Gaps = 1/68 (1%)
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
+SDPY+V + QT+K+ + P WNEE T IK PN P+++ +D + +M
Sbjct: 26 SSDPYIVLNVADQTLKTRVVKKNCNPVWNEEMTVAIK-DPNVPIRLTVFDWDKFTGDDKM 84
Query: 166 GNAGVDLE 173
G+A +D++
Sbjct: 85 GDANIDIQ 92
>At5g04220 calcium lipid binding protein - like
Length = 540
Score = 50.8 bits (120), Expect = 3e-06
Identities = 34/116 (29%), Positives = 58/116 (49%), Gaps = 13/116 (11%)
Query: 105 GTSDPYVVFQMDSQTV---KSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAP 161
GTSDPYV + + + K+ IK P WNE F +K P ++ LQ+ +D + V
Sbjct: 280 GTSDPYVKLSLTGEKLPAKKTTIKKRNLNPEWNEHFKLIVKDPNSQVLQLEVFDWDKVGG 339
Query: 162 HKRMGNAGVDLEWLCDGDVHEILVEL-----EGMGGG-----GKVQLEVKYKTFDE 207
H R+G + L+ + G+ E ++L M G G+++++++Y F E
Sbjct: 340 HDRLGMQMIPLQKINPGERKEFNLDLIKNSNVVMDSGDKKKRGRLEVDLRYVPFRE 395
Score = 37.7 bits (86), Expect = 0.028
Identities = 16/41 (39%), Positives = 24/41 (58%)
Query: 107 SDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNK 147
S+PY V + K+ + T++P WNEEF F ++ PP K
Sbjct: 439 SNPYAVVLFRGEKKKTKMLKKTRDPRWNEEFQFTLEEPPVK 479
>At3g19830 unknown protein
Length = 666
Score = 50.1 bits (118), Expect = 6e-06
Identities = 56/224 (25%), Positives = 92/224 (41%), Gaps = 29/224 (12%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTK------EPTWNEEFTFNIKLPPNKPLQVAAWDANL 158
G +DPYV+ ++ Q ++S T +P WN++F F + P + LQ+ D L
Sbjct: 404 GKTDPYVILRIGDQVIRSKKNSQTTVIGAPGQPIWNQDFQFLVSNPREQVLQIEVNDC-L 462
Query: 159 VAPHKRMGNAGVDLEWLCDGDVHEILVELE------GMGGGGKVQLEVKYKTFDEIDDEK 212
+G VDLE L D + V L G G G++ L + YK + E +++
Sbjct: 463 GFADMAIGIGEVDLESLPDTVPTDRFVSLRGGWSLFGKGSTGEILLRLTYKAYVEDEEDD 522
Query: 213 KWWKMPFVSDFLKINGFDSALKKVTGSNTVQTRQFVEYAFGQLKSFN-------KAYLQK 265
K +D DS S+ VQ + GQ N Q
Sbjct: 523 KRNAKAIYADASDDEMSDSE----EPSSFVQNDKIPSDDIGQESFMNVLSALILSEEFQG 578
Query: 266 GQMSDTDNDEYDTEGSGELNESPFV----LEMPPREAGSSEASN 305
S+T N++ D +G ++ P + E P++AG+ + S+
Sbjct: 579 IVSSETGNNKVD-DGESSVSPVPSMSGADSESRPKDAGNGDVSD 621
>At1g70800 unknown protein
Length = 174
Score = 50.1 bits (118), Expect = 6e-06
Identities = 27/78 (34%), Positives = 44/78 (55%), Gaps = 3/78 (3%)
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
+SDP+VV M Q +KS P WNEE T I+ PN+P+++ +D + +M
Sbjct: 34 SSDPFVVITMGPQKLKSFTVKNNCNPEWNEELTLAIE-DPNEPVKLMVYDKDTFTADDKM 92
Query: 166 GNAGVDLEWLCDGDVHEI 183
G+A +D++ DVH++
Sbjct: 93 GDAQIDMKPFL--DVHKL 108
>At1g20080 unknown protein
Length = 535
Score = 50.1 bits (118), Expect = 6e-06
Identities = 34/117 (29%), Positives = 54/117 (46%), Gaps = 12/117 (10%)
Query: 105 GTSDPYVVFQMDSQTV---KSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAP 161
G SDPYV + V K+ +K P WNEEF +K P ++ LQ+ +D V
Sbjct: 277 GGSDPYVKLTLSGDKVPGKKTVVKHSNLNPEWNEEFDLVVKEPESQELQLIVYDWEQVGK 336
Query: 162 HKRMGNAGVDLEWLCDGDVHEILVEL---------EGMGGGGKVQLEVKYKTFDEID 209
H ++G + L+ L + + +EL G++ +EV+YK F + D
Sbjct: 337 HDKIGMNVIQLKDLTPEEPKLMTLELLKSMEPKEPVSEKSRGQLVVEVEYKPFKDDD 393
>At1g73580 hypothetical protein
Length = 168
Score = 49.7 bits (117), Expect = 7e-06
Identities = 26/88 (29%), Positives = 47/88 (52%), Gaps = 5/88 (5%)
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
+SDPYVV ++ Q +K+ + P W E+ +F + PN PL + +D + + +M
Sbjct: 28 SSDPYVVLKLGRQKLKTKVVKQNVNPQWQEDLSFTV-TDPNLPLTLIVYDHDFFSKDDKM 86
Query: 166 GNAGVDLEWLCDGDVHEILVELEGMGGG 193
G+A +DL+ + + +EL G+ G
Sbjct: 87 GDAEIDLK----PYIEALRMELSGLPDG 110
>At1g50260 hypothetical protein
Length = 778
Score = 49.7 bits (117), Expect = 7e-06
Identities = 33/121 (27%), Positives = 57/121 (46%), Gaps = 13/121 (10%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTK------EPTWNEEFTFNIKLPPNKPLQVAAWDANL 158
G +DPY + ++ Q ++S T +P WN++F F + P + LQ+ D L
Sbjct: 424 GKTDPYAILRLGDQVIRSKRNSQTTVIGAPGQPIWNQDFQFLVSNPREQVLQIEVND-RL 482
Query: 159 VAPHKRMGNAGVDLEWLCDGDVHEILVELE------GMGGGGKVQLEVKYKTFDEIDDEK 212
+G VDL +L D + +V L G G G++ L + YK++ E +++
Sbjct: 483 GFADMAIGTGEVDLRFLQDTVPTDRIVVLRGGWSLFGKGSAGEILLRLTYKSYVEEEEDD 542
Query: 213 K 213
K
Sbjct: 543 K 543
>At2g21010 hypothetical protein
Length = 221
Score = 48.9 bits (115), Expect = 1e-05
Identities = 32/116 (27%), Positives = 54/116 (45%), Gaps = 13/116 (11%)
Query: 105 GTSDPYVVFQMDSQTV---KSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAP 161
G +PYV ++ + K+ +K P WNEEF F+++ P + L+ + +
Sbjct: 2 GMINPYVQIELSEDKISSKKTTVKHKNLNPEWNEEFKFSVRDPKTQVLEFNVYGWEKIGK 61
Query: 162 HKRMGNAGVDLEWLCDGDVHEILVEL--------EGMGG--GGKVQLEVKYKTFDE 207
H +MG + L+ L + +EL EG G GK+++E+ YK F E
Sbjct: 62 HDKMGMNVLALKELAPDERKAFTLELRKTLDGGEEGQPGKYRGKLEVELLYKPFTE 117
Score = 33.1 bits (74), Expect = 0.70
Identities = 14/39 (35%), Positives = 22/39 (55%)
Query: 107 SDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPP 145
++PYV + K+ K+P WNEEF+F ++ PP
Sbjct: 153 TNPYVHIYFKGEERKTKNVKKNKDPKWNEEFSFMLEEPP 191
>At1g70790 unknown protein
Length = 185
Score = 48.9 bits (115), Expect = 1e-05
Identities = 25/88 (28%), Positives = 48/88 (54%), Gaps = 3/88 (3%)
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
TSDPYVV + +Q +K+ + P WNE+ T +IK N P+++ +D + + +M
Sbjct: 26 TSDPYVVITLANQKLKTRVINNNCNPVWNEQLTLSIK-DVNDPIRLTVFDKDRFSGDDKM 84
Query: 166 GNAGVDLEWLCDGDVHEILVELEGMGGG 193
G+A +D + H++ ++ + + G
Sbjct: 85 GDAEIDFRPFL--EAHQMELDFQKLPNG 110
>At2g20990 unknown protein
Length = 541
Score = 48.5 bits (114), Expect = 2e-05
Identities = 31/117 (26%), Positives = 58/117 (49%), Gaps = 14/117 (11%)
Query: 105 GTSDPYVVFQMDSQTV---KSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAP 161
G +DP+V ++ + K+ +K P WNEEF F+++ P + L+ + +D V
Sbjct: 279 GGADPFVKIKLSEDKIPSKKTTVKHKNLNPEWNEEFKFSVRDPQTQVLEFSVYDWEQVGN 338
Query: 162 HKRMGNAGVDLEWLCDGDVHEILVEL-EGMGGG----------GKVQLEVKYKTFDE 207
++MG + L+ + + +EL + + GG GK+++E+ YK F E
Sbjct: 339 PEKMGMNVLALKEMVPDEHKAFTLELRKTLDGGEDGQPPDKYRGKLEVELLYKPFTE 395
Score = 34.3 bits (77), Expect = 0.31
Identities = 25/103 (24%), Positives = 48/103 (46%), Gaps = 6/103 (5%)
Query: 107 SDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPP-NKPLQVAAWDAN----LVAP 161
++PYV + K+ ++P WNEEFTF ++ PP + L V + L+ P
Sbjct: 438 TNPYVRIYFKGEERKTKHVKKNRDPRWNEEFTFMLEEPPVREKLHVEVLSTSSRIGLLHP 497
Query: 162 HKRMGNAGVDLEWLCDGDVHEILVELEGMGGGGKVQLEVKYKT 204
+ +G + + + + L GK+Q+E++++T
Sbjct: 498 KETLGYVDIPVVDVVNNKRMNQKFHLID-SKNGKIQIELEWRT 539
>At3g07940 putative GTPase activating protein
Length = 385
Score = 47.4 bits (111), Expect = 4e-05
Identities = 23/68 (33%), Positives = 36/68 (52%), Gaps = 1/68 (1%)
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
TSDPYV+ + Q+VK+ + P WNE +I P PL+V +D + + M
Sbjct: 248 TSDPYVILALGQQSVKTRVIKNNLNPVWNETLMLSIP-EPMPPLKVLVYDKDTFSTDDFM 306
Query: 166 GNAGVDLE 173
G A +D++
Sbjct: 307 GEAEIDIQ 314
>At1g03370 unknown protein
Length = 1859
Score = 46.6 bits (109), Expect = 6e-05
Identities = 30/94 (31%), Positives = 45/94 (46%), Gaps = 5/94 (5%)
Query: 99 AYTTPPGTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANL 158
A P G DPY+VF + +T S+IK+ P WNE F F+ P L V +D +
Sbjct: 1374 AAVDPSGHCDPYIVFTSNGKTRTSSIKFQKSNPQWNEIFEFDAMADPPSVLNVEVFDFD- 1432
Query: 159 VAPHKR---MGNAGVDLEWLCDGDVHEILVELEG 189
P +G+A V+ D+ ++ V L+G
Sbjct: 1433 -GPFDEAVSLGHAEVNFVRSNISDLADVWVPLQG 1465
Score = 33.5 bits (75), Expect = 0.53
Identities = 20/75 (26%), Positives = 33/75 (43%), Gaps = 1/75 (1%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDPYV Q+ Q ++ + P W E+F+F + N L V+ D +
Sbjct: 846 GFSDPYVRLQLGKQRSRTKVVKKNLNPKWTEDFSFGVD-DLNDELVVSVLDEDKYFNDDF 904
Query: 165 MGNAGVDLEWLCDGD 179
+G V + + D +
Sbjct: 905 VGQVRVSVSLVFDAE 919
>At4g18550 lipase-like protein
Length = 419
Score = 46.2 bits (108), Expect = 8e-05
Identities = 49/183 (26%), Positives = 79/183 (42%), Gaps = 47/183 (25%)
Query: 614 LNPERIGGDFKQEVQVHSGFLSAYDS-------VRTRIISLIRLAIGYVDDHFEPLHKWH 666
+N +I G+ +VQ+H G+ S Y S +T + +G + + ++ +
Sbjct: 171 VNAIKIFGERNDQVQIHQGWYSIYMSQDERSPFTKTNARDQVLREVGRLLEKYKD-EEVS 229
Query: 667 IYVTGHSLGGALATLLALELSSNQLTNMVDYVEEPKTHHIRQIMIANYGRISAMALSCPI 726
I + GHSLG ALATL A ++ +N PK+ + SCP
Sbjct: 230 ITICGHSLGAALATLSATDIVANG-------YNRPKSRPDK---------------SCP- 266
Query: 727 FDNNAHLRGAISITMYNFGSPRVGNKRFAEVYN--EKIKDSWRVVNHRDIIPTVPRLMGY 784
+T + F SPRVG+ F ++++ E I+ R N D+IP P + GY
Sbjct: 267 ------------VTAFVFASPRVGDSDFRKLFSGLEDIR-VLRTRNLPDVIPIYPPI-GY 312
Query: 785 CHV 787
V
Sbjct: 313 SEV 315
>At1g23140 unknown protein
Length = 165
Score = 46.2 bits (108), Expect = 8e-05
Identities = 23/68 (33%), Positives = 38/68 (55%), Gaps = 1/68 (1%)
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
TSDP+VV M SQ +K+ + P W++E T I PN+ + + +D + H M
Sbjct: 25 TSDPFVVVTMGSQKLKTRGVENSCNPEWDDELTLGIN-DPNQHVTLEVYDKDTFTSHDPM 83
Query: 166 GNAGVDLE 173
G+A +D++
Sbjct: 84 GDAEIDIK 91
>At5g37740 unknown protein
Length = 168
Score = 45.4 bits (106), Expect = 1e-04
Identities = 19/63 (30%), Positives = 37/63 (58%), Gaps = 1/63 (1%)
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
+SDPY+V Q +K+ + + P WN++ T ++ PN P+++ +D +L++ +M
Sbjct: 25 SSDPYIVVHCGKQKLKTRVVKHSVNPEWNDDLTLSV-TDPNLPIKLTVYDYDLLSADDKM 83
Query: 166 GNA 168
G A
Sbjct: 84 GEA 86
>At1g48590 unknown protein
Length = 185
Score = 43.5 bits (101), Expect = 5e-04
Identities = 20/63 (31%), Positives = 35/63 (54%), Gaps = 1/63 (1%)
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
+SDPYVV +M Q +K+ + + P WNE+ T ++ PN + + +D + +M
Sbjct: 46 SSDPYVVVKMAKQKLKTRVIYKNVNPEWNEDLTLSVS-DPNLTVLLTVYDYDTFTKDDKM 104
Query: 166 GNA 168
G+A
Sbjct: 105 GDA 107
>At5g47710 unknown protein
Length = 166
Score = 43.1 bits (100), Expect = 7e-04
Identities = 20/68 (29%), Positives = 37/68 (54%), Gaps = 1/68 (1%)
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
+SDPYV+ ++ +++ K+ + P WNEE F +K P L + +D + +M
Sbjct: 25 SSDPYVIVKLGNESAKTKVINNCLNPVWNEELNFTLK-DPAAVLALEVFDKDRFKADDKM 83
Query: 166 GNAGVDLE 173
G+A + L+
Sbjct: 84 GHASLSLQ 91
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,235,151
Number of Sequences: 26719
Number of extensions: 904754
Number of successful extensions: 3055
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 2901
Number of HSP's gapped (non-prelim): 171
length of query: 876
length of database: 11,318,596
effective HSP length: 108
effective length of query: 768
effective length of database: 8,432,944
effective search space: 6476500992
effective search space used: 6476500992
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0082a.8


