

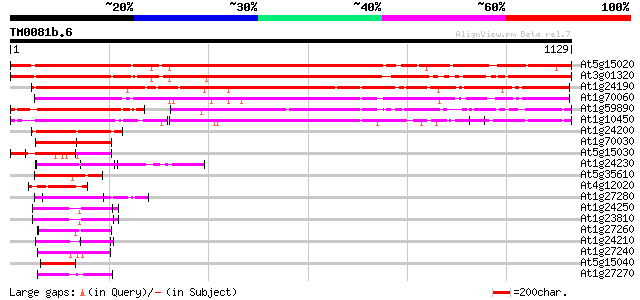
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0081b.6
(1129 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g15020 transcriptional regulatory - like protein 1214 0.0
At3g01320 unknown protein 1132 0.0
At1g24190 hypothetical protein 915 0.0
At1g70060 hypothetical protein 881 0.0
At1g59890 hypothetical protein 492 e-139
At1g10450 unknown protein 414 e-115
At1g24200 hypothetical protein 186 6e-47
At1g70030 unknown protein 174 3e-43
At5g15030 putative protein 138 2e-32
At1g24230 hypothetical protein 115 2e-25
At5g35610 putative protein 112 1e-24
At4g12020 putative disease resistance protein 103 6e-22
At1g27280 hypothetical protein 100 4e-21
At1g24250 hypothetical protein 100 6e-21
At1g23810 hypothetical protein 99 2e-20
At1g27260 hypothetical protein 93 1e-18
At1g24210 unknown protein 92 2e-18
At1g27240 hypothetical protein 86 9e-17
At5g15040 putative protein 85 2e-16
At1g27270 hypothetical protein 84 3e-16
>At5g15020 transcriptional regulatory - like protein
Length = 1343
Score = 1214 bits (3141), Expect = 0.0
Identities = 661/1184 (55%), Positives = 835/1184 (69%), Gaps = 107/1184 (9%)
Query: 1 MKRARDDIYSASASQFKRPFASSRGDSYGQSQVPGGGGGGGGAGGGGEATTSQKLTTNDA 60
MKR RDDIY A+ SQFKRP SSRG+SY QS + GGG G G +QKLTT+DA
Sbjct: 1 MKRIRDDIY-ATGSQFKRPLGSSRGESYEQSPITGGGSIGEGG------INTQKLTTDDA 53
Query: 61 LSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFGFNTFL 120
L+YLK+VK+MFQDQR+KYD+FLEVMKDFKAQ+TDT+GVI+RVKELFKGHN+LIFGFNTFL
Sbjct: 54 LTYLKEVKEMFQDQRDKYDMFLEVMKDFKAQKTDTSGVISRVKELFKGHNNLIFGFNTFL 113
Query: 121 PKGYEITLDEDEAPAKKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIG 180
PKG+EITLD+ EAP+KKTVEFEEAISFVNKIK RFQ +E VYKSFL+ILNMYRK++KDI
Sbjct: 114 PKGFEITLDDVEAPSKKTVEFEEAISFVNKIKTRFQHNELVYKSFLEILNMYRKDNKDIT 173
Query: 181 EVYSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQHAPFGRNSLQRFNERNSMTPMMRQMQ 240
EVY+EV+TLF+DH DLLEEFTRFLPD S AP T+ A R+ QR+++R S P++R+M
Sbjct: 174 EVYNEVSTLFEDHSDLLEEFTRFLPD-SLAPHTE-AQLLRSQAQRYDDRGSGPPLVRRMF 231
Query: 241 VDKQRYRRDRLPSHDRDRDLSVEHPEMDDDKTMINLHKEQRKR------DRRIRDQDERD 294
++K R RR+R + DRD SV+ +++DDK+M+ +H++QRKR +RR RD ++ +
Sbjct: 232 MEKDR-RRERTVASRGDRDHSVDRSDLNDDKSMVKMHRDQRKRVDKDNRERRSRDLEDGE 290
Query: 295 PDLDNSRDLTSQRFRDKKKTVKKAEG------------------MYGEAFSFCEKVKEKL 336
+ DN Q F +K+K+ ++ EG MY +AF FCEKVKE+L
Sbjct: 291 AEQDNL-----QHFSEKRKSSRRMEGFEAYSGPASHSEKNNLKSMYNQAFLFCEKVKERL 345
Query: 337 SSSDDYQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKDFLERCENIEGF--L 394
S DDYQ FLKCLN+F+NGII++ DLQNLV+D+LGK DLMDEF F ERCE+I+GF L
Sbjct: 346 CSQDDYQAFLKCLNMFSNGIIQRKDLQNLVSDVLGKFPDLMDEFNQFFERCESIDGFQHL 405
Query: 395 AGVMSKKSLSTDAHLSRSSKLEDKDKDQKREMDGAKEKDRYKEKYMGKSIQELDLSDCKR 454
AGVMSKKSL ++ +LSRS K E+KD++ KR+++ AKEK+R K+KYMGKSIQELDLSDC+R
Sbjct: 406 AGVMSKKSLGSEENLSRSVKGEEKDREHKRDVEAAKEKERSKDKYMGKSIQELDLSDCER 465
Query: 455 CTPSYRLLPSDYPIPTASQRSELGAQVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCE 514
CTPSYRLLP DYPIP+ R + GA VLNDHWVSVTSGSEDYSFKHMR+NQYEESLFRCE
Sbjct: 466 CTPSYRLLPPDYPIPSVRHRQKSGAAVLNDHWVSVTSGSEDYSFKHMRRNQYEESLFRCE 525
Query: 515 DDRFELDLLLESVSSASKRAEELYNNINENKISVEALSRIEDHFTVLNLRCIERLYGDHG 574
DDRFELD+LLESV SA+K AEEL N I + KIS E RIEDHFT LNLRCIERLYGDHG
Sbjct: 526 DDRFELDMLLESVGSAAKSAEELLNIIIDKKISFEGSFRIEDHFTALNLRCIERLYGDHG 585
Query: 575 LDVIDILRKNPTHALPVILTRLKQKQEEWNRCRSDFNKVWAEIYAKNHYKSLDHRSFYFK 634
LDV D++RKNP ALPVILTRLKQKQ+EW +CR FN VWA++YAKNHYKSLDHRSFYFK
Sbjct: 586 LDVTDLIRKNPAAALPVILTRLKQKQDEWTKCREGFNVVWADVYAKNHYKSLDHRSFYFK 645
Query: 635 QQDSKNLSTKSLVAEIKEIKEKLQKEDHIIQSIAAENRQPLIPHLEFEYSDGGIHEDLYK 694
QQDSKNLS K+LV+E+K++KEK QKED ++ SI+A RQP+IPHLE++Y D IHEDL+K
Sbjct: 646 QQDSKNLSAKALVSEVKDLKEKSQKEDDVVLSISAGYRQPIIPHLEYDYLDRAIHEDLFK 705
Query: 695 LVQYSCEEVFSSKELLNKIMRLWSTFLEPMLGVTSQSHGTERVEDRKAGHSSRNFAASNV 754
LVQ+SCEE+ S+KE K+++LW+ FLE ML V ++ G++ VED R F +
Sbjct: 706 LVQFSCEEICSTKEQTGKVLKLWANFLELMLDVAPRAKGSDSVEDVVETQHQRAFTS--- 762
Query: 755 GGDGSPHRDSISTNSRLPKSDKNEVDGRVTEVKNIHRTSVAANDKENGSV----GGELVC 810
G+ + D+IS SR K N ++H +S + E G + G+
Sbjct: 763 -GEANESSDAISLVSRQLKFATN---------GDVHASSGVSKHGETGLLNRDSSGKENL 812
Query: 811 RDDQLMDKGLKKVECSDKAGFSKQFAS---------DEQGVKNNPSIAIRGENSLNRTNL 861
+D L +K + C++K ++ + DE+ ++ S EN+ +
Sbjct: 813 KDGDLANKDV--ATCAEKPQKDQEIGNGAAKRSGDVDERVATSSSSFPSGVENNNGKVGS 870
Query: 862 DVSPGCVSAPSRPTDADDSVAKSQTVNLPLVEGGDIAAPVPVANGVLVENSKVKS-HEES 920
S G S+P++A D V Q +G DI + + NG+ + SK S ++ES
Sbjct: 871 RDSSGSRGILSKPSEAIDKVDSIQH-----TQGVDIGRIIVLGNGLQSDTSKANSNYDES 925
Query: 921 SGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETG 980
GP K+EKEEGELSP GDSE++FV Y D ++ AK++H++E
Sbjct: 926 GGPSKIEKEEGELSPVGDSEDNFVVYEDRELKATAKTEHSVEAE---------------- 969
Query: 981 GDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDH-EEDDIEHDDVDGKAESEGEAEG 1039
G+ND DADDED ++ SEAGED SG+ES GDEC Q+D+ E++ EHD++DGKAESEGEAEG
Sbjct: 970 GENDEDADDEDGDDASEAGEDASGTESIGDECSQDDNGVEEEGEHDEIDGKAESEGEAEG 1029
Query: 1040 MCDAQGGGDSSSLPLSERFLSSVKPLTKHVSAVSFAEE-MKDSRVFYGNDDFYALFRLHQ 1098
M ++ D P SER L SVKPL+KH++A + +E KDSRVFYGNDDFY LFRLH+
Sbjct: 1030 M-ESHLIEDKGLFPSSERVLLSVKPLSKHIAAAALVDEKKKDSRVFYGNDDFYVLFRLHR 1088
Query: 1099 ------------ILYERILSAKINSMSAEMKWK-AKDASSPDPY 1129
ILYERILSAK +EMK + KD SPDPY
Sbjct: 1089 VSAIDSYDLLSHILYERILSAKTYCSGSEMKLRNTKDTCSPDPY 1132
>At3g01320 unknown protein
Length = 1324
Score = 1132 bits (2929), Expect = 0.0
Identities = 643/1167 (55%), Positives = 784/1167 (67%), Gaps = 114/1167 (9%)
Query: 1 MKRARDDIYSASASQFKRPFASSRGDSYGQSQVPGGGGGGGGAGGGGEATTSQKLTTNDA 60
MKR RDD+Y AS SQF+RP SSRG GQS V G G GG SQKLTTNDA
Sbjct: 1 MKRIRDDVY-ASGSQFRRPLGSSRGQLCGQSPVHGSGDTEEEEEGGSRRV-SQKLTTNDA 58
Query: 61 LSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFGFNTFL 120
LSYL++VK+MFQDQREKYD FLEVMKDFKAQRTDT GVIARVKELFKGHN+LI+GFNTFL
Sbjct: 59 LSYLREVKEMFQDQREKYDRFLEVMKDFKAQRTDTGGVIARVKELFKGHNNLIYGFNTFL 118
Query: 121 PKGYEITL-DEDEAPAKKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDI 179
PKGYEITL +ED+A KKTVEFE+AI+FVNKIK RF+ DEHVYKSFL+ILNMYRKE+K+I
Sbjct: 119 PKGYEITLIEEDDALPKKTVEFEQAINFVNKIKMRFKHDEHVYKSFLEILNMYRKENKEI 178
Query: 180 GEVYSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQHAPFGRNSLQRFNERNSMTPMMRQM 239
EVY+EV+ LF+ H DLLE+FTRFLP ++ PS A R+ Q++++R S P++ QM
Sbjct: 179 KEVYNEVSILFQGHLDLLEQFTRFLP--ASLPSHSAAQHSRSQAQQYSDRGSDPPLLHQM 236
Query: 240 QVDKQRYRRDRLPSHDRDRDLSVEHPEMDDDKTMINLHKEQRKR------DRRIRDQDER 293
QV+K+R RR+R + D SVE +++DDKTM+ + +EQRKR RR RD D+R
Sbjct: 237 QVEKER-RRERAVA--LRGDYSVERYDLNDDKTMVKIQREQRKRLDKENRARRGRDLDDR 293
Query: 294 DPDLDNSRDLTSQRFRDKKKTVKKAEG------------------MYGEAFSFCEKVKEK 335
+ DN F +K+K+ ++AE MY +AF FCEKVK++
Sbjct: 294 EAGQDNLH-----HFPEKRKSSRRAEALEAYSGSASHSEKDNLKSMYKQAFVFCEKVKDR 348
Query: 336 LSSSDDYQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKDFLERCENIEGF-- 393
L S DDYQTFLKCLNIF+NGII++ DLQNLV+DLLGK DLMDEF F ERCE+I G
Sbjct: 349 LCSQDDYQTFLKCLNIFSNGIIQRKDLQNLVSDLLGKFPDLMDEFNQFFERCESITGTEI 408
Query: 394 -----LAGVMSKKSLSTDAHLSRSSKLEDKDKDQKREMDGAKEKDRYKEKYMGKSIQELD 448
LAGVMSKK S++ LSR K+E+K+ + K E++ KE ++ K++YMGKSIQELD
Sbjct: 409 HGFQRLAGVMSKKLFSSEEQLSRPMKVEEKESEHKPELEAVKETEQCKKEYMGKSIQELD 468
Query: 449 LSDCKRCTPSYRLLPSDYPIPTASQRSELGAQVLNDHWVSVTSGSEDYSFKHMRKNQYEE 508
LSDC+ CTPSYRLLP+DYPIP ASQRSELGA+VLNDHWVSVTSGSEDYSFKHMR+NQYEE
Sbjct: 469 LSDCECCTPSYRLLPADYPIPIASQRSELGAEVLNDHWVSVTSGSEDYSFKHMRRNQYEE 528
Query: 509 SLFRCEDDRFELDLLLESVSSASKRAEELYNNINENKISVEALSRIEDHFTVLNLRCIER 568
SLFRCEDDRFELD+LLESVSSA++ AE L N I E KIS RIEDHFT LNLRCIER
Sbjct: 529 SLFRCEDDRFELDMLLESVSSAARSAESLLNIITEKKISFSGSFRIEDHFTALNLRCIER 588
Query: 569 LYGDHGLDVIDILRKNPTHALPVILTRLKQKQEEWNRCRSDFNKVWAEIYAKNHYKSLDH 628
LYGDHGLDVIDIL KNP ALPVILTRLKQKQ EW +CR DF+KVWA +YAKNHYKSLDH
Sbjct: 589 LYGDHGLDVIDILNKNPATALPVILTRLKQKQGEWKKCRDDFDKVWANVYAKNHYKSLDH 648
Query: 629 RSFYFKQQDSKNLSTKSLVAEIKEIKEKLQKEDHIIQSIAAENRQPLIPHLEFEYSDGGI 688
RSFYFKQQDSKNLS KSL+AEIKE+KEK Q +D ++ SI+A RQP+ P+LE+EY + I
Sbjct: 649 RSFYFKQQDSKNLSAKSLLAEIKELKEKSQNDDDVLLSISAGYRQPINPNLEYEYLNRAI 708
Query: 689 HEDLYKLVQYSCEEVFSSKELLNKIMRLWSTFLEPMLGVTSQSHGTERVEDRKAGHSSRN 748
HED++K+VQ+SCEE+ S+KE L+K++RLW FLE +LGV ++ GT+ VED + +
Sbjct: 709 HEDMFKVVQFSCEELCSTKEQLSKVLRLWENFLEAVLGVPPRAKGTDLVEDVVINPKTLD 768
Query: 749 FAASNVGGDGSPHRDSISTNSRLPKSDKNEVDGRVTEVKNIHRTSVAANDKENGSVG--- 805
+S P + G T + AAN EN S G
Sbjct: 769 V-----------------NHSTSPNGEAAVSSGGDTARLASRKLKSAANGDENSSSGTFK 811
Query: 806 -GELVCRDDQLMDKGLKKVECSDKAGFSKQFASDEQGVKNNPSIAIRGENSLNRTNLDVS 864
G + D + L+ VE +++ G + ++ + G + R +
Sbjct: 812 HGIGLLNKDSTGKENLEDVEIANRDGVACSAVKPQKEQET-------GNEAEKRFGKPI- 863
Query: 865 PGCVSAPSRPTDADDSVAKSQTVNLPLVEGGDIAAPVPVANGVLVENSKVKSHEESSGPC 924
P D + A S I+ P N N V E GP
Sbjct: 864 ---------PMDISERAAIS-----------SISIPSGAEN-----NHCVVGKEVLPGPS 898
Query: 925 KVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDND 984
+ EKEEGELSPNGD E++F Y+D +S +K ++ S + E E E +N+
Sbjct: 899 RNEKEEGELSPNGDFEDNFGVYKDHGVKSTSKPEN--------SAEAEVEADAEV--ENE 948
Query: 985 ADADDEDSENVSEAGEDVSGSESAGDECFQ-EDHEEDDIEHDDVDGKAESEGEAEGMCDA 1043
DADD DSEN SEA SG+ES GD C Q ED EE++ EHD++DGKAESEGEAEGM
Sbjct: 949 DDADDVDSENASEA----SGTESGGDVCSQDEDREEENGEHDEIDGKAESEGEAEGMDPH 1004
Query: 1044 QGGGDSSSLPLSERFLSSVKPLTKHVSAVSFAEEMKDSRVFYGNDDFYALFRLHQILYER 1103
G+S LP SER L SV+PL+KHV+AV E KD +VFYGNDDFY LFRLHQILYER
Sbjct: 1005 LLEGESELLPQSERVLLSVRPLSKHVAAVLCDERTKDLQVFYGNDDFYVLFRLHQILYER 1064
Query: 1104 ILSAKINSMSAEMKWK-AKDASSPDPY 1129
IL AK N E+K K KD ++ DPY
Sbjct: 1065 ILYAKRNCSGGELKSKNLKDTNAGDPY 1091
>At1g24190 hypothetical protein
Length = 1263
Score = 915 bits (2366), Expect = 0.0
Identities = 551/1140 (48%), Positives = 727/1140 (63%), Gaps = 96/1140 (8%)
Query: 45 GGGEATTSQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKE 104
GGG A QKLTTNDAL+YLK VKD FQDQR KYD FLEVMK+FK+QR DTAGVI RVKE
Sbjct: 3 GGGSA---QKLTTNDALAYLKAVKDKFQDQRGKYDEFLEVMKNFKSQRVDTAGVITRVKE 59
Query: 105 LFKGHNHLIFGFNTFLPKGYEITL--DEDEAPAKKTVEFEEAISFVNKIKKRFQSDEHVY 162
LFKGH LI GFNTFLPKG+EITL ++ + P KK VEFEEAISFVNKIK RFQ D+ VY
Sbjct: 60 LFKGHQELILGFNTFLPKGFEITLQPEDGQPPLKKRVEFEEAISFVNKIKTRFQGDDRVY 119
Query: 163 KSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFLPDTSAA---PSTQHAPFG 219
KSFLDILNMYR++ K I EVY EVA LF+DH DLL EFT FLPDTSA PS + +
Sbjct: 120 KSFLDILNMYRRDSKSITEVYQEVAILFRDHSDLLVEFTHFLPDTSATASIPSVKTSVRE 179
Query: 220 RNSLQRFNERNSMTPM------MRQMQVDKQRYRRDRLPSHDRDRDLSVEHPEMDDDKTM 273
R + +TP + D++R + H R + EH + D +
Sbjct: 180 RGVSLADKKDRIITPHPDHDYGTEHIDQDRERPIKKENKEHMRGTNKENEHRDARDFEP- 238
Query: 274 INLHKEQ---RKRDRRIRDQDERDPDLDNSRDLTSQRFRDKKKTVKKAEGMYGEAFSFCE 330
+ KEQ +K+ IR D + + N L+ K A Y + + +
Sbjct: 239 -HSKKEQFLNKKQKLHIRGDDPAE--ISNQSKLSGAVPSSSTYDEKGAMKSYSQDLAIVD 295
Query: 331 KVKEKLSSSDDYQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKDFLERCEN- 389
+VKEKL++S+ YQ FL+CLN+F+ II + +LQ+LV +L+G + DLMD F +FL +CE
Sbjct: 296 RVKEKLNASE-YQEFLRCLNLFSKEIISRPELQSLVGNLIGVYPDLMDSFIEFLVQCEKN 354
Query: 390 ------------IEGFLAGVMSKKSLSTDAHLSRSSKLEDKDKDQKREMDGAKEKDRYKE 437
EG L+G+++KKSL ++ + S D+D++ KR+ DG +++D KE
Sbjct: 355 EKRQICNLLNLLAEGLLSGILTKKSLWSEGKYPQPSLDNDRDQEHKRD-DGLRDRDHEKE 413
Query: 438 KY--------MGKSIQELDLSDCKRCTPSYRLLPSDYPIPTASQRSELGAQVLNDHWVSV 489
+ K I ELDLS+C++CTPSYRLLP +YPI ASQ++E+G VLNDHWVSV
Sbjct: 414 RLEKAAANLKWAKPISELDLSNCEQCTPSYRLLPKNYPISIASQKTEIGKLVLNDHWVSV 473
Query: 490 TSGSEDYSFKHMRKNQYEESLFRCEDDRFELDLLLESVSSASKRAEELYNNINENKISVE 549
TSGSEDYSF HMRKNQYEESLF+CEDDRFELD+LLESV+S +K EEL IN N++
Sbjct: 474 TSGSEDYSFSHMRKNQYEESLFKCEDDRFELDMLLESVNSTTKHVEELLTKINSNELKTN 533
Query: 550 ALSRIEDHFTVLNLRCIERLYGDHGLDVIDILRKNPTHALPVILTRLKQKQEEWNRCRSD 609
+ R+EDH T LNLRCIERLYGDHGLDV+D+L+KN + ALPVILTRLKQKQEEW RCRSD
Sbjct: 534 SPIRVEDHLTALNLRCIERLYGDHGLDVMDVLKKNVSLALPVILTRLKQKQEEWARCRSD 593
Query: 610 FNKVWAEIYAKNHYKSLDHRSFYFKQQDSKNLSTKSLVAEIKEIKEKLQKEDHIIQSIAA 669
F+KVWAEIYAKN+YKSLDHRSFYFKQQDSK LS K+L+AEIKEI EK ++ED + + AA
Sbjct: 594 FDKVWAEIYAKNYYKSLDHRSFYFKQQDSKKLSMKALLAEIKEITEK-KREDDSLLAFAA 652
Query: 670 ENRQPLIPHLEFEYSDGGIHEDLYKLVQYSCEEVFSSKELLNKIMRLWSTFLEPMLGVTS 729
NR + P LEF+Y D +HEDLY+L++YSC E+ S+ E L+K+M++W+TF+E + GV S
Sbjct: 653 GNRLSISPDLEFDYPDHDLHEDLYQLIKYSCAEMCST-EQLDKVMKIWTTFVEQIFGVPS 711
Query: 730 QSHGTERVED--RKAGHSSRNFAASNVGGDGSPHRDSISTNSRLPKSDKNEVDGRVTEVK 787
+ G E ED + + ++ ++S +GSPH + +SR KS + + E
Sbjct: 712 RPQGAEDQEDVVKSMNQNVKSGSSSAGESEGSPHNYASVADSRRSKSSR-----KANEHS 766
Query: 788 NIHRTSVAANDKENGSVG--GELVCRDDQLMDKGLKKVECSDKAGFSKQFASDEQGVKNN 845
+ +TS N + +G+ G + +C Q +K LK V SD+ SKQ S E+ +
Sbjct: 767 QLGQTS---NSERDGAAGRTSDALCETAQ-HEKMLKNVVTSDEKPESKQAVSIERA---H 819
Query: 846 PSIAIRGENSLNRTN-----LDVSPGCVSAPSRPTDADDSVAKSQTVNLPLVEGGDIAAP 900
S A+ + L+++N + ++ C + T + K N P +E G+
Sbjct: 820 DSTALAVDGLLDQSNGGSSIVHMTGHCNNNLKPVTCGTELELKMNDGNGPKLEVGNKKL- 878
Query: 901 VPVANGVLVENSKVKSHEESSGPCKVEKEEGELSPNGDSEED-FVAYRDSNAQSMAKSKH 959
+ NG+ VE + S +E +G KVE+EEGELSPNGD EED F Y ++ ++ +K+
Sbjct: 879 --LTNGIAVE---ITSDQEMAGTSKVEREEGELSPNGDFEEDNFAVYAKTDFETFSKAND 933
Query: 960 NIERRKYESRDRE-EECGPETGGDNDADADD------EDSENVSEAGEDVSGSESAGDEC 1012
+ R RE E ET +NDA+ D+ EDS N E G DVSG+ES G E
Sbjct: 934 STGNNISGDRSREGEPSCLETRAENDAEGDENAARSSEDSRNEYENG-DVSGTESGGGE- 991
Query: 1013 FQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGDS-SSLPLSERFLSSVKPLTKHV-S 1070
EDD+++++ K ESEGEAE M DA ++ S+LP+S RFL VKPL K+V S
Sbjct: 992 ----DPEDDLDNNN---KGESEGEAECMADAHDAEENGSALPVSARFLLHVKPLVKYVPS 1044
Query: 1071 AVSF----AEEMKDSRVFYGNDDFYALFRLHQILYERILSAKINSMSAEMKWKAKDASSP 1126
A++ + +K+S+VFYGND FY LFRLH+ILYERILSAK+NS S E KW+ + +P
Sbjct: 1045 AIALHDKDKDSLKNSQVFYGNDSFYVLFRLHRILYERILSAKVNSSSPEGKWRTSNTKNP 1104
>At1g70060 hypothetical protein
Length = 1386
Score = 881 bits (2277), Expect = 0.0
Identities = 541/1183 (45%), Positives = 717/1183 (59%), Gaps = 165/1183 (13%)
Query: 50 TTSQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGH 109
+++QKLTTNDAL+YLK VKD FQD+R+KYD FLEVMKDFKAQR DT GVI RVKELFKG+
Sbjct: 5 SSAQKLTTNDALAYLKAVKDKFQDKRDKYDEFLEVMKDFKAQRVDTTGVILRVKELFKGN 64
Query: 110 NHLIFGFNTFLPKGYEITL-DEDEAPA--KKTVEFEEAISFVNKIKKRFQSDEHVYKSFL 166
LI GFNTFLPKG+EITL ED+ PA KK VEFEEAISFVNKIK RFQ D+ VYKSFL
Sbjct: 65 RELILGFNTFLPKGFEITLRPEDDQPAAPKKPVEFEEAISFVNKIKTRFQGDDRVYKSFL 124
Query: 167 DILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQHA---PFGRNSL 223
DILNMYRKE+K I EVY EVA LF+DH DLL EFT FLPDTSA ST + P +
Sbjct: 125 DILNMYRKENKSITEVYHEVAILFRDHHDLLGEFTHFLPDTSATASTNDSVKVPVRDRGI 184
Query: 224 QRFNERNSMTPMMRQMQVDKQRYRRDRLPSHDRDRDLSVEHPEMDDDKTMINLHKEQRKR 283
+ P MRQ+ +DK +DR+ + +R L E+ ++D +++++ KE+ +R
Sbjct: 185 KSL-------PTMRQIDLDK----KDRIITSHPNRALKTENMDVDHERSLLKDSKEEVRR 233
Query: 284 -DRRIRDQDERDPDLDNSRDLTSQR---FRDKKKTVKKAE--------GMYGEAFS---- 327
D++ D+RD D S + F KKK ++K + G+ FS
Sbjct: 234 IDKKNDFMDDRDRKDYRGLDHDSHKEHFFNSKKKLIRKDDDSAEMSDQAREGDKFSGAIP 293
Query: 328 -----------------------FCEKVKEKLSSSDDYQTFLKCLNIFNNGIIKKNDLQN 364
F ++VK KL ++D+ Q FL+CLN+++ II + +LQ+
Sbjct: 294 SSSTYDEKGFIIDFLESHSQELAFVDRVKAKLDTADN-QEFLRCLNLYSKEIISQPELQS 352
Query: 365 LVTDLLGKHSDLMDEFKDFLERCENIEGFLAGVMSKKSLST----------------DAH 408
LV+DL+G + DLMD FK FL +C+ +G L+G++SK ST +
Sbjct: 353 LVSDLIGVYPDLMDAFKVFLAQCDKNDGLLSGIVSKSKSSTFYNILLTYLFGQSLWSEGK 412
Query: 409 LSRSSKLEDKDKDQKRE-MDGAKEKDRYKEKY--------MGKSIQELDLSDCKRCTPSY 459
+ +K DKD D++RE ++ +E+DR KE+ K I ELDLS+C++CTPSY
Sbjct: 413 CPQPTKSLDKDTDREREKIERYRERDREKERLEKVAASQKWAKPISELDLSNCEQCTPSY 472
Query: 460 RLLPSD-----------YPIPTASQRSELGAQVLNDHWVSVTSGSEDYSFKHMRKNQYEE 508
R LP + YPIP ASQ+ E+G+QVLNDHWVSVTSGSEDYSFKHMRKNQYEE
Sbjct: 473 RRLPKNLNVHTYFVLLQYPIPIASQKMEIGSQVLNDHWVSVTSGSEDYSFKHMRKNQYEE 532
Query: 509 SLFRCEDDRFELDLLLESVSSASKRAEELYNNINENKISVEALSRIEDHFTVLNLRCIER 568
SLF+CEDDRFELD+LLESV SA+ R EEL IN N++ + IEDH T LNLRCIER
Sbjct: 533 SLFKCEDDRFELDMLLESVISATNRVEELLAKINSNELKTDTPICIEDHLTALNLRCIER 592
Query: 569 LYGDHGLDVIDILRKNPTHALPVILTRLKQKQEEWNRCRSDFNKVWAEIYAKNHYKSLDH 628
LY DHGLDV+D+L+KN ALPVILTRLKQKQEEW RCR++FNKVWA+IY KN+++SLDH
Sbjct: 593 LYSDHGLDVLDLLKKNAYLALPVILTRLKQKQEEWARCRTEFNKVWADIYTKNYHRSLDH 652
Query: 629 RSFYFKQQDSKNLSTKSLVAEIKEIKEKLQKEDHIIQSIAAENRQPLIPHLEFEYSDGGI 688
RSFYFKQQDSKNLSTK+L+AEIKEI EK + ED + ++AA NR+ + ++ F+Y D +
Sbjct: 653 RSFYFKQQDSKNLSTKALLAEIKEISEKKRGEDDALLALAAGNRRTISSNMSFDYPDPDL 712
Query: 689 HEDLYKLVQYSCEEVFSSKELLNKIMRLWSTFLEPMLGVTSQSHGTERVEDRKAGHSSRN 748
HEDLY+L++YSC E+ S+ E L+K+M++W+ FLEP+ GV S+ G E ED
Sbjct: 713 HEDLYQLIKYSCGEMCST-EQLDKVMKVWTEFLEPIFGVPSRPQGAEDRED--------- 762
Query: 749 FAASNVGGDGSPHRDSIS--TNSRLPKSDKNEVDGRVTEVKNIHRTSVAANDKENGSVGG 806
A + D D++S + + S ++ +V E + + S D +
Sbjct: 763 -AVKSTNHDREDQEDAVSPQNGASIANSMRSNGPRKVNESNQVRQASELDKDVTSSKTSD 821
Query: 807 ELVCRDDQLMDKGLKKVECSDKAGFSKQFASDEQGVKNN--PSIAIRGENSLNRTNLDV- 863
L+ D+ DK K + D+ +KQ S E+ +N P + + + ++L V
Sbjct: 822 ALLSCDNTQNDKMPKNLTTPDERAETKQAVSIERAHNSNALPLDGLLPQRNGKISSLSVA 881
Query: 864 ----------SPGCVSAPSRPT-DADDSVAKSQTVNLPLVEGGDIAAPVPVANGVLVENS 912
SPG ++ +P + K VN P VE GD PV + NG +
Sbjct: 882 DEEWYPFLLYSPGLSNSNPKPALTSGTEELKPNYVNGPRVEIGD--NPV-IPNGTVA--- 935
Query: 913 KVKSHEESSGPCKVEKEEGELSPNGDSEED-FVAYRDSNAQSMAKSKHNIERRKYESRDR 971
E +G KVE+EEGELSP GD EED + + +++ ++++KSK N
Sbjct: 936 -----EWFAGEAKVEREEGELSPTGDFEEDNYAVHGENDMEALSKSKEN----------- 979
Query: 972 EEECGPETGGDNDADADDEDSENVSEAGEDVSGSESA-GDECFQEDHEEDDIEHDDVDGK 1030
T D A + S N S G DVSG++S G++C++ EDDI+H+ K
Sbjct: 980 -----DATADDASAPRSSDGSGNTSHNG-DVSGTDSGDGEDCYR----EDDIDHN----K 1025
Query: 1031 AESEGEA-EGMCDAQGG--GDSSSLPLSERFLSSVKPLTKHVSAVSFAEEMKDSR----V 1083
ESEGEA EGM D GD L +S + L VKPL K+V + ++ DSR V
Sbjct: 1026 VESEGEAEEGMSDGHDDTEGDMPVLSISVKNLLHVKPLAKYVPPALYDKDNDDSRKNSQV 1085
Query: 1084 FYGNDDFYALFRLHQILYERILSAKINSMSAEMKWKAKDASSP 1126
FYGND FY LFRLHQILY+RILSAKINS S + KWK + ++P
Sbjct: 1086 FYGNDSFYVLFRLHQILYDRILSAKINSSSPDRKWKTSNPTNP 1128
>At1g59890 hypothetical protein
Length = 1125
Score = 492 bits (1267), Expect = e-139
Identities = 329/835 (39%), Positives = 444/835 (52%), Gaps = 118/835 (14%)
Query: 325 AFSFCEKVKEKLSSSDD-YQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKDF 383
A F ++K + D Y+ FL LN++ N++ VT L H DL+ EF F
Sbjct: 130 AIEFVNRIKARFGGDDRAYKKFLDILNMYRKETKSINEVYQEVTLLFQDHEDLLGEFVHF 189
Query: 384 L---------------------ERCENIEGFLAGVMSKKSLST--DAHLSRSSKLEDKDK 420
L +R G KK + D + S + ED D+
Sbjct: 190 LPDFRGSVSVNDPLFQRNTIPRDRNSTFPGMHPKHFEKKIKRSRHDEYTELSDQREDGDE 249
Query: 421 DQKR-EMDGAKEKDRYKEKYMGKSIQELDLSDCKRCTPSYRLLPSDYPIPTASQRSELGA 479
+ D +K+ +M K+I ELDL+DC +CTPSYR LP DYPI S R+ LG
Sbjct: 250 NLVAYSADIGSQKNLLSTNHMAKAINELDLTDCAQCTPSYRRLPDDYPIQIPSYRNSLGE 309
Query: 480 QVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDLLLESVSSASKRAEELYN 539
+VLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELD+LLESVS+A KR E L
Sbjct: 310 KVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESVSAAIKRVESLLE 369
Query: 540 NINENKISVEALSRIEDHFTVLNLRCIERLYGDHGLDVIDILRKNPTHALPVILTRLKQK 599
IN N IS+E I +H L+ +CIERLYGD+GLDV+D L+KN ALPVILTRLKQK
Sbjct: 370 KINNNTISIETPICIREH---LSGKCIERLYGDYGLDVMDFLKKNSHIALPVILTRLKQK 426
Query: 600 QEEWNRCRSDFNKVWAEIYAKNHYKSLDHRSFYFKQQDSKNLSTKSLVAEIKEIKEKLQK 659
QEEW RCR+DF KVWAE+YAKNH+KSLDHRSFYFKQQDSKNLSTK LVAEIK+I E+ K
Sbjct: 427 QEEWARCRADFRKVWAEVYAKNHHKSLDHRSFYFKQQDSKNLSTKGLVAEIKDISERKHK 486
Query: 660 EDHIIQSIAAENRQPLIPHLEFEYSDGGIHEDLYKLVQYSCEEVFSSKELLNKIMRLWST 719
ED ++++IA + P +EF Y+D +H DLYKL++Y CEE+ ++ E +K+M+LW T
Sbjct: 487 ED-LLRAIAVGTKPSFTPDVEFIYTDTKVHTDLYKLIKYYCEEICAT-EQSDKVMKLWVT 544
Query: 720 FLEPMLGVTSQSHGTERVEDRKAGHSSRNFAASNVGGDGSPHRDSISTNSRLPKSDKNEV 779
FLEPM GV S+S E ++D + + D H D+ +N
Sbjct: 545 FLEPMFGVPSRSETIETMKD-----------VAKI-EDNQEHHDASEA------VKENTC 586
Query: 780 DGRV-TEVKNIHRTSVAANDKENGSVGGELVCRDDQL-MDKGLKKVECSDKAGFSKQFAS 837
DG + + +K + T +KEN + G +D + + +++ + D A + + +
Sbjct: 587 DGSMASNLKPL--TPPKMPNKENPMIQGSSFAQDLPVNTGESIQQDKLHDVAAITNEDSQ 644
Query: 838 DEQGVKNNPSIAIRGENSLNRTNLDVSPGCVSAPSRPTDADDSVAKSQTVNLPLVEGGDI 897
+ V + + G + +R + DVS G + ++
Sbjct: 645 PSKLVSTRNDLIMEGVENRSRVS-DVSMG-----GHKVEREEG----------------- 681
Query: 898 AAPVPVANGVLVENSKVKSHEESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKS 957
E S +S E+ + +V KE G L P ++ ++ D + A
Sbjct: 682 ------------ELSPTESCEQEN--FEVYKENG-LEPVQKLPDNEISNTDREPKEGACG 726
Query: 958 KHNIERRKYESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDH 1017
+ R D + + + ++ D+ S+ + A S+ G E+H
Sbjct: 727 TEAVTRSNALPEDDDNKITQKL-----SEGDENASKFIVSA------SKFGGQVSSDEEH 775
Query: 1018 EEDDIEHDDVDGKAESEGEAEGMCDAQGGGDSSSLPLSERFLSSVKPLTKHVSAVSFAEE 1077
+ + E+ EA GM ++ G D S SER+L VKPL KHV A E
Sbjct: 776 KGAESEN-----------EAGGMVNSNEGEDGSFFTFSERYLQPVKPLAKHVPGTLQASE 824
Query: 1078 ---MKDSRVFYGNDDFYALFRLHQILYERILSAKINSMSAEMKWKAKDASSPDPY 1129
DSRVFYGND Y LFRLHQ+LYERI SAKI+S E KWKA D++S D Y
Sbjct: 825 CDTRNDSRVFYGNDSLYVLFRLHQMLYERIQSAKIHS---ERKWKAPDSTSTDSY 876
Score = 249 bits (635), Expect = 8e-66
Identities = 139/270 (51%), Positives = 173/270 (63%), Gaps = 20/270 (7%)
Query: 1 MKRARDDIYSASASQFKRPFASSRGDSYGQSQVPGGGGGGGGAGGGGEATTSQKLTTNDA 60
MKR R+++Y Q + P SSRG++ G+ GGG GG LTT DA
Sbjct: 1 MKRVREEVYVEP--QMRGPTVSSRGETNGRPSTISGGGTTGG------------LTTVDA 46
Query: 61 LSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFGFNTFL 120
L+YLK VKDMFQD +EKY+ FL VMKDFKAQR DT GVIARVK+LFKG++ L+ GFNTFL
Sbjct: 47 LTYLKAVKDMFQDNKEKYETFLGVMKDFKAQRVDTNGVIARVKDLFKGYDDLLLGFNTFL 106
Query: 121 PKGYEITLDEDEAPAKKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIG 180
PKGY+ITL ++ KK V+F+ AI FVN+IK RF D+ YK FLDILNMYRKE K I
Sbjct: 107 PKGYKITLQPEDEKPKKPVDFQVAIEFVNRIKARFGGDDRAYKKFLDILNMYRKETKSIN 166
Query: 181 EVYSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQHAPFGRNSLQRFNERNSMTPMMRQMQ 240
EVY EV LF+DH DLL EF FLPD + S F RN++ R +RNS P M
Sbjct: 167 EVYQEVTLLFQDHEDLLGEFVHFLPDFRGSVSVNDPLFQRNTIPR--DRNSTFPGMHPKH 224
Query: 241 VDKQRYRRDRLPSHDRDRDLSVEHPEMDDD 270
+K + +R R HD +LS + + D++
Sbjct: 225 FEK-KIKRSR---HDEYTELSDQREDGDEN 250
>At1g10450 unknown protein
Length = 1173
Score = 414 bits (1063), Expect = e-115
Identities = 273/731 (37%), Positives = 375/731 (50%), Gaps = 126/731 (17%)
Query: 322 YGEAFSFCEKVKEKLSSSDD-YQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEF 380
+ +A F K+K + + Y+ FL LN++ +++ VT L H DL+ EF
Sbjct: 166 FKDAIGFVTKIKTRFGDDEHAYKRFLDILNLYRKEKKSISEVYEEVTMLFKGHEDLLMEF 225
Query: 381 KDFLERC-ENIEGFLAGVMSKKSLSTDAHLS-----RSSKLED----------------- 417
+FL C E+ V K +T A S + KLED
Sbjct: 226 VNFLPNCPESAPSTKNAVPRHKGTATTAMHSDKKRKQRCKLEDYSGHSDQREDGDENLVT 285
Query: 418 ----------------------------KDKDQKREMDGAKEKDRYKE--KYMGKSIQEL 447
D+ +K G+++ +K KY+G I EL
Sbjct: 286 CSADSPVGEGQPGYFRDYENREDTETDTADRTEKSAASGSQDIGNHKSTTKYVGTPINEL 345
Query: 448 DLSDCKRCTPSYRLLPSDYPIPTASQRSELGAQVLNDHWVSVTSGSEDYSFKHMRKNQYE 507
DLS+C +CTPSYRLLP DY + S R+ LG + LNDH VSVTSGSEDYSF HMRKNQYE
Sbjct: 346 DLSECTQCTPSYRLLPKDYAVEIPSYRNTLGKKTLNDHLVSVTSGSEDYSFSHMRKNQYE 405
Query: 508 ESLFRCEDDRFELDLLLESVSSASKRAEELYNNINENKISVEALSRIEDHFTVLNLRCIE 567
ESLFRCEDDR+E+D+LL SVSSA K+ E L +N N ISV++ IE H + +NLRCIE
Sbjct: 406 ESLFRCEDDRYEMDMLLGSVSSAIKQVEILLEKMNNNTISVDSTICIEKHLSAMNLRCIE 465
Query: 568 RLYGDHGLDVIDILRKNPTHALPVILTRLKQKQEEWNRCRSDFNKVWAEIYAKNHYKSLD 627
RLYGD+GLDV+D+L+KN ALPVILTRLKQKQEEW RC SDF KVWAE+YAKNH+KSLD
Sbjct: 466 RLYGDNGLDVMDLLKKNMHSALPVILTRLKQKQEEWARCHSDFQKVWAEVYAKNHHKSLD 525
Query: 628 HRSFYFKQQDSKNLSTKSLVAEIKEIKEKLQKEDHIIQSIAAENRQPLIPHLEFEYSDGG 687
HRSFYFKQQDSKNLSTK LVAE+K+I EK +ED ++Q+IA P LEF Y D
Sbjct: 526 HRSFYFKQQDSKNLSTKCLVAEVKDISEKKHQED-LLQAIAVRVMPLFTPDLEFNYCDTQ 584
Query: 688 IHEDLYKLVQYSCEEVFSSKELLNKIMRLWSTFLEPMLGVTSQSHGTERVED-------- 739
IHEDLY L++Y CEE+ ++ E +K+M+LW TFLEP+ G+ S+S +ED
Sbjct: 585 IHEDLYLLIKYYCEEICAT-EQSDKVMKLWITFLEPIFGILSRSQDNLALEDVSKLKNNR 643
Query: 740 -----------------RKAGHSSRNFAASNVGGDGSPHRDSISTNSRLPKSDKNEVDGR 782
RK S + + N GS R+ +S N ++ + +++
Sbjct: 644 ELQDACLAVKETASGSNRKHPISPKRLSKDNTKMQGSSSREDVSANIKVKTAQPDKLQDD 703
Query: 783 VTEVKNIHRTSVAANDKENGSVGGELVCRDDQLM-DKGLKKVECSD------KAGFSKQF 835
+ ++S + K +DQ+M D+G V + + G
Sbjct: 704 AAMTNEVIQSSKFVSPK------------NDQIMEDEGNHMVNAASVEKHELEEGELSPT 751
Query: 836 ASDEQGVKNNPSIAIRGENSLN---------RTNLDVSPGCVSAPSRPTDADDSVAKSQT 886
AS EQ + + G+N+ R+N D T A+D K +
Sbjct: 752 ASREQS-----NFEVNGQNAFKPLQKVTDNVRSNKDKQSCDKKGAKNKTRAEDD--KQEN 804
Query: 887 VNLPLVEGGDIAAPVPVANGVLVENSKVKSHEESSGPCKVE---KEEGELSPNGDSEEDF 943
+ ++ A+ +LV +KV HEE++ GE++ E+
Sbjct: 805 CH-------KLSENNKTASEMLVSGTKVSCHEENNRVMNCNGRGSVAGEMANGNQGEDGS 857
Query: 944 VAYRDSNAQSM 954
A+ + Q++
Sbjct: 858 FAFSERFLQTV 868
Score = 211 bits (536), Expect = 2e-54
Identities = 134/327 (40%), Positives = 181/327 (54%), Gaps = 53/327 (16%)
Query: 1 MKRARDDIYSASASQFKRPFASSRGDSYGQSQVPGGGGGGGGAGGGGEATTSQKLTTNDA 60
MKRAR+D+++ + Q ++P SSRG+ T++ T DA
Sbjct: 52 MKRAREDVHTDT--QKRKPEVSSRGE------------------------TNKLPRTIDA 85
Query: 61 LSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFGFNTFL 120
L+YLK VKD+F D +EKY+ FLE+MK+FKAQ DT GVI R+K LFKG+ L+ GFNTFL
Sbjct: 86 LTYLKAVKDIFHDNKEKYESFLELMKEFKAQTIDTNGVIERIKVLFKGYRDLLLGFNTFL 145
Query: 121 PKGYEITLDEDEAPAKKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIG 180
PKGY+ITL +E K V+F++AI FV KIK RF DEH YK FLDILN+YRKE K I
Sbjct: 146 PKGYKITLLPEEEKPKIRVDFKDAIGFVTKIKTRFGDDEHAYKRFLDILNLYRKEKKSIS 205
Query: 181 EVYSEVATLFKDHRDLLEEFTRFLPD-TSAAPSTQHAPFGRNSLQRFNERNSMTPMMRQM 239
EVY EV LFK H DLL EF FLP+ +APST +N++ R + + T M
Sbjct: 206 EVYEEVTMLFKGHEDLLMEFVNFLPNCPESAPST------KNAVPR--HKGTATTAMHSD 257
Query: 240 QVDKQRYRRDRLPSHDRDRDLSVEHPEMDDDKTMINLHKEQRKRDRR---IRDQDERDPD 296
+ KQR + + H R+ D D+ ++ + + + RD + R+
Sbjct: 258 KKRKQRCKLEDYSGHSDQRE--------DGDENLVTCSADSPVGEGQPGYFRDYENREDT 309
Query: 297 LDNSRDLT-------SQRFRDKKKTVK 316
++ D T SQ + K T K
Sbjct: 310 ETDTADRTEKSAASGSQDIGNHKSTTK 336
Score = 108 bits (271), Expect = 1e-23
Identities = 79/211 (37%), Positives = 108/211 (50%), Gaps = 21/211 (9%)
Query: 925 KVEKEEGELSPNGDSEEDFVAYRDSNA-QSMAKSKHNIERRKYESRDREEECGPETGGDN 983
K E EEGELSP E+ NA + + K N+ K +++ + G N
Sbjct: 740 KHELEEGELSPTASREQSNFEVNGQNAFKPLQKVTDNVRSNK------DKQSCDKKGAKN 793
Query: 984 DADADDEDSENVSEAGED-VSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCD 1042
A+D+ EN + E+ + SE HEE++ + +G+ GE
Sbjct: 794 KTRAEDDKQENCHKLSENNKTASEMLVSGTKVSCHEENN-RVMNCNGRGSVAGEM----- 847
Query: 1043 AQGG-GDSSSLPLSERFLSSVKPLTKHVSAVSFAEE---MKDSRVFYGNDDFYALFRLHQ 1098
A G G+ S SERFL +VKP+ KH+S A E DS+VFYGND +Y LFRLHQ
Sbjct: 848 ANGNQGEDGSFAFSERFLQTVKPVAKHLSWPLQASETCSQNDSQVFYGNDSYYVLFRLHQ 907
Query: 1099 ILYERILSAKINSMSAEMKWKAKDASSPDPY 1129
+LYERI +AK +S E KWKA D ++PD Y
Sbjct: 908 MLYERIQTAKKHS---EKKWKAADNTTPDSY 935
>At1g24200 hypothetical protein
Length = 196
Score = 186 bits (472), Expect = 6e-47
Identities = 103/184 (55%), Positives = 121/184 (64%), Gaps = 8/184 (4%)
Query: 44 GGGGEATTSQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVK 103
G GG + K TTNDAL YL+ VK FQ QREKYD FL++M D+K QR D +GVI R+K
Sbjct: 3 GEGG----AHKPTTNDALKYLRAVKAKFQGQREKYDEFLQIMIDYKTQRIDISGVIIRMK 58
Query: 104 ELFKGHNHLIFGFNTFLPKGYEITLDEDEAPAKKTVEFEEAISFVNKIKKRFQSDEHVYK 163
EL K L+ GFN FLP GY IT E + KK VE EAISF+NKIK RFQ D+ VYK
Sbjct: 59 ELLKEQQGLLLGFNAFLPNGYMITHHEQPSQ-KKPVELGEAISFINKIKTRFQGDDRVYK 117
Query: 164 SFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQHAPFGRNSL 223
S LDILNMYRK+ K I VY EVA LF DH +LL EFT FLP A P + + N +
Sbjct: 118 SVLDILNMYRKDRKPITAVYREVAILFLDHNNLLVEFTHFLP---ATPDKGYRNYSTNKV 174
Query: 224 QRFN 227
+F+
Sbjct: 175 TKFS 178
Score = 32.7 bits (73), Expect = 1.2
Identities = 26/89 (29%), Positives = 39/89 (43%), Gaps = 6/89 (6%)
Query: 297 LDNSRDLTSQRFRDKKKTVKKAEGMYGEAFSFCEKVKEKLSSSDD-YQTFLKCLNIFNNG 355
L N +T +KK V+ GEA SF K+K + D Y++ L LN++
Sbjct: 75 LPNGYMITHHEQPSQKKPVE-----LGEAISFINKIKTRFQGDDRVYKSVLDILNMYRKD 129
Query: 356 IIKKNDLQNLVTDLLGKHSDLMDEFKDFL 384
+ V L H++L+ EF FL
Sbjct: 130 RKPITAVYREVAILFLDHNNLLVEFTHFL 158
>At1g70030 unknown protein
Length = 160
Score = 174 bits (441), Expect = 3e-43
Identities = 87/155 (56%), Positives = 113/155 (72%), Gaps = 2/155 (1%)
Query: 52 SQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNH 111
++KLTT DAL +L VK +QD RE YD FL +MKDF+ QR T VI++VKELFKG
Sbjct: 4 NKKLTTTDALDFLHLVKTKYQDNREIYDSFLTIMKDFRGQRAKTCDVISKVKELFKGQPE 63
Query: 112 LIFGFNTFLPKGYEITLDEDEAPA-KKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILN 170
L+ GFNTFLP G+EITL +DE + K F+EA FVNK+K RFQ+++ V+ SFL++L
Sbjct: 64 LLLGFNTFLPTGFEITLSDDELTSNSKFAHFDEAYEFVNKVKTRFQNND-VFNSFLEVLK 122
Query: 171 MYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFLP 205
++KE+K + E+Y EVA LF+ HRDLLEEF FLP
Sbjct: 123 THKKENKSVAELYQEVAILFQGHRDLLEEFHLFLP 157
Score = 48.9 bits (115), Expect = 2e-05
Identities = 24/71 (33%), Positives = 40/71 (55%)
Query: 135 AKKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHR 194
A K + +A+ F++ +K ++Q + +Y SFL I+ +R + +V S+V LFK
Sbjct: 3 ANKKLTTTDALDFLHLVKTKYQDNREIYDSFLTIMKDFRGQRAKTCDVISKVKELFKGQP 62
Query: 195 DLLEEFTRFLP 205
+LL F FLP
Sbjct: 63 ELLLGFNTFLP 73
Score = 37.0 bits (84), Expect = 0.064
Identities = 22/75 (29%), Positives = 34/75 (45%)
Query: 310 DKKKTVKKAEGMYGEAFSFCEKVKEKLSSSDDYQTFLKCLNIFNNGIIKKNDLQNLVTDL 369
D + T + EA+ F KVK + ++D + +FL+ L +L V L
Sbjct: 82 DDELTSNSKFAHFDEAYEFVNKVKTRFQNNDVFNSFLEVLKTHKKENKSVAELYQEVAIL 141
Query: 370 LGKHSDLMDEFKDFL 384
H DL++EF FL
Sbjct: 142 FQGHRDLLEEFHLFL 156
>At5g15030 putative protein
Length = 271
Score = 138 bits (348), Expect = 2e-32
Identities = 72/132 (54%), Positives = 87/132 (65%), Gaps = 12/132 (9%)
Query: 1 MKRARDDIYSASASQFKRPFASSRGDSYGQSQVPGGGGGGGGAGGGGEATTSQKLTTNDA 60
MKR D S SQ + P S R + GQS PG G T LTT+DA
Sbjct: 150 MKRIGDKEIDGSGSQNQHPVGSPRNELQGQSPKPGNGN------------TKDALTTDDA 197
Query: 61 LSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFGFNTFL 120
L+YLK++KD+F DQ+ KY LFLE+M DFKAQRTDT+ VIARVK+L KGHNHLI FN FL
Sbjct: 198 LAYLKEIKDVFHDQKYKYHLFLEIMSDFKAQRTDTSVVIARVKDLLKGHNHLILVFNKFL 257
Query: 121 PKGYEITLDEDE 132
P G+EITLD+++
Sbjct: 258 PHGFEITLDDED 269
Score = 69.3 bits (168), Expect = 1e-11
Identities = 56/199 (28%), Positives = 93/199 (46%), Gaps = 26/199 (13%)
Query: 33 VPGGGGGGGGAGGGGEATTSQKLTTNDALSYLKQVKDMF--QDQREKYDLFLEVMKDFKA 90
VP GG S +T +DA +YL+QVK+ F D+R+KY +F +V+ DFKA
Sbjct: 60 VPRSPRHGGNIERSIYLDPSLGMTISDARAYLQQVKNTFIDHDERDKYAMFRKVLFDFKA 119
Query: 91 Q-----RTDTAGVIARVKEL------FKGHNHLI-------FGFNTFLPKGYEITLDEDE 132
Q R+D+A + +K+L F I G P G + +
Sbjct: 120 QRIVADRSDSARFCSHLKDLSDLFQIFSERMKRIGDKEIDGSGSQNQHPVGSPRNELQGQ 179
Query: 133 AP------AKKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEV 186
+P K + ++A++++ +IK F ++ Y FL+I++ ++ + D V + V
Sbjct: 180 SPKPGNGNTKDALTTDDALAYLKEIKDVFHDQKYKYHLFLEIMSDFKAQRTDTSVVIARV 239
Query: 187 ATLFKDHRDLLEEFTRFLP 205
L K H L+ F +FLP
Sbjct: 240 KDLLKGHNHLILVFNKFLP 258
>At1g24230 hypothetical protein
Length = 245
Score = 115 bits (287), Expect = 2e-25
Identities = 61/165 (36%), Positives = 97/165 (57%), Gaps = 5/165 (3%)
Query: 54 KLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLI 113
K T +D SY+ +K+ F+D+ KY FLE++ D+ A+R D +AR+ EL K H +L+
Sbjct: 81 KETFHDVRSYIYSLKESFRDEPAKYAQFLEILNDYSARRVDAPSAVARMTELMKDHRNLV 140
Query: 114 FGFNTFLPKG-YEITLDEDEAPAKKTVEFEEAISFVNKIKKRFQ-SDEHVYKSFLDILNM 171
GF+ L G + T E E K + +F++K+K RFQ +D HVY+SFL+IL M
Sbjct: 141 LGFSVLLSTGDTKTTPLEAEPDNNKRI---RVANFISKLKARFQGNDGHVYESFLEILTM 197
Query: 172 YRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQHA 216
Y++ +K + ++Y EV L + H DL+ EF+ T+ ++ A
Sbjct: 198 YQQGNKSVNDLYQEVVALLQGHEDLVMEFSNVFKRTTGPSGSKSA 242
Score = 94.4 bits (233), Expect = 3e-19
Identities = 59/163 (36%), Positives = 85/163 (51%), Gaps = 16/163 (9%)
Query: 52 SQKLTTNDALSYLKQVKDMF-QDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHN 110
S T ++A SY+ VK+ F DQ KY FL++M D +A R D A V+ R++EL K H
Sbjct: 7 SPAFTIDEATSYINAVKEAFGADQPAKYREFLDIMLDLRANRVDLATVVPRMRELLKDHV 66
Query: 111 HLIFGFNTFLPKGYEITLDEDEAPAKKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILN 170
+L+ FN FLP A AK+T F + S++ +K+ F+ + Y FL+ILN
Sbjct: 67 NLLLRFNAFLP-----------AEAKET--FHDVRSYIYSLKESFRDEPAKYAQFLEILN 113
Query: 171 MYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFLP--DTSAAP 211
Y D + + L KDHR+L+ F+ L DT P
Sbjct: 114 DYSARRVDAPSAVARMTELMKDHRNLVLGFSVLLSTGDTKTTP 156
Score = 62.4 bits (150), Expect = 1e-09
Identities = 59/254 (23%), Positives = 103/254 (40%), Gaps = 31/254 (12%)
Query: 142 EEAISFVNKIKKRFQSDEHV-YKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEF 200
+EA S++N +K+ F +D+ Y+ FLDI+ R D+ V + L KDH +LL F
Sbjct: 13 DEATSYINAVKEAFGADQPAKYREFLDIMLDLRANRVDLATVVPRMRELLKDHVNLLLRF 72
Query: 201 TRFLPDTSAAPSTQHAPFGRNSLQRFNERNSMTPMMRQMQVDKQRYRRDRLPSHDRDRDL 260
FLP + + + + F + + ++ D R D + R +L
Sbjct: 73 NAFLPAEAKETFHDVRSYIYSLKESFRDEPAKYAQFLEILNDYSARRVDAPSAVARMTEL 132
Query: 261 SVEHPEMDDDKTMINLHKEQRKRDRRIRDQDERDPDLDNSRDLTSQRFRDKKKTVKKAEG 320
+H + +++ + D + L+ D K ++ A
Sbjct: 133 MKDHRNLVLGFSVL------------LSTGDTKTTPLEAEPD--------NNKRIRVA-- 170
Query: 321 MYGEAFSFCEKVKEKLSSSDD--YQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMD 378
+F K+K + +D Y++FL+ L ++ G NDL V LL H DL+
Sbjct: 171 ------NFISKLKARFQGNDGHVYESFLEILTMYQQGNKSVNDLYQEVVALLQGHEDLVM 224
Query: 379 EFKDFLERCENIEG 392
EF + +R G
Sbjct: 225 EFSNVFKRTTGPSG 238
>At5g35610 putative protein
Length = 155
Score = 112 bits (280), Expect = 1e-24
Identities = 64/149 (42%), Positives = 93/149 (61%), Gaps = 17/149 (11%)
Query: 51 TSQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHN 110
++ K+T ++ YLK VK+ Q++RE Y FL+VM + AQR D +GV + VKELFK
Sbjct: 2 SASKITKSNPRKYLKIVKNKLQNKREIYVRFLQVMTAYSAQRIDPSGVKSVVKELFKEDQ 61
Query: 111 HLIFGFNTFLPKGYE-----------ITLDEDEAPAKK--TVEFEEAISFVNKIKKRFQS 157
I GFNTFLPKG+E I L+ ++ P KK +E+ EA+ FV K+K
Sbjct: 62 EPISGFNTFLPKGFEIKPECDQNGFKIKLECEQTPPKKYVDIEYSEALDFVRKVK----D 117
Query: 158 DEHVYKSFLDILNMYRKEHKDIGEVYSEV 186
D+ +YKSF+ I++MY+K++K + EV EV
Sbjct: 118 DDRIYKSFVTIMDMYKKKNKSLDEVCREV 146
Score = 37.7 bits (86), Expect = 0.037
Identities = 17/59 (28%), Positives = 30/59 (50%)
Query: 147 FVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFLP 205
++ +K + Q+ +Y FL ++ Y + D V S V LFK+ ++ + F FLP
Sbjct: 14 YLKIVKNKLQNKREIYVRFLQVMTAYSAQRIDPSGVKSVVKELFKEDQEPISGFNTFLP 72
>At4g12020 putative disease resistance protein
Length = 1895
Score = 103 bits (257), Expect = 6e-22
Identities = 59/119 (49%), Positives = 77/119 (64%), Gaps = 21/119 (17%)
Query: 39 GGGGAGGGGEATTSQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGV 98
GGG GG +TT DAL+YLK VKD F+D EKYD FLEV+ D K Q DT+GV
Sbjct: 297 GGGNMGG---------VTTGDALNYLKAVKDKFEDS-EKYDTFLEVLNDCKHQGVDTSGV 346
Query: 99 IARVKELFKGHNHLIFGFNTFLPKGYEIT-LDEDEAPAKKTVEFEEAISFVNKIKKRFQ 156
IAR+K+LFKGH+ L+ GFNT+L K Y+IT L ED+ P I F++K++ ++
Sbjct: 347 IARLKDLFKGHDDLLLGFNTYLSKEYQITILPEDDFP----------IDFLDKVEGPYE 395
Score = 41.2 bits (95), Expect = 0.003
Identities = 30/126 (23%), Positives = 54/126 (42%), Gaps = 2/126 (1%)
Query: 143 EAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTR 202
+A++++ +K +F+ E Y +FL++LN + + D V + + LFK H DLL F
Sbjct: 308 DALNYLKAVKDKFEDSEK-YDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLLGFNT 366
Query: 203 FLPDTSAAPSTQHAPFGRNSLQRFNERNSMTPMMRQ-MQVDKQRYRRDRLPSHDRDRDLS 261
+L F + L + MT Q +Q + + PS + S
Sbjct: 367 YLSKEYQITILPEDDFPIDFLDKVEGPYEMTYQQAQTVQANANMQPQTEYPSSSAVQSFS 426
Query: 262 VEHPEM 267
P++
Sbjct: 427 SGQPQI 432
Score = 38.1 bits (87), Expect = 0.029
Identities = 19/64 (29%), Positives = 34/64 (52%)
Query: 323 GEAFSFCEKVKEKLSSSDDYQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKD 382
G+A ++ + VK+K S+ Y TFL+ LN + + + + + DL H DL+ F
Sbjct: 307 GDALNYLKAVKDKFEDSEKYDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLLGFNT 366
Query: 383 FLER 386
+L +
Sbjct: 367 YLSK 370
>At1g27280 hypothetical protein
Length = 225
Score = 100 bits (250), Expect = 4e-21
Identities = 54/143 (37%), Positives = 85/143 (58%), Gaps = 3/143 (2%)
Query: 50 TTSQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGH 109
+ S T +DA+SY+ VK+ F D+ KY F ++ D + + D AG I RV+EL K H
Sbjct: 72 SVSPDSTIDDAVSYINTVKEAFHDEPAKYYEFFQLFYDIRYRLIDVAGGITRVEELLKAH 131
Query: 110 NHLIFGFNTFLPKGYE--ITLDEDEAPAKKTVEFEEAISFVNKIKKRFQSDE-HVYKSFL 166
+L+ N FLP + + L ++ A + + SF+ K+K+RFQ D+ HVY+SFL
Sbjct: 132 KNLLVRLNAFLPPEAQRILHLKIEQRAASDINKRKRVASFIGKLKERFQGDDRHVYESFL 191
Query: 167 DILNMYRKEHKDIGEVYSEVATL 189
+IL MY++ +K + ++Y EV L
Sbjct: 192 EILTMYQEGNKSVNDLYQEVGFL 214
Score = 66.6 bits (161), Expect = 8e-11
Identities = 49/217 (22%), Positives = 101/217 (45%), Gaps = 11/217 (5%)
Query: 67 VKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFGFNTFLPKGYEI 126
+++++ D+ EK++ L +++D+ R D A + A + EL + H +L+ F F P
Sbjct: 1 MEEVYHDEPEKFNHILHLIRDYFNHRDDRARITACMGELMRDHLNLLVRFFDFFPAEASE 60
Query: 127 TLDEDEAPAKKTVE----FEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEV 182
L + + A +V ++A+S++N +K+ F + Y F + R D+
Sbjct: 61 GLAQLQTIAATSVSPDSTIDDAVSYINTVKEAFHDEPAKYYEFFQLFYDIRYRLIDVAGG 120
Query: 183 YSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQHAPFGRNSLQRFNERNSMTPMMRQMQVD 242
+ V L K H++LL FLP A H + + N+R + + ++
Sbjct: 121 ITRVEELLKAHKNLLVRLNAFLP--PEAQRILHLKIEQRAASDINKRKRVASFIGKL--- 175
Query: 243 KQRYRRDRLPSHDRDRDLSVEHPEMDDDKTMINLHKE 279
K+R++ D H + L + + +K++ +L++E
Sbjct: 176 KERFQGD--DRHVYESFLEILTMYQEGNKSVNDLYQE 210
>At1g24250 hypothetical protein
Length = 252
Score = 100 bits (248), Expect = 6e-21
Identities = 62/161 (38%), Positives = 83/161 (51%), Gaps = 22/161 (13%)
Query: 47 GEATTSQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELF 106
G + + T +DA SYL VK+ F D+ KY L+++KDFKA+R D A VIARV+EL
Sbjct: 108 GRSLPRPEPTIDDATSYLIAVKEAFHDEPAKYGEMLKLLKDFKARRVDAACVIARVEELM 167
Query: 107 KGHNHLIFGFNTFLPKGYEITLDEDEAPAKKTVEFEEAISFVNKIKKRFQSD-EHVYKSF 165
K H +L+FGF FL SF K+K RFQ D V S
Sbjct: 168 KDHLNLLFGFCVFL---------------------SATTSFTTKLKARFQGDGSQVVDSV 206
Query: 166 LDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFLPD 206
L I+ MY + +K + Y EV L + H DL+ E ++ L D
Sbjct: 207 LQIMRMYGEGNKSKHDAYQEVVALVQGHDDLVMELSQILTD 247
Score = 60.5 bits (145), Expect = 5e-09
Identities = 46/195 (23%), Positives = 81/195 (40%), Gaps = 24/195 (12%)
Query: 46 GGEATTSQKLTTNDALSYLKQVKDMFQDQRE-KYDLFLEVMKDFKAQRTDTAGVIARVKE 104
GG + + L D +Y+ V+ Q+ ++ +F+ + + F A R AR+++
Sbjct: 3 GGSLSPASSL--EDVKAYVNAVEVALQEMEPARFGMFVRLFRGFTAPRIGMPTFSARMQD 60
Query: 105 LFKGHNHLIFGFNTFLPKGYEITLDEDEAPAKKTV---------------------EFEE 143
L K H L G N LP Y++T+ + + V ++
Sbjct: 61 LLKDHPSLCLGLNVLLPPEYQLTIPPEASEEFHKVVGRSVPVPPKVVGRSLPRPEPTIDD 120
Query: 144 AISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRF 203
A S++ +K+ F + Y L +L ++ D V + V L KDH +LL F F
Sbjct: 121 ATSYLIAVKEAFHDEPAKYGEMLKLLKDFKARRVDAACVIARVEELMKDHLNLLFGFCVF 180
Query: 204 LPDTSAAPSTQHAPF 218
L T++ + A F
Sbjct: 181 LSATTSFTTKLKARF 195
>At1g23810 hypothetical protein
Length = 241
Score = 98.6 bits (244), Expect = 2e-20
Identities = 58/163 (35%), Positives = 84/163 (50%), Gaps = 22/163 (13%)
Query: 47 GEATTSQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELF 106
G + + T +DA SYL VK+ F D+ KY+ L+++ DFKA+R + A VIARV+EL
Sbjct: 97 GRSVPRPEPTIDDATSYLIAVKEAFHDEPAKYEEMLKLLNDFKARRVNAASVIARVEELM 156
Query: 107 KGHNHLIFGFNTFLPKGYEITLDEDEAPAKKTVEFEEAISFVNKIKKRFQSD-EHVYKSF 165
K H++L+FGF FL SF K+K +FQ D V S
Sbjct: 157 KDHSNLLFGFCVFL---------------------SATTSFTTKLKAKFQGDGSQVVDSV 195
Query: 166 LDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFLPDTS 208
L I+ MY + +K + Y E+ L + H DL+ E ++ D S
Sbjct: 196 LQIMRMYGEGNKSKHDAYQEIVALVQGHDDLVMELSQIFTDPS 238
Score = 63.5 bits (153), Expect = 6e-10
Identities = 47/184 (25%), Positives = 81/184 (43%), Gaps = 13/184 (7%)
Query: 46 GGEATTSQKLTTNDALSYLKQVKDMFQD-QREKYDLFLEVMKDFKAQRTDTAGVIARVKE 104
GG + + L D +Y+ VK ++ + KY FL + + A+R A AR+++
Sbjct: 3 GGSKSPASSL--EDGKAYVNAVKVALEEAEPAKYQEFLRLFHEVIARRMGMATFSARMQD 60
Query: 105 LFKGHNHLIFGFNTFLPKGYEITLDEDEAPAKKTV----------EFEEAISFVNKIKKR 154
L K H L G N L Y+ + + + V ++A S++ +K+
Sbjct: 61 LLKDHPSLCLGLNVMLAPEYQRAIPPEASEEFHKVVGRSVPRPEPTIDDATSYLIAVKEA 120
Query: 155 FQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQ 214
F + Y+ L +LN ++ + V + V L KDH +LL F FL T++ +
Sbjct: 121 FHDEPAKYEEMLKLLNDFKARRVNAASVIARVEELMKDHSNLLFGFCVFLSATTSFTTKL 180
Query: 215 HAPF 218
A F
Sbjct: 181 KAKF 184
>At1g27260 hypothetical protein
Length = 222
Score = 92.8 bits (229), Expect = 1e-18
Identities = 54/150 (36%), Positives = 75/150 (50%), Gaps = 22/150 (14%)
Query: 56 TTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFG 115
T DA SYL VK F D+ KY+ L+++ D +A+R D A IA V+EL K H L+ G
Sbjct: 90 TIEDATSYLNSVKRAFHDEPAKYEELLKLLNDIEARRVDAASFIASVEELMKDHQTLLNG 149
Query: 116 FNTFLPKGYEITLDEDEAPAKKTVEFEEAISFVNKIKKRFQSD-EHVYKSFLDILNMYRK 174
F+ FL + F+ K+K +FQ D HV S L IL MY +
Sbjct: 150 FSVFL---------------------SAEMKFIRKLKAKFQGDGSHVADSVLQILRMYSE 188
Query: 175 EHKDIGEVYSEVATLFKDHRDLLEEFTRFL 204
+K E + EV L +DH DL+ E + +
Sbjct: 189 GNKSKSEAFQEVVPLVQDHEDLVMELIKII 218
Score = 87.8 bits (216), Expect = 3e-17
Identities = 50/155 (32%), Positives = 78/155 (50%), Gaps = 10/155 (6%)
Query: 58 NDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFGFN 117
N A +Y+ V+D F+D+ KY FL +++D +A+R D A + EL K H L+ GFN
Sbjct: 2 NKAFAYIIAVRDRFRDEPAKYRQFLSLLRDRRARRIDKATFFVGLVELIKDHLDLLLGFN 61
Query: 118 TFLPKGYEITLDE--------DEAPAKKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDIL 169
LP ++I + P + T+ E+A S++N +K+ F + Y+ L +L
Sbjct: 62 ALLPARFQIPITPAGFQNVVGRSVPPETTI--EDATSYLNSVKRAFHDEPAKYEELLKLL 119
Query: 170 NMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFL 204
N D + V L KDH+ LL F+ FL
Sbjct: 120 NDIEARRVDAASFIASVEELMKDHQTLLNGFSVFL 154
Score = 39.7 bits (91), Expect = 0.010
Identities = 21/71 (29%), Positives = 33/71 (45%)
Query: 143 EAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTR 202
+A +++ ++ RF+ + Y+ FL +L R D + + L KDH DLL F
Sbjct: 3 KAFAYIIAVRDRFRDEPAKYRQFLSLLRDRRARRIDKATFFVGLVELIKDHLDLLLGFNA 62
Query: 203 FLPDTSAAPST 213
LP P T
Sbjct: 63 LLPARFQIPIT 73
>At1g24210 unknown protein
Length = 155
Score = 92.0 bits (227), Expect = 2e-18
Identities = 56/153 (36%), Positives = 80/153 (51%), Gaps = 20/153 (13%)
Query: 52 SQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNH 111
S LT +DA SY+ VK+ F D+ KY F++++ R D VIARV+EL K H
Sbjct: 7 SPALTKDDAHSYIIAVKETFHDEPTKYQEFIKLLNGVCDHRVDKYSVIARVEELMKDHQD 66
Query: 112 LIFGFNTFLPKGYEITLDEDEAPAKKTVEFEEAISFVNKIKKRFQS-DEHVYKSFLDILN 170
L+ GF+ FLP V E+ F+NK+K RFQS D HV + ++
Sbjct: 67 LLLGFSVFLP----------------PVSVED---FINKLKTRFQSLDTHVVGAIRGLMK 107
Query: 171 MYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRF 203
M+++ + EV EV + H DL+E+F RF
Sbjct: 108 MFKEGKMSVKEVQEEVIDVLFYHEDLIEDFLRF 140
Score = 43.9 bits (102), Expect = 5e-04
Identities = 23/67 (34%), Positives = 36/67 (53%)
Query: 142 EEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFT 201
++A S++ +K+ F + Y+ F+ +LN D V + V L KDH+DLL F+
Sbjct: 13 DDAHSYIIAVKETFHDEPTKYQEFIKLLNGVCDHRVDKYSVIARVEELMKDHQDLLLGFS 72
Query: 202 RFLPDTS 208
FLP S
Sbjct: 73 VFLPPVS 79
>At1g27240 hypothetical protein
Length = 186
Score = 86.3 bits (212), Expect = 9e-17
Identities = 61/170 (35%), Positives = 86/170 (49%), Gaps = 23/170 (13%)
Query: 56 TTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFG 115
T +DA SY+ VK+ F D+ KY+ F+++M D + D A IA++ EL KGH L+ G
Sbjct: 11 TLSDAHSYITAVKEAFHDEPTKYEEFIKLMNDIRDHGVDKASGIAKLTELIKGHPRLLRG 70
Query: 116 FNTFLP----------KGYEITLDEDEAP-------AKKTVEFEEAI-----SFVNKIKK 153
+ F P K I D+ P AK T + I +F+N +K
Sbjct: 71 LSFFFPQVNRDIHHEAKRTIILKDKATIPPEAAYRGAKSTYTKIKQIEPDWENFMNMLKT 130
Query: 154 RFQS-DEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTR 202
RF+S D HV +SFL I+ MY + K EV EV L H DL+++F R
Sbjct: 131 RFRSLDTHVVESFLKIMIMYDEGKKSEKEVQEEVVDLLYYHEDLIDKFFR 180
Score = 39.3 bits (90), Expect = 0.013
Identities = 33/125 (26%), Positives = 60/125 (47%), Gaps = 17/125 (13%)
Query: 258 RDLSVEHPEMDDDKTMINLHKEQRKRDRRIRDQDERDPDLDNSRDLTSQRFRDKKKTVKK 317
R LS P+++ D +H E KR ++D+ P+ +R K T K
Sbjct: 69 RGLSFFFPQVNRD-----IHHEA-KRTIILKDKATIPPEA---------AYRGAKSTYTK 113
Query: 318 AEGMYGEAFSFCEKVKEKLSSSDDY--QTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSD 375
+ + + +F +K + S D + ++FLK + +++ G + ++Q V DLL H D
Sbjct: 114 IKQIEPDWENFMNMLKTRFRSLDTHVVESFLKIMIMYDEGKKSEKEVQEEVVDLLYYHED 173
Query: 376 LMDEF 380
L+D+F
Sbjct: 174 LIDKF 178
>At5g15040 putative protein
Length = 87
Score = 85.1 bits (209), Expect = 2e-16
Identities = 39/70 (55%), Positives = 49/70 (69%)
Query: 62 SYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFGFNTFLP 121
+Y +VKD F DQ EKYD+F ++ D KA+R A++KELFK HN LI GFNTFLP
Sbjct: 6 AYFMEVKDTFHDQIEKYDMFKNILLDLKARRIGRHTAFAQLKELFKEHNELIIGFNTFLP 65
Query: 122 KGYEITLDED 131
GY+I LD+D
Sbjct: 66 TGYKIALDDD 75
Score = 30.8 bits (68), Expect = 4.6
Identities = 16/63 (25%), Positives = 29/63 (45%)
Query: 143 EAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTR 202
E ++ ++K F Y F +IL + ++++ LFK+H +L+ F
Sbjct: 3 ECRAYFMEVKDTFHDQIEKYDMFKNILLDLKARRIGRHTAFAQLKELFKEHNELIIGFNT 62
Query: 203 FLP 205
FLP
Sbjct: 63 FLP 65
>At1g27270 hypothetical protein
Length = 241
Score = 84.3 bits (207), Expect = 3e-16
Identities = 54/152 (35%), Positives = 76/152 (49%), Gaps = 22/152 (14%)
Query: 56 TTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFG 115
T +DA SYL VK+ F D+ KY +++ D KA+R + A VIAR++EL K H +L+ G
Sbjct: 110 TMDDATSYLNAVKEAFHDEPAKYMEITKLLTDLKARRINAASVIARMEELLKDHLNLLLG 169
Query: 116 FNTFLPKGYEITLDEDEAPAKKTVEFEEAISFVNKIKKRFQSD-EHVYKSFLDILNMYRK 174
F FL +P ++ F+ K+K RF D V S L IL M+ +
Sbjct: 170 FCVFL------------SPTRR---------FITKLKARFLGDGSQVVDSVLQILRMHSE 208
Query: 175 EHKDIGEVYSEVATLFKDHRDLLEEFTRFLPD 206
+K E EV L + H DLL E + D
Sbjct: 209 GNKSKDEASQEVRALIQGHEDLLMELSEIFSD 240
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,967,593
Number of Sequences: 26719
Number of extensions: 1307191
Number of successful extensions: 15064
Number of sequences better than 10.0: 483
Number of HSP's better than 10.0 without gapping: 185
Number of HSP's successfully gapped in prelim test: 313
Number of HSP's that attempted gapping in prelim test: 8633
Number of HSP's gapped (non-prelim): 2992
length of query: 1129
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1019
effective length of database: 8,379,506
effective search space: 8538716614
effective search space used: 8538716614
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0081b.6


