

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0081b.1
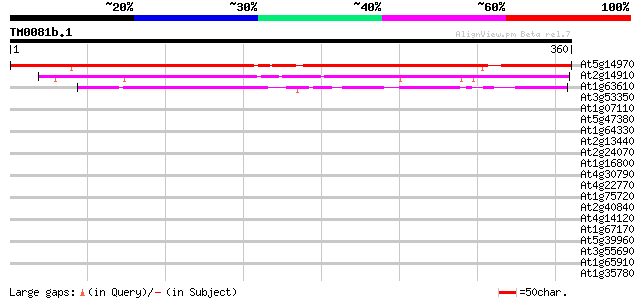
(360 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g14970 seed maturation -like protein 360 e-100
At2g14910 unknown protein 199 3e-51
At1g63610 unknown protein 100 2e-21
At3g53350 unknown protein 34 0.14
At1g07110 fructose-6-phosphate,2-kinase/fructose-2, 6-bisphospha... 33 0.18
At5g47380 putative protein 33 0.24
At1g64330 unknown protein 32 0.41
At2g13440 similar to glucose inhibited division protein A from p... 31 0.91
At2g24070 unknown protein 31 1.2
At1g16800 hypothetical protein 31 1.2
At4g30790 putative protein 30 1.6
At4g22770 putative DNA binding protein 30 1.6
At1g75720 hypothetical protein 30 1.6
At2g40840 4-alpha-glucanotransferase 30 2.0
At4g14120 unknown protein 30 2.7
At1g67170 unknown protein 30 2.7
At5g39960 putative protein 29 3.5
At3g55690 unknown protein 29 3.5
At1g65910 unknown protein 29 3.5
At1g35780 unknown protein 29 3.5
>At5g14970 seed maturation -like protein
Length = 355
Score = 360 bits (924), Expect = e-100
Identities = 210/370 (56%), Positives = 263/370 (70%), Gaps = 25/370 (6%)
Query: 1 MAALSWRPF-ILSRLTDLSPNPLHPPKPPPLFLRRRRCF-----LTSCYADGFSSSSSSS 54
MAA S R F +LSR+TDLS L +PPP R + ++S S S
Sbjct: 1 MAAASARAFFMLSRVTDLSKKKLILHQPPPSSSPHRLPYAPNRAVSSSAVISCLSGGGVS 60
Query: 55 SSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLLPS 114
S D VSTR+S DRGF VIAN++ RI+PLD SVISKG+SD+A+DSMKQTIS+MLGLLPS
Sbjct: 61 SDDSYVSTRRSKLDRGFAVIANLVNRIQPLDTSVISKGLSDSAKDSMKQTISSMLGLLPS 120
Query: 115 DHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGARDSDCEKRSEI 174
D FSV+VT+S+ PL+RLL+SSIITGYTLWNAEYR+SL RN DI P R + E +S
Sbjct: 121 DQFSVSVTISEQPLYRLLISSIITGYTLWNAEYRVSLRRNFDI--PIDPRKEE-EDQSSK 177
Query: 175 LEVKGGGEDGGEIEVASDLGLKDLENCSSSPRVFGDLPPQALNYIQQLQSELTNVKEELN 234
V+ G E G ++ DLG E SP+VFGDL P+AL+YIQ LQSEL+++KEEL+
Sbjct: 178 DNVRFGSEKG----MSEDLGNCVEEFERLSPQVFGDLSPEALSYIQLLQSELSSMKEELD 233
Query: 235 ARKQEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVD 294
++K++ +++E ++G RN+LL+YLRSLDPEMVTELS+ SS EVE+I++QLVQN+L R F D
Sbjct: 234 SQKKKALRIECEKGNRNDLLDYLRSLDPEMVTELSQLSSPEVEEIVNQLVQNVLERLFED 293
Query: 295 DASGSFME----QSVEGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENR 350
+ +FM+ ++ EG GD V TSRDYLAKLLFWCMLLGHHLRGLENR
Sbjct: 294 QTTSNFMQNPGIRTTEG--------GDGTGRKVDTSRDYLAKLLFWCMLLGHHLRGLENR 345
Query: 351 LQLSCVVGLL 360
L LSCVVGLL
Sbjct: 346 LHLSCVVGLL 355
>At2g14910 unknown protein
Length = 386
Score = 199 bits (505), Expect = 3e-51
Identities = 135/366 (36%), Positives = 203/366 (54%), Gaps = 29/366 (7%)
Query: 19 PNPLHPPKPP-------PLFLRRRRCFLTSCYADGFSSSSSSSSSDDVVSTRKSTFDRGF 71
P LH P P P F RR R + + S++ SS+ DD S T
Sbjct: 14 PQLLHKPTKPLPFLFLLPRFNRRFRSLTITSSSTTSSNNFSSNCGDDGFSLDDFTLHSDS 73
Query: 72 T-----VIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKH 126
V++++++ IEPLD S+I K V D+MK+TIS MLGLLPSD F V +
Sbjct: 74 RSPKKCVLSDLIQEIEPLDVSLIQKDVPVTTLDAMKRTISGMLGLLPSDRFQVHIESLWE 133
Query: 127 PLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGARDSDCEKRSEILEVKGGGEDGGE 186
PL +LLVSS++TGYTL NAEYR+ L +NLD++ G DS + +E +++G D
Sbjct: 134 PLSKLLVSSMMTGYTLRNAEYRLFLEKNLDMSG--GGLDSHASENTE-YDMEGTFPDEDH 190
Query: 187 IEVASDLGLKDLENCSSSPRVFGDLPPQALNYIQQLQSELTNVKEELNARKQEMMQLEYD 246
+ D ++L + G + +A YI +LQS+L++VK+EL +++ L+
Sbjct: 191 VSSKRDSRTQNLSE-TIDEEGLGRVSSEAQEYILRLQSQLSSVKKELQEMRRKNAALQMQ 249
Query: 247 RGI---RNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNIL--------SRFFVDD 295
+ + +N+LL+YLRSL PE V ELS P++ EV++ IH +V +L S+F +
Sbjct: 250 QFVGEEKNDLLDYLRSLQPEKVAELSEPAAPEVKETIHSVVHGLLATLSPKMHSKFPASE 309
Query: 296 A--SGSFMEQSVEGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQL 353
+ + +S E + + +F + +RDYLA+LLFWCMLLGH+LRGLE R++L
Sbjct: 310 VPPTETVKAKSDEDCAELVENTSLQFQPLISLTRDYLARLLFWCMLLGHYLRGLEYRMEL 369
Query: 354 SCVVGL 359
V+ L
Sbjct: 370 MEVLSL 375
>At1g63610 unknown protein
Length = 340
Score = 99.8 bits (247), Expect = 2e-21
Identities = 84/318 (26%), Positives = 151/318 (47%), Gaps = 55/318 (17%)
Query: 44 ADGFSSSSSSSSSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMKQ 103
A G SS SS+ SS TR+ R ++ ++ ++P + K ++M+Q
Sbjct: 61 AYGSSSDSSADSSTPPNGTRQPKSRRD--ILLEYVQNVKPEFMEMFVKRAPKHVVEAMRQ 118
Query: 104 TISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGA 163
T++ M+G LP F+VTVT L +L++S ++TGY NA+YR+ L ++L+ +
Sbjct: 119 TVTNMIGTLPPQFFAVTVTSVAENLAQLMMSVLMTGYMFRNAQYRLELQQSLEQVALPEP 178
Query: 164 RDSDCEKRSEILEVKGGGED---GGEIEVASDLGLKDLENCSSSPRVFGDLPPQALNYIQ 220
RD KGG ED G + V+ + + N S ++ A YI+
Sbjct: 179 RDQ-----------KGGDEDYAPGTQKNVSGE--VIRWNNVSGPEKI------DAKKYIE 219
Query: 221 QLQSELTNVKEELNARKQEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDII 280
L++E+ + ++ + +N +LEYL+SL+P+ + EL+ + +V +
Sbjct: 220 LLEAEIEELNRQVGRKSANQ---------QNEILEYLKSLEPQNLKELTSTAGEDVAVAM 270
Query: 281 HQLVQNILSRFFVDDASGSFMEQSVEGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLL 340
+ V+ +L+ V D + ++ N+ TS LAKLL+W M++
Sbjct: 271 NTFVKRLLA---VSDPN------QMKTNVTE-------------TSAADLAKLLYWLMVV 308
Query: 341 GHHLRGLENRLQLSCVVG 358
G+ +R +E R + V+G
Sbjct: 309 GYSIRNIEVRFDMERVLG 326
>At3g53350 unknown protein
Length = 396
Score = 33.9 bits (76), Expect = 0.14
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 10/65 (15%)
Query: 215 ALNYIQQLQSELTNVKEELNARKQEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSV 274
A+N +Q+L+S+L + EL K E+ LE + +R L+ E V SS+
Sbjct: 161 AINEVQKLKSKLFESESELEQSKYEVRSLE----------KLVRQLEEERVNSRDSSSSM 210
Query: 275 EVEDI 279
EVE++
Sbjct: 211 EVEEL 215
>At1g07110 fructose-6-phosphate,2-kinase/fructose-2,
6-bisphosphatase (fkfbp)
Length = 744
Score = 33.5 bits (75), Expect = 0.18
Identities = 22/67 (32%), Positives = 36/67 (52%), Gaps = 4/67 (5%)
Query: 228 NVKEELNARKQEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNI 287
N+ EE +RK++ ++ Y RG + L+ ++ L+P ++ EL R V I HQ V
Sbjct: 647 NMPEEYESRKKDKLRYRYPRG--ESYLDVIQRLEP-VIIELER-QRAPVVVISHQAVLRA 702
Query: 288 LSRFFVD 294
L +F D
Sbjct: 703 LYAYFAD 709
>At5g47380 putative protein
Length = 598
Score = 33.1 bits (74), Expect = 0.24
Identities = 21/85 (24%), Positives = 44/85 (51%), Gaps = 12/85 (14%)
Query: 227 TNVKEELNARKQEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQN 286
+NVKEEL+A K+E + G + + L P+++ ++ +S+ +D++ L+ N
Sbjct: 474 SNVKEELDASKREFL------GANVVVKMQKKVLLPKIIERFTKEASLSFDDLMRWLIDN 527
Query: 287 ILSRFFVDDASGSFMEQSVEGNIDN 311
D+ G +++ V+G +N
Sbjct: 528 ------ADEKLGESIQKCVQGKPNN 546
>At1g64330 unknown protein
Length = 555
Score = 32.3 bits (72), Expect = 0.41
Identities = 49/201 (24%), Positives = 78/201 (38%), Gaps = 41/201 (20%)
Query: 43 YADGFSSSSSSSSSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMK 102
+ G + SSSSSSSD + RG I + +++E + + + A D K
Sbjct: 89 HGKGENDSSSSSSSDSDSDKKSKRNGRGENEIELLKKQMEDANLEIADLKMKLATTDEHK 148
Query: 103 QTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMS----LTRNLDIA 158
+ + S+H + L +L S I G E S L L++A
Sbjct: 149 EAVE-------SEHQEI--------LKKLKESDEICGNLRVETEKLTSENKELNEKLEVA 193
Query: 159 SPCGARDSDCEKRSEILEVKGGGEDGGEIEVASDLGLKDLENCSSSPRVFGDLPPQALNY 218
G +SD ++ E ++ + DG E E+AS KD E+ L
Sbjct: 194 ---GETESDLNQKLEDVKKE---RDGLEAELASK--AKDHES--------------TLEE 231
Query: 219 IQQLQSELTNVKEELNARKQE 239
+ +LQ + + EL KQE
Sbjct: 232 VNRLQGQKNETEAELEREKQE 252
>At2g13440 similar to glucose inhibited division protein A from
prokaryotes
Length = 723
Score = 31.2 bits (69), Expect = 0.91
Identities = 18/37 (48%), Positives = 23/37 (61%), Gaps = 1/37 (2%)
Query: 21 PLHPPKPPPLFLRRRRCFLTSCYADGFSSSSSSSSSD 57
P H P+ +FLR R+ FL A FSSSSS ++SD
Sbjct: 35 PFHSPRLC-VFLRPRQLFLNRPLAASFSSSSSGATSD 70
>At2g24070 unknown protein
Length = 560
Score = 30.8 bits (68), Expect = 1.2
Identities = 26/81 (32%), Positives = 37/81 (45%), Gaps = 6/81 (7%)
Query: 39 LTSCYADGFSSSSSSSSSDDVVSTRK------STFDRGFTVIANMLRRIEPLDNSVISKG 92
L SCY +G SSS++S+D ST S+ R + A R P +S S
Sbjct: 252 LFSCYDNGRLEVSSSTTSEDSSSTESLKHFSTSSLPRLHPMSAPGSRTASPSRSSFSSSS 311
Query: 93 VSDAARDSMKQTISTMLGLLP 113
S++ S + +S M GL P
Sbjct: 312 SSNSRGMSPSRGVSPMRGLSP 332
>At1g16800 hypothetical protein
Length = 1939
Score = 30.8 bits (68), Expect = 1.2
Identities = 23/85 (27%), Positives = 39/85 (45%), Gaps = 11/85 (12%)
Query: 238 QEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSVEVE-----------DIIHQLVQN 286
QEM + EY+ LL LR LD E VT + ++++E +++ + +
Sbjct: 113 QEMYEKEYECSSVGPLLVVLRKLDEERVTRHLQEINLKIEKGTYDPDHHHAEVVSVMYEV 172
Query: 287 ILSRFFVDDASGSFMEQSVEGNIDN 311
++ FF DD S + +IDN
Sbjct: 173 LMFPFFFDDMSLCTEFEKFIESIDN 197
>At4g30790 putative protein
Length = 1148
Score = 30.4 bits (67), Expect = 1.6
Identities = 30/136 (22%), Positives = 55/136 (40%), Gaps = 29/136 (21%)
Query: 149 MSLTRNLDIASPCGARDSDCEKR------------SEILEVKGGGEDGGEIEVASDLGL- 195
+S NL PC AR+ E S +LE + E GG+ A ++G+
Sbjct: 722 VSCVSNLTSKQPCKAREGMDENMVDSSQVLSQPLDSSMLESQQNNEKGGKDSEAGEMGVF 781
Query: 196 -----------KDLENCSSSPRVF-----GDLPPQALNYIQQLQSELTNVKEELNARKQE 239
K L++ ++ R GD+ + N + + ++L+ ++ +LN +E
Sbjct: 782 LSNSSTAESPQKSLDDNVATGRGLDAKDSGDIILELRNELMEKSNKLSEMESKLNGAMEE 841
Query: 240 MMQLEYDRGIRNNLLE 255
+ L + LLE
Sbjct: 842 VSNLSRELETNQKLLE 857
>At4g22770 putative DNA binding protein
Length = 334
Score = 30.4 bits (67), Expect = 1.6
Identities = 27/97 (27%), Positives = 35/97 (35%), Gaps = 8/97 (8%)
Query: 12 SRLTDLSPNPLHPPKPPPLFLRRRRCFLTSCYADGFSSSSSSSSSDDVVSTRKSTFD-RG 70
S ++ PN + PP PPP + F S DGFSS RK D
Sbjct: 35 SETSNTPPNSVAPPPPPP----PQNSFTPSAAMDGFSSGPIKKRRG---RPRKYGHDGAA 87
Query: 71 FTVIANMLRRIEPLDNSVISKGVSDAARDSMKQTIST 107
T+ N + P + VI + R MK T
Sbjct: 88 VTLSPNPISSAAPTTSHVIDFSTTSEKRGKMKPATPT 124
>At1g75720 hypothetical protein
Length = 197
Score = 30.4 bits (67), Expect = 1.6
Identities = 13/35 (37%), Positives = 22/35 (62%)
Query: 221 QLQSELTNVKEELNARKQEMMQLEYDRGIRNNLLE 255
Q+++ L+ +KEEL KQE+ +L D G+ L+
Sbjct: 78 QMRNSLSCLKEELERTKQELQKLRVDPGVNETKLD 112
>At2g40840 4-alpha-glucanotransferase
Length = 858
Score = 30.0 bits (66), Expect = 2.0
Identities = 25/105 (23%), Positives = 47/105 (43%), Gaps = 18/105 (17%)
Query: 208 FGDLPPQ-ALNYIQQLQSELTNVKEELNARK----------QEMMQLEYDRGIRNNLLEY 256
FGD P A N++ + Q ++ KE+ N K ++ + LE + +R ++ +
Sbjct: 525 FGDFWPFIASNFLNETQKDMYEFKEDCNTEKKIVAKLKSLAEKSLLLENEDKVRRDVFDI 584
Query: 257 LRSL----DPEMVTELSRPSSVEVEDIIHQL---VQNILSRFFVD 294
LR++ DPE + ++E L +N+L R + D
Sbjct: 585 LRNVVLIKDPEDARKFYPRFNIEDTSSFQDLDDHSKNVLKRLYYD 629
>At4g14120 unknown protein
Length = 368
Score = 29.6 bits (65), Expect = 2.7
Identities = 32/124 (25%), Positives = 52/124 (41%), Gaps = 22/124 (17%)
Query: 121 VTVSKHPLHRLLVSSIITGYTLWNAEYRM-SLTRNLDIASPCGARDSDCEKRSEILEVKG 179
VTV L R+ S I+ W+ + +LT+ L++ SPC +S+ + V G
Sbjct: 77 VTVKNLCLRRVFSPSSISSD--WDFHVKGGNLTKELNVESPCDTPNSNVMREGSKYLVDG 134
Query: 180 GGEDGGEIEVASDLGLKDLENCSSSPRVFGDLPPQALNYIQQLQSELTNVKEELNARKQE 239
GG++EV CS + PP + EL+ VK E + QE
Sbjct: 135 LVATGGDLEVV---------ECSQT------TPPD----LGMFTGELSLVKNEAGSINQE 175
Query: 240 MMQL 243
+ ++
Sbjct: 176 ISEV 179
>At1g67170 unknown protein
Length = 359
Score = 29.6 bits (65), Expect = 2.7
Identities = 28/108 (25%), Positives = 48/108 (43%), Gaps = 14/108 (12%)
Query: 219 IQQLQSELTNVKEELNARKQEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSVEVED 278
I L SEL N+++E +Q +Y++ N+ LE L++++ +T + EVE
Sbjct: 177 IPALMSELENLRQEY---QQCRATYDYEKKFYNDHLESLQAMEKNYMT-----MAREVEK 228
Query: 279 IIHQLVQNILSRFFVDDASGSFMEQSVEGNID--NHPDNGDEFSDTVG 324
+ QL+ N S D +G ++ ID H + D G
Sbjct: 229 LQAQLMNNANS----DRRAGGPYGNNINAEIDASGHQSGNGYYEDAFG 272
>At5g39960 putative protein
Length = 614
Score = 29.3 bits (64), Expect = 3.5
Identities = 16/60 (26%), Positives = 30/60 (49%)
Query: 52 SSSSSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGL 111
S + S+ V++ R RG VI N + R+ +NS + K + +A ++ I + G+
Sbjct: 411 SMTHSEVVIARRAVEEGRGLVVIVNKMDRLRGRENSEMYKKIKEAVPIEIQTVIPQITGI 470
>At3g55690 unknown protein
Length = 293
Score = 29.3 bits (64), Expect = 3.5
Identities = 28/99 (28%), Positives = 43/99 (43%), Gaps = 2/99 (2%)
Query: 22 LHPPKPPPLFLRRRRCFLTSCYADGFSSSSSSSSSDDVVSTRKSTFDRGFTVIANMLRRI 81
L PP P + + + F+SS+ S+S +D T T D +A + RR+
Sbjct: 13 LPPPFPAVFEQPMEKLTFPNEFPYEFASSTFSTSPEDSTETEDETTDDEDDFLAGLTRRL 72
Query: 82 EPLDNSVISKG-VSDAARDSMKQTISTMLGL-LPSDHFS 118
+ S V+D ++ K T ST GL P+ FS
Sbjct: 73 ALSTQRLSSPSFVTDKSQMKPKVTESTQSGLGSPNGPFS 111
>At1g65910 unknown protein
Length = 631
Score = 29.3 bits (64), Expect = 3.5
Identities = 28/103 (27%), Positives = 41/103 (39%), Gaps = 7/103 (6%)
Query: 217 NYIQQLQSELTNVKEELNARKQEMMQLEYDRGIRNNLLEYLRSLDPEM--VTELSRPSSV 274
N I++ K N+ + Y G+ NL + PE +T+L SS
Sbjct: 161 NKIEEQHHGTKKNKGTTNSEQSTSSTCLYSDGMYENLENSGYPVSPETGGLTQLGNNSSS 220
Query: 275 EVEDIIHQLVQNILSRFFVDDASGSFMEQSVEGNIDNHPDNGD 317
++E I +N S+F D S +F QS G I P D
Sbjct: 221 DMETI-----ENKWSQFMSHDTSFNFPPQSQYGTISYPPSKVD 258
>At1g35780 unknown protein
Length = 286
Score = 29.3 bits (64), Expect = 3.5
Identities = 20/80 (25%), Positives = 32/80 (40%), Gaps = 9/80 (11%)
Query: 28 PPLFLRRRRCFLTSCYADGFSSSSSSSSSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNS 87
PP ++ R Y D F S + D + T K DR FT ++ N+
Sbjct: 177 PPPEIKLRPTVRALAYKDNFDLGESDTKPDGELKTAKKIADRKFTDLSG---------NN 227
Query: 88 VISKGVSDAARDSMKQTIST 107
V VS + + ++ +ST
Sbjct: 228 VFKSDVSSPSSATAERLLST 247
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,392,124
Number of Sequences: 26719
Number of extensions: 371009
Number of successful extensions: 1887
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 1859
Number of HSP's gapped (non-prelim): 42
length of query: 360
length of database: 11,318,596
effective HSP length: 101
effective length of query: 259
effective length of database: 8,619,977
effective search space: 2232574043
effective search space used: 2232574043
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0081b.1


