

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0079c.8
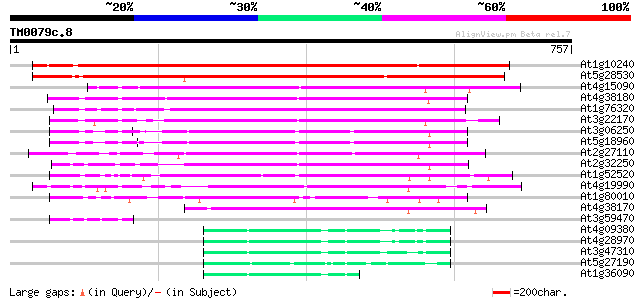
(757 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g10240 unknown protein 603 e-172
At5g28530 far-red impaired response protein (FAR1) - like 537 e-153
At4g15090 unknown protein 284 1e-76
At4g38180 unknown protein (At4g38180) 282 4e-76
At1g76320 putative phytochrome A signaling protein 253 4e-67
At3g22170 far-red impaired response protein, putative 251 8e-67
At3g06250 unknown protein 231 9e-61
At5g18960 FAR1 - like protein 216 3e-56
At2g27110 Mutator-like transposase 214 1e-55
At2g32250 Mutator-like transposase 194 2e-49
At1g52520 F6D8.26 183 4e-46
At4g19990 putative protein 177 2e-44
At1g80010 hypothetical protein 173 4e-43
At4g38170 hypothetical protein 166 4e-41
At3g59470 unknown protein 67 4e-11
At4g09380 putative protein 65 1e-10
At4g28970 putative protein 64 4e-10
At3g47310 putative protein 59 8e-09
At5g27190 putative protein 59 1e-08
At1g36090 hypothetical protein 57 3e-08
>At1g10240 unknown protein
Length = 680
Score = 603 bits (1554), Expect = e-172
Identities = 306/645 (47%), Positives = 435/645 (67%), Gaps = 10/645 (1%)
Query: 32 SSTCQEPQLEPLSL-ANNSADWIPYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKST 90
SST + P LSL A ++A IPY GQIF + AY+FY FA++ GFSIRR
Sbjct: 27 SSTEESPDDNNLSLEAVHNA--IPYLGQIFLTHDTAYEFYSTFAKRCGFSIRRHRT---- 80
Query: 91 KNQSENNPLGIYKREFVCHRAGITKQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEE 150
+ G+ +R FVCHRAG T +SE + QR R++SRC C A + ++ T
Sbjct: 81 -EGKDGVGKGLTRRYFVCHRAGNTPIKTLSEGKPQRNRRSSRCGCQAYLRISKLTELGST 139
Query: 151 KWVVKYFHNHHNHQLLDDKEVQFLPAYRSIPIVDQDRIMLLSKAGCSISLIIRVLELEKG 210
+W V F NHHNH+LL+ +V+FLPAYRSI D+ RI++ SK G S+ ++R+LELEK
Sbjct: 140 EWRVTGFANHHNHELLEPNQVRFLPAYRSISDADKSRILMFSKTGISVQQMMRLLELEKC 199
Query: 211 IDTCTLPFLERDIRNFIQSQSSIGKESDASNVLKLCKSLKDRDDSFKYDFTLDENNKLEH 270
++ LPF E+D+RN +QS + E + + L++C+S+K++D +FK++FTLD N+KLE+
Sbjct: 200 VEPGFLPFTEKDVRNLLQSFKKLDPEDENIDFLRMCQSIKEKDPNFKFEFTLDANDKLEN 259
Query: 271 IIWAFGDSIRAYEAFGDVVVFDTTYRINRYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIP 330
I W++ SI++YE FGD VVFDTT+R++ +MPLGIWVGV+N+G FFGCVLLRDE +
Sbjct: 260 IAWSYASSIQSYELFGDAVVFDTTHRLSAVEMPLGIWVGVNNYGVPCFFGCVLLRDENLR 319
Query: 331 SFSWALKSFLYFVNGKHPKTILTDQDVALKEAIAMELPNTKHAFCIWHIVAKLSSWFSFS 390
S+SWAL++F F+NGK P+TILTD ++ LKEAIA E+P TKHA CIW +V K SWF+
Sbjct: 320 SWSWALQAFTGFMNGKAPQTILTDHNMCLKEAIAGEMPATKHALCIWMVVGKFPSWFNAG 379
Query: 391 LGSRYNDFKYEFHKVYHLECEDDFEHKWTLMIAQFGLGMDKHIDLLYSNRQFWALAYLKD 450
LG RYND+K EF+++YHLE ++FE W M+ FGL ++HI+ LY++R W+L YL+
Sbjct: 380 LGERYNDWKAEFYRLYHLESVEEFELGWRDMVNSFGLHTNRHINNLYASRSLWSLPYLRS 439
Query: 451 FFFAGMTTTGRSESINSYIKRFLHVRTSLTDFVNQVGIAVNIRNQAGEEARMRQKYHNPH 510
F AGMT TGRS++IN++I+RFL +T L FV QV + V+ ++QA E+ M+Q N
Sbjct: 440 HFLAGMTLTGRSKAINAFIQRFLSAQTRLAHFVEQVAVVVDFKDQATEQQTMQQNLQNIS 499
Query: 511 IRTGFPIEEQAASMLTPYAFKLLQHEIELSTKYASTEEGENSYIVRFHTKVDGGRIVKWI 570
++TG P+E AAS+LTP+AF LQ ++ L+ YAS + E Y+VR HTK+DGGR V W+
Sbjct: 500 LKTGAPMESHAASVLTPFAFSKLQEQLVLAAHYASFQMDE-GYLVRHHTKLDGGRKVYWV 558
Query: 571 EEDKSINCSCKEFEFSGILCRHAIRVLVMKNYFHLPTKYLPFRWRQESSLVPTSSHIINC 630
++ I+CSC+ FEFSG LCRHA+RVL N F +P +YLP RWR+ S+ +
Sbjct: 559 PQEGIISCSCQLFEFSGFLCRHALRVLSTGNCFQVPDRYLPLRWRRISTSFSKTFRSNAE 618
Query: 631 NDGSSFE-FRSLVQWLQVESLKTKDREEVAIRELERVIRIITDMP 674
+ G + ++LV L ES K+K+R ++A + ++ I + P
Sbjct: 619 DHGERVQLLQNLVSTLVSESAKSKERLDIATEQTSILLSRIREQP 663
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 537 bits (1384), Expect = e-153
Identities = 280/650 (43%), Positives = 413/650 (63%), Gaps = 22/650 (3%)
Query: 31 NSSTCQEPQLEPLSLANNSADWI--PYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYK 88
+ ST + ++E + D + PY GQIF ++ +A+++Y FARKSGFSIR+ +
Sbjct: 45 DESTITKSEIESTPTPTSQYDTVFTPYVGQIFTTDDEAFEYYSTFARKSGFSIRKA---R 101
Query: 89 STKNQSENNPLGIYKREFVCHRAGITKQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGF 148
ST++Q+ LG+Y+R+FVC+R+G + K + VE R+RK+ RC C K+ +T +
Sbjct: 102 STESQN----LGVYRRDFVCYRSGFNQPRKKANVEHPRERKSVRCGCDGKLYLTKEVVDG 157
Query: 149 EEKWVVKYFHNHHNHQLLDDKEVQFLPAYRSIPIVDQDRIMLLSKAGCSISLIIRVLELE 208
W V F N HNH+LL+D +V+ LPAYR I DQ+RI+LLSKAG ++ I+++LELE
Sbjct: 158 VSHWYVSQFSNVHNHELLEDDQVRLLPAYRKIQQSDQERILLLSKAGFPVNRIVKLLELE 217
Query: 209 KGIDTCTLPFLERDIRNFIQS-QSSIG---------KESDASNVLKLCKSLKDRDDSFKY 258
KG+ + LPF+E+D+RNF+++ + S+ +ESD +L+ CK L +RD F Y
Sbjct: 218 KGVVSGQLPFIEKDVRNFVRACKKSVQENDAFMTEKRESDTLELLECCKGLAERDMDFVY 277
Query: 259 DFTLDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRINRYDMPLGIWVGVDNHGNSIF 318
D T DEN K+E+I WA+GDS+R Y FGDVVVFDT+YR Y + LG++ G+DN+G ++
Sbjct: 278 DCTSDENQKVENIAWAYGDSVRGYSLFGDVVVFDTSYRSVPYGLLLGVFFGIDNNGKAML 337
Query: 319 FGCVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQDVALKEAIAMELPNTKHAFCIWH 378
GCVLL+DE SF+WAL++F+ F+ G+HP+TILTD D LK+AI E+PNT H + H
Sbjct: 338 LGCVLLQDESCRSFTWALQTFVRFMRGRHPQTILTDIDTGLKDAIGREMPNTNHVVFMSH 397
Query: 379 IVAKLSSWFSFSLGSRYNDFKYEFHKVYHLECEDDFEHKWTLMIAQFGLGMDKHIDLLYS 438
IV+KL+SWFS +LGS Y +F+ F + D+FE +W L++ +FGL D+H LLYS
Sbjct: 398 IVSKLASWFSQTLGSHYEEFRAGFDMLCRAGNVDEFEQQWDLLVTRFGLVPDRHAALLYS 457
Query: 439 NRQFWALAYLKDFFFAGMTTTGRSESINSYIKRFLHVRTSLTDFVNQVGIAVNIRNQAGE 498
R W +++ F A T+ + SI+S++KR + T + + + + V+ +
Sbjct: 458 CRASWLPCCIREHFVAQTMTSEFNLSIDSFLKRVVDGATCMQLLLEESALQVSAAASLAK 517
Query: 499 EARMRQKYHNPHIRTGFPIEEQAASMLTPYAFKLLQHEIELSTKYASTEEGENSYIVRFH 558
+ R Y P ++T P+E+ A +LTPYAF +LQ+E+ LS +YA E +IV +
Sbjct: 518 QILPRFTY--PSLKTCMPMEDHARGILTPYAFSVLQNEMVLSVQYAVAEMANGPFIVHHY 575
Query: 559 TKVDGGRIVKWIEEDKSINCSCKEFEFSGILCRHAIRVLVMKNYFHLPTKYLPFRWRQES 618
K++G V W E++ I CSCKEFE SGILCRH +RVL +KN FH+P +Y RWRQES
Sbjct: 576 KKMEGECCVIWNPENEEIQCSCKEFEHSGILCRHTLRVLTVKNCFHIPEQYFLLRWRQES 635
Query: 619 SLVPT-SSHIINCNDGSSFEFRSLVQWLQVESLKTKDREEVAIRELERVI 667
V T + + D S+ F SL + L ES+ +KDR + A +EL +I
Sbjct: 636 PHVATENQNGQGIGDDSAQTFHSLTETLLTESMISKDRLDYANQELSLLI 685
>At4g15090 unknown protein
Length = 768
Score = 284 bits (726), Expect = 1e-76
Identities = 168/593 (28%), Positives = 299/593 (50%), Gaps = 14/593 (2%)
Query: 105 EFVCHRAGITKQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQ 164
+F C R G+T +S+ S S R+ + +C A M V R G KW++ F HNH+
Sbjct: 37 KFACSRYGVTPESE-SSGSSSRRSTVKKTDCKASMHVKRRPDG---KWIIHEFVKDHNHE 92
Query: 165 LLDDKEVQFLPAYRSIPIVDQDRIMLLSKAGCSISLIIRVLELEKGIDTCTLPFLERDIR 224
LL F R++ + +++ I +L + + + G L+ D+
Sbjct: 93 LLPALAYHFR-IQRNVKLAEKNNIDILHAVSERTKKMYVEMSRQSGGYKNIGSLLQTDVS 151
Query: 225 NFIQSQSSIG-KESDASNVLKLCKSLKDRDDSFKYDFTLDENNKLEHIIWAFGDSIRAYE 283
+ + + +E D+ +L+ K +K + F Y L+E+ +L ++ WA S Y
Sbjct: 152 SQVDKGRYLALEEGDSQVLLEYFKRIKKENPKFFYAIDLNEDQRLRNLFWADAKSRDDYL 211
Query: 284 AFGDVVVFDTTYRINRYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFV 343
+F DVV FDTTY +PL +++GV++H + GC L+ DE + +F W +K++L +
Sbjct: 212 SFNDVVSFDTTYVKFNDKLPLALFIGVNHHSQPMLLGCALVADESMETFVWLIKTWLRAM 271
Query: 344 NGKHPKTILTDQDVALKEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGSRYNDFKYEFH 403
G+ PK ILTDQD L A++ LPNT+H F +WH++ K+ +FS + R+ +F +F+
Sbjct: 272 GGRAPKVILTDQDKFLMSAVSELLPNTRHCFALWHVLEKIPEYFSHVM-KRHENFLLKFN 330
Query: 404 K-VYHLECEDDFEHKWTLMIAQFGLGMDKHIDLLYSNRQFWALAYLKDFFFAGMTTTGRS 462
K ++ +D+F+ +W M++QFGL D+ + L+ +RQ W ++ D F AGM+T+ RS
Sbjct: 331 KCIFRSWTDDEFDMRWWKMVSQFGLENDEWLLWLHEHRQKWVPTFMSDVFLAGMSTSQRS 390
Query: 463 ESINSYIKRFLHVRTSLTDFVNQVGIAVNIRNQAGEEARMRQKYHNPHIRTGFPIEEQAA 522
ES+NS+ +++H + +L +F+ Q G+ + R + A + P +++ P E+Q A
Sbjct: 391 ESVNSFFDKYIHKKITLKEFLRQYGVILQNRYEEESVADFDTCHKQPALKSPSPWEKQMA 450
Query: 523 SMLTPYAFKLLQHEIELSTKYASTEEGENSYIVRFHT---KVDGGRIVKWIEEDKSINCS 579
+ T FK Q E+ +E E+ + F + D +V W + + C
Sbjct: 451 TTYTHTIFKKFQVEVLGVVACHPRKEKEDENMATFRVQDCEKDDDFLVTWSKTKSELCCF 510
Query: 580 CKEFEFSGILCRHAIRVLVMKNYFHLPTKYLPFRWRQESS---LVPTSSHIINCNDGSSF 636
C+ FE+ G LCRHA+ +L M + +P +Y+ RW +++ L + I
Sbjct: 511 CRMFEYKGFLCRHALMILQMCGFASIPPQYILKRWTKDAKSGVLAGEGADQIQTRVQRYN 570
Query: 637 EFRSLVQWLQVESLKTKDREEVAIRELERVIRIITDMPEEQEHAVDMEPNVPN 689
+ S L E +++ +A+R L ++ DM + + + + N
Sbjct: 571 DLCSRATELSEEGCVSEENYNIALRTLVETLKNCVDMNNARNNITESNSQLNN 623
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 282 bits (722), Expect = 4e-76
Identities = 177/575 (30%), Positives = 286/575 (48%), Gaps = 18/575 (3%)
Query: 51 DWIPYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHR 110
D PY+G F SE A FY +AR+ GFS R +S ++ + I +R+FVC +
Sbjct: 70 DLEPYDGLEFESEEAAKAFYNSYARRIGFSTRVSSSRRSRRDGA------IIQRQFVCAK 123
Query: 111 AGITK--QSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQLLDD 168
G + + + E +R R +R C A + V + G KW+V F HNH+L+
Sbjct: 124 EGFRNMNEKRTKDREIKRPRTITRVGCKASLSVKMQDSG---KWLVSGFVKDHNHELVPP 180
Query: 169 KEVQFLPAYRSIPIVDQDRIMLLSKAGCSISLIIRVLELEKGIDTCTLPFLERDIRNFIQ 228
+V L ++R I + I L AG I+ L E G + F E D RN+++
Sbjct: 181 DQVHCLRSHRQISGPAKTLIDTLQAAGMGPRRIMSALIKEYG-GISKVGFTEVDCRNYMR 239
Query: 229 SQSSIGKESDASNVLKLCKSLKDRDDSFKYDFTLDENNKLEHIIWAFGDSIRAYEAFGDV 288
+ E + +L + + + +F Y E+ + ++ WA +I + FGD
Sbjct: 240 NNRQKSIEGEIQLLLDYLRQMNADNPNFFYSVQGSEDQSVGNVFWADPKAIMDFTHFGDT 299
Query: 289 VVFDTTYRINRYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFVNGKHP 348
V FDTTYR NRY +P + GV++HG I FGC + +E SF W ++L ++ P
Sbjct: 300 VTFDTTYRSNRYRLPFAPFTGVNHHGQPILFGCAFIINETEASFVWLFNTWLAAMSAHPP 359
Query: 349 KTILTDQDVALKEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGSRYNDFKYEFHKVYHL 408
+I TD D ++ AI P +H FC WHI+ K S + ++ F+ +FHK +L
Sbjct: 360 VSITTDHDAVIRAAIMHVFPGARHRFCKWHILKKCQEKLS-HVFLKHPSFESDFHKCVNL 418
Query: 409 -ECEDDFEHKWTLMIAQFGLGMDKHIDLLYSNRQFWALAYLKDFFFAGMTTTGRSESINS 467
E +DFE W ++ ++ L + + +YS+R+ W YL+D FFA M+ T RS+SINS
Sbjct: 419 TESVEDFERCWFSLLDKYELRDHEWLQAIYSDRRQWVPVYLRDTFFADMSLTHRSDSINS 478
Query: 468 YIKRFLHVRTSLTDFVNQVGIAVNIRNQAGEEARMRQKYHNPHIRTGFPIEEQAASMLTP 527
Y +++ T+L+ F A+ R + +A P ++T P+E+QA+ + T
Sbjct: 479 YFDGYINASTNLSQFFKLYEKALESRLEKEVKADYDTMNSPPVLKTPSPMEKQASELYTR 538
Query: 528 YAFKLLQHEIELSTKYASTEEGENSYIVRFHTKVDG----GRIVKWIEEDKSINCSCKEF 583
F Q E+ + + +++ ++ +V + G VK+ + NCSC+ F
Sbjct: 539 KLFMRFQEELVGTLTFMASKADDDGDLVTYQVAKYGEAHKAHFVKFNVLEMRANCSCQMF 598
Query: 584 EFSGILCRHAIRVLVMKNYFHLPTKYLPFRWRQES 618
EFSGI+CRH + V + N LP Y+ RW + +
Sbjct: 599 EFSGIICRHILAVFRVTNLLTLPPYYILKRWTRNA 633
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 253 bits (645), Expect = 4e-67
Identities = 159/557 (28%), Positives = 273/557 (48%), Gaps = 22/557 (3%)
Query: 60 FNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGITKQSKV 119
F + DAY FY +A+ GF + +S ++ +F C R G +QS
Sbjct: 3 FETHEDAYLFYKDYAKSVGFGTAKLSSRRSRASKE------FIDAKFSCIRYGSKQQSD- 55
Query: 120 SEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQLLDDKEVQFLPAYRS 179
++ R + + C A M V R G KW V F HNH LL + + + ++R+
Sbjct: 56 ---DAINPRASPKIGCKASMHVKRRPDG---KWYVYSFVKEHNHDLLPE-QAHYFRSHRN 108
Query: 180 IPIVDQDRIMLLSKAGCSISLIIRVLELEKGIDTCTLPFLERDIRN-FIQSQSSIGKESD 238
+V + L K ++ + L F++ +RN + + + D
Sbjct: 109 TELVKSNDSRLRRKKNTPLTDCKHLSAYHD------LDFIDGYMRNQHDKGRRLVLDTGD 162
Query: 239 ASNVLKLCKSLKDRDDSFKYDFTLDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRIN 298
A +L+ +++ + F + E++ L ++ W I Y++F DVV F+T+Y ++
Sbjct: 163 AEILLEFLMRMQEENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFVS 222
Query: 299 RYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQDVA 358
+Y +PL ++VGV++H + GC LL D+ + ++ W ++S+L + G+ PK +LTDQ+ A
Sbjct: 223 KYKVPLVLFVGVNHHVQPVLLGCGLLADDTVYTYVWLMQSWLVAMGGQKPKVMLTDQNNA 282
Query: 359 LKEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGSRYNDFKYEFHKVYHLECEDDFEHKW 418
+K AIA LP T+H +C+WH++ +L + + K F +Y E++F+ +W
Sbjct: 283 IKAAIAAVLPETRHCYCLWHVLDQLPRNLDYWSMWQDTFMKKLFKCIYRSWSEEEFDRRW 342
Query: 419 TLMIAQFGLGMDKHIDLLYSNRQFWALAYLKDFFFAGMTTTGRSESINSYIKRFLHVRTS 478
+I +F L + LY R+FWA +++ FAG++ RSES+NS R++H TS
Sbjct: 343 LKLIDKFHLRDVPWMRSLYEERKFWAPTFMRGITFAGLSMRCRSESVNSLFDRYVHPETS 402
Query: 479 LTDFVNQVGIAVNIRNQAGEEARMRQKYHNPHIRTGFPIEEQAASMLTPYAFKLLQHEIE 538
L +F+ G+ + R + +A + P +++ P E+Q + + F+ Q E+
Sbjct: 403 LKEFLEGYGLMLEDRYEEEAKADFDAWHEAPELKSPSPFEKQMLLVYSHEIFRRFQLEVL 462
Query: 539 LSTKYASTEEGENSYIVRFHTKVDGGR-IVKWIEEDKSINCSCKEFEFSGILCRHAIRVL 597
+ T+E E D + +V W E I CSC+ FE+ G LCRHAI VL
Sbjct: 463 GAAACHLTKESEEGTTYSVKDFDDEQKYLVDWDEFKSDIYCSCRSFEYKGYLCRHAIVVL 522
Query: 598 VMKNYFHLPTKYLPFRW 614
M F +P Y+ RW
Sbjct: 523 QMSGVFTIPINYVLQRW 539
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 251 bits (642), Expect = 8e-67
Identities = 168/627 (26%), Positives = 294/627 (46%), Gaps = 70/627 (11%)
Query: 54 PYEGQIFNSEGDAYDFYCLFARKSGFS--IRRDHIYKSTKNQSENNPLGIYKREFVCHRA 111
P G F S G+AY FY ++R GF+ I+ K+T+ +F C R
Sbjct: 70 PLNGMEFESHGEAYSFYQEYSRAMGFNTAIQNSRRSKTTRE--------FIDAKFACSRY 121
Query: 112 GI------------TKQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHN 159
G +QSK +R ++ +C A M V R G KWV+ F
Sbjct: 122 GTKREYDKSFNRPRARQSKQDPENMAGRRTCAKTDCKASMHVKRRPDG---KWVIHSFVR 178
Query: 160 HHNHQLLDDKEVQFLPAYRSIPIVDQDRIMLLSKAGCSISLIIRVLELEKGIDTCTLPFL 219
HNH+LL + V +Q R + + A ++ + T+ L
Sbjct: 179 EHNHELLPAQAVS-----------EQTRKIYAAMA-------------KQFAEYKTVISL 214
Query: 220 ERDIRNFIQSQSSIGKES-DASNVLKLCKSLKDRDDSFKYDFTLDENNKLEHIIWAFGDS 278
+ D ++ + ++ E+ D +L ++ + +F Y L ++ +++++ W S
Sbjct: 215 KSDSKSSFEKGRTLSVETGDFKILLDFLSRMQSLNSNFFYAVDLGDDQRVKNVFWVDAKS 274
Query: 279 IRAYEAFGDVVVFDTTYRINRYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKS 338
Y +F DVV DTTY N+Y MPL I+VGV+ H + GC L+ DE ++SW +++
Sbjct: 275 RHNYGSFCDVVSLDTTYVRNKYKMPLAIFVGVNQHYQYMVLGCALISDESAATYSWLMET 334
Query: 339 FLYFVNGKHPKTILTDQDVALKEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGSRYNDF 398
+L + G+ PK ++T+ DV + + PNT+H +WH++ K+S + ++++F
Sbjct: 335 WLRAIGGQAPKVLITELDVVMNSIVPEIFPNTRHCLFLWHVLMKVSENLG-QVVKQHDNF 393
Query: 399 KYEFHK-VYHLECEDDFEHKWTLMIAQFGLGMDKHIDLLYSNRQFWALAYLKDFFFAGMT 457
+F K +Y ++DF KW +A+FGL D+ + LY +R+ WA Y+ D AGM+
Sbjct: 394 MPKFEKCIYKSGKDEDFARKWYKNLARFGLKDDQWMISLYEDRKKWAPTYMTDVLLAGMS 453
Query: 458 TTGRSESINSYIKRFLHVRTSLTDFVNQVGIAVNIRNQAGEEARMRQKYHNPHIRTGFPI 517
T+ R++SIN++ +++H +TS+ +FV + R + +A P +++ P
Sbjct: 454 TSQRADSINAFFDKYMHKKTSVQEFVKVYDTVLQDRCEEEAKADSEMWNKQPAMKSPSPF 513
Query: 518 EEQAASMLTPYAFKLLQHEIELSTKYASTEEGENSYIVRFHT---KVDGGRIVKWIEEDK 574
E+ + + TP FK Q E+ + + EE ++ F + + +V W +
Sbjct: 514 EKSVSEVYTPAVFKKFQIEVLGAIACSPREENRDATCSTFRVQDFENNQDFMVTWNQTKA 573
Query: 575 SINCSCKEFEFSGILCRHAIRVLVMKNYFHLPTKYLPFRWRQESSLVPTSSHIINCNDGS 634
++C C+ FE+ G LCRH + VL + +P++Y+ RW + D
Sbjct: 574 EVSCICRLFEYKGYLCRHTLNVLQCCHLSSIPSQYILKRWTK---------------DAK 618
Query: 635 SFEFRSLVQWLQVESLKTKDREEVAIR 661
S F Q LQ L+ D E A++
Sbjct: 619 SRHFSGEPQQLQTRLLRYNDLCERALK 645
>At3g06250 unknown protein
Length = 764
Score = 231 bits (590), Expect = 9e-61
Identities = 161/572 (28%), Positives = 266/572 (46%), Gaps = 65/572 (11%)
Query: 54 PYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGI 113
PY G FNS +A FY +A GF +R +++S + S I R FVC + G
Sbjct: 190 PYAGLEFNSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGS------ITSRRFVCSKEGF 243
Query: 114 TKQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQLLDDKEVQF 173
+ SR CGA M + + G W+V + HNH L K+
Sbjct: 244 --------------QHPSRMGCGAYMRIKRQDSG---GWIVDRLNKDHNHDLEPGKKNAG 286
Query: 174 LPAYRSIPIVDQDRIMLLSKAGCSISLIIRVLELEKGIDTCTLPFLERDIRNFIQS--QS 231
+ I D ++ G+D+ L L D+ N I S ++
Sbjct: 287 MKK-----ITD---------------------DVTGGLDSVDLIELN-DLSNHISSTREN 319
Query: 232 SIGKESDASNVLKLCKSLKDRDDSFKYDFTLDENNKLEHIIWAFGDSIRAYEAFGDVVVF 291
+IGKE +L +S + D F Y LD N I WA S A FGD VVF
Sbjct: 320 TIGKEWYPV-LLDYFQSKQAEDMGFFYAIELDSNGSCMSIFWADSRSRFACSQFGDAVVF 378
Query: 292 DTTYRINRYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFVNGKHPKTI 351
DT+YR Y +P ++G ++H + G L+ DE +FSW +++L ++G+ P+++
Sbjct: 379 DTSYRKGDYSVPFATFIGFNHHRQPVLLGGALVADESKEAFSWLFQTWLRAMSGRRPRSM 438
Query: 352 LTDQDVALKEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGSRYNDFKYEFHK-VYHLEC 410
+ DQD+ +++A+A P T H F W I +K +L S N+FKYE+ K +Y +
Sbjct: 439 VADQDLPIQQAVAQVFPGTHHRFSAWQIRSKERE----NLRSFPNEFKYEYEKCLYQSQT 494
Query: 411 EDDFEHKWTLMIAQFGLGMDKHIDLLYSNRQFWALAYLKDFFFAGMTTTGRSESINSYIK 470
+F+ W+ ++ ++GL + + +Y R+ W AYL+ FF G+ G + + +
Sbjct: 495 TVEFDTMWSSLVNKYGLRDNMWLREIYEKREKWVPAYLRASFFGGIHVDG---TFDPFYG 551
Query: 471 RFLHVRTSLTDFVNQVGIAVNIRNQAGEEARMRQKYHNPHIRTGFPIEEQAASMLTPYAF 530
L+ TSL +F+++ + R + + P ++T P+EEQ + T F
Sbjct: 552 TSLNSLTSLREFISRYEQGLEQRREEERKEDFNSYNLQPFLQTKEPVEEQCRRLYTLTIF 611
Query: 531 KLLQHEIELSTKYASTEEGENSYIVRFHTKVDGG----RIVKWIEEDKSINCSCKEFEFS 586
++ Q E+ S Y + E I RF + G V + + + +CSC+ FE+
Sbjct: 612 RIFQSELAQSYNYLGLKTYEEGAISRFLVRKCGNENEKHAVTFSASNLNASCSCQMFEYE 671
Query: 587 GILCRHAIRVLVMKNYFHLPTKYLPFRWRQES 618
G+LCRH ++V + + LP++Y+ RW + +
Sbjct: 672 GLLCRHILKVFNLLDIRELPSRYILHRWTKNA 703
Score = 50.1 bits (118), Expect = 5e-06
Identities = 32/112 (28%), Positives = 49/112 (43%), Gaps = 23/112 (20%)
Query: 54 PYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGI 113
PY G F++ +A D+Y +A ++GF +R +Y+S + + + R FVC + G
Sbjct: 28 PYVGLEFDTAEEARDYYNSYATRTGFKVRTGQLYRSRTDGT------VSSRRFVCSKEGF 81
Query: 114 TKQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQL 165
+ SR C A + V R G KWV+ HNH L
Sbjct: 82 --------------QLNSRTGCPAFIRVQRRDTG---KWVLDQIQKEHNHDL 116
>At5g18960 FAR1 - like protein
Length = 788
Score = 216 bits (551), Expect = 3e-56
Identities = 158/573 (27%), Positives = 257/573 (44%), Gaps = 64/573 (11%)
Query: 54 PYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGI 113
PY G F S +A FY +A GF +R +++S + S I R FVC R G
Sbjct: 211 PYAGLEFGSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGS------ITSRRFVCSREGF 264
Query: 114 TKQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQLLDDKEVQF 173
+ SR CGA M + + G W+V + HNH L K+
Sbjct: 265 --------------QHPSRMGCGAYMRIKRQDSG---GWIVDRLNKDHNHDLEPGKKND- 306
Query: 174 LPAYRSIPIVDQDRIMLLSKAGCSISLIIRVLELEKGIDTCTLPFLERDIRNFIQS--QS 231
+ IP + G+D+ L L N I+ ++
Sbjct: 307 -AGMKKIPD-----------------------DGTGGLDSVDLIELNDFGNNHIKKTREN 342
Query: 232 SIGKESDASNVLKLCKSLKDRDDSFKYDFTLDENN-KLEHIIWAFGDSIRAYEAFGDVVV 290
IGKE +L +S + D F Y LD NN I WA + A FGD VV
Sbjct: 343 RIGKEWYPL-LLDYFQSRQTEDMGFFYAVELDVNNGSCMSIFWADSRARFACSQFGDSVV 401
Query: 291 FDTTYRINRYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFVNGKHPKT 350
FDT+YR Y +P +G ++H + GC ++ DE +F W +++L ++G+ P++
Sbjct: 402 FDTSYRKGSYSVPFATIIGFNHHRQPVLLGCAMVADESKEAFLWLFQTWLRAMSGRRPRS 461
Query: 351 ILTDQDVALKEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGSRYNDFKYEFHK-VYHLE 409
I+ DQD+ +++A+ P H + W I K +L ++FKYE+ K +Y +
Sbjct: 462 IVADQDLPIQQALVQVFPGAHHRYSAWQIREKERE----NLIPFPSEFKYEYEKCIYQTQ 517
Query: 410 CEDDFEHKWTLMIAQFGLGMDKHIDLLYSNRQFWALAYLKDFFFAGMTTTGRSESINSYI 469
+F+ W+ +I ++GL D + +Y R+ W AYL+ FFAG+ G +I +
Sbjct: 518 TIVEFDSVWSALINKYGLRDDVWLREIYEQRENWVPAYLRASFFAGIPING---TIEPFF 574
Query: 470 KRFLHVRTSLTDFVNQVGIAVNIRNQAGEEARMRQKYHNPHIRTGFPIEEQAASMLTPYA 529
L T L +F+++ A+ R + + P ++T P+EEQ + T
Sbjct: 575 GASLDALTPLREFISRYEQALEQRREEERKEDFNSYNLQPFLQTKEPVEEQCRRLYTLTV 634
Query: 530 FKLLQHEIELSTKYASTEEGENSYIVRFHTKVDGG----RIVKWIEEDKSINCSCKEFEF 585
F++ Q+E+ S Y + E I RF + G V + + + +CSC+ FE
Sbjct: 635 FRIFQNELVQSYNYLCLKTYEEGAISRFLVRKCGNESEKHAVTFSASNLNSSCSCQMFEH 694
Query: 586 SGILCRHAIRVLVMKNYFHLPTKYLPFRWRQES 618
G+LCRH ++V + + LP++Y+ RW + +
Sbjct: 695 EGLLCRHILKVFNLLDIRELPSRYILHRWTKNA 727
Score = 51.2 bits (121), Expect = 2e-06
Identities = 33/119 (27%), Positives = 53/119 (43%), Gaps = 23/119 (19%)
Query: 54 PYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGI 113
PY G F++ +A +FY +A ++GF +R +Y+S + + + R FVC + G
Sbjct: 43 PYVGLEFDTAEEAREFYNAYAARTGFKVRTGQLYRSRTDGT------VSSRRFVCSKEGF 96
Query: 114 TKQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQLLDDKEVQ 172
+ SR C A + V R G KWV+ HNH+L + V+
Sbjct: 97 --------------QLNSRTGCTAFIRVQRRDTG---KWVLDQIQKEHNHELGGEGSVE 138
>At2g27110 Mutator-like transposase
Length = 851
Score = 214 bits (546), Expect = 1e-55
Identities = 173/629 (27%), Positives = 278/629 (43%), Gaps = 57/629 (9%)
Query: 26 DNGNTNSSTC--QEPQLEPLSLANNSADWIPYEGQIFNSEGDAYDFYCLFARKSGFSIRR 83
D G+ S C Q L + + P G FNSE +A FY ++R+ GF
Sbjct: 19 DEGDVEPSDCSGQNNMDNSLGVQDEIGIAEPCVGMEFNSEKEAKSFYDEYSRQLGF---- 74
Query: 84 DHIYKSTKNQSENNPLGIYKREFVCHRAGITKQSKVSEVESQRKRKTSRCNCGAKMLVTT 143
T + REFVC + + ++SE +C A + +
Sbjct: 75 ------TSKLLPRTDGSVSVREFVCSSSSKRSKRRLSE------------SCDAMVRIEL 116
Query: 144 RTIGFEEKWVVKYFHNHHNHQLLDDKEVQFLPAYRSIPIVDQDRIMLLSKAGCSISLIIR 203
+ EKWVV F H H L + L R A S
Sbjct: 117 QG---HEKWVVTKFVKEHTHGLASSNMLHCLRPRRHF-------------ANSEKSSYQE 160
Query: 204 VLELEKGIDTCTLPFLERDIRNF---IQSQSSIGKESDASNVLKLCKSLKDRDDSFKYDF 260
+ + G+ ++ R RN ++ +IG+ DA N+L+ K ++ + F Y
Sbjct: 161 GVNVPSGMMYVSMDANSRGARNASMATNTKRTIGR--DAHNLLEYFKRMQAENPGFFYAV 218
Query: 261 TLDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRINRYDMPLGIWVGVDNHGNSIFFG 320
LDE+N++ ++ WA S AY FGD V DT YR N++ +P + GV++HG +I FG
Sbjct: 219 QLDEDNQMSNVFWADSRSRVAYTHFGDTVTLDTRYRCNQFRVPFAPFTGVNHHGQAILFG 278
Query: 321 CVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQDVALKEAIAMELPNTKHAFCIWHIV 380
C L+ DE SF W K+FL + + P +++TDQD A++ A P +H W ++
Sbjct: 279 CALILDESDTSFIWLFKTFLTAMRDQPPVSLVTDQDRAIQIAAGQVFPGARHCINKWDVL 338
Query: 381 AKLSSWFSFSLGSRYNDFKYEFHK-VYHLECEDDFEHKWTLMIAQFGLGMDKHIDLLYSN 439
+ + + Y F+ E + + E ++FE W+ +I ++ LG + ++ LY+
Sbjct: 339 REGQEKLA-HVCLAYPSFQVELYNCINFTETIEEFESSWSSVIDKYDLGRHEWLNSLYNA 397
Query: 440 RQFWALAYLKDFFFAGM-TTTGRSESINSYIKRFLHVRTSLTDFVNQVGIAVNIRNQAGE 498
R W Y +D FFA + + G S S+ +++ +T+L F A+ +
Sbjct: 398 RAQWVPVYFRDSFFAAVFPSQGYS---GSFFDGYVNQQTTLPMFFRLYERAMESWFEMEI 454
Query: 499 EARMRQKYHNPHIRTGFPIEEQAASMLTPYAFKLLQHE-IELSTKYASTEEGE---NSYI 554
EA + P ++T P+E QAA++ T F Q E +E A+ E + +++
Sbjct: 455 EADLDTVNTPPVLKTPSPMENQAANLFTRKIFGKFQEELVETFAHTANRIEDDGTTSTFR 514
Query: 555 VRFHTKVDGGRIVKWIEEDKSINCSCKEFEFSGILCRHAIRVLVMKNYFHLPTKYLPFRW 614
V + IV + + NCSC+ FE SGILCRH + V + N LP Y+ RW
Sbjct: 515 VANFENDNKAYIVTFCYPEMRANCSCQMFEHSGILCRHVLTVFTVTNILTLPPHYILRRW 574
Query: 615 -RQESSLVPTSSHII-NCNDGSSFEFRSL 641
R S+V H+ N +D S + L
Sbjct: 575 TRNAKSMVELDEHVSENGHDSSIHRYNHL 603
>At2g32250 Mutator-like transposase
Length = 684
Score = 194 bits (493), Expect = 2e-49
Identities = 143/567 (25%), Positives = 246/567 (43%), Gaps = 58/567 (10%)
Query: 57 GQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGITKQ 116
G F S+ AY FY +AR GF I I S +++ + + + C R G TK+
Sbjct: 41 GMDFESKEAAYYFYREYARSVGFGIT---IKASRRSKRSGKFIDV---KIACSRFG-TKR 93
Query: 117 SKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQLLDDKEVQFLPA 176
K + + + KT C G M +EKWV+ F HNH++ D
Sbjct: 94 EKATAINPRSCPKTG-CKAGLHMKRKE-----DEKWVIYNFVKEHNHEICPDD------F 141
Query: 177 YRSIPIVDQDRIMLLSKAGCSISLIIRVLELEKGIDTCTLPFLERDIRNFIQSQSSIGKE 236
Y S+ ++ L K G ++L +E
Sbjct: 142 YVSVRGKNKPAGALAIKKGLQLAL----------------------------------EE 167
Query: 237 SDASNVLKLCKSLKDRDDSFKYDFTLDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYR 296
D +L+ ++D+ F Y D + ++ ++ W + Y +F DVV+FDT Y
Sbjct: 168 EDLKLLLEHFMEMQDKQPGFFYAVDFDSDKRVRNVFWLDAKAKHDYCSFSDVVLFDTFYV 227
Query: 297 INRYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQD 356
N Y +P ++GV +H + GC L+ + ++SW +++L V G+ P ++TDQD
Sbjct: 228 RNGYRIPFAPFIGVSHHRQYVLLGCALIGEVSESTYSWLFRTWLKAVGGQAPGVMITDQD 287
Query: 357 VALKEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGSRYNDFKYEFHKVYHLECEDD-FE 415
L + + P+ +H FC+W +++K+S + S+ + F F D+ FE
Sbjct: 288 KLLSDIVVEVFPDVRHIFCLWSVLSKISEMLN-PFVSQDDGFMESFGNCVASSWTDEHFE 346
Query: 416 HKWTLMIAQFGLGMDKHIDLLYSNRQFWALAYLKDFFFAGMTTTGRSESINSYIKRFLHV 475
+W+ MI +F L ++ + LL+ +R+ W Y AG++ RS SI S+ ++++
Sbjct: 347 RRWSNMIGKFELNENEWVQLLFRDRKKWVPHYFHGICLAGLSGPERSGSIASHFDKYMNS 406
Query: 476 RTSLTDFVNQVGIAVNIRNQAGEEARMRQKYHNPHIRTGFPIEEQAASMLTPYAFKLLQH 535
+ DF + R + + + P +R+ E+Q + + T AFK Q
Sbjct: 407 EATFKDFFELYMKFLQYRCDVEAKDDLEYQSKQPTLRSSLAFEKQLSLIYTDAAFKKFQA 466
Query: 536 EIELSTKYASTEEGENSYIVRFHTKVDGGR---IVKWIEEDKSINCSCKEFEFSGILCRH 592
E+ +E E+ F + R V E CSC FE+ G LC+H
Sbjct: 467 EVPGVVSCQLQKEREDGTTAIFRIEDFEERQNFFVALNNELLDACCSCHLFEYQGFLCKH 526
Query: 593 AIRVLVMKNYFHLPTKYLPFRWRQESS 619
AI VL + +P++Y+ RW ++ +
Sbjct: 527 AILVLQSADVSRVPSQYILKRWSKKGN 553
>At1g52520 F6D8.26
Length = 703
Score = 183 bits (464), Expect = 4e-46
Identities = 163/654 (24%), Positives = 290/654 (43%), Gaps = 64/654 (9%)
Query: 54 PYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGI 113
P G F S DAY++Y +A + GF +R + + +++ + Y C G
Sbjct: 85 PAVGMEFESYDDAYNYYNCYASEVGFRVRVKNSWFKRRSKEK------YGAVLCCSSQGF 138
Query: 114 TKQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQLLDDKEVQF 173
+ + V+ V RK +R C A ++ R + ++W V HNH LL K +
Sbjct: 139 KRINDVNRV-----RKETRTGCPA--MIRMRQVD-SKRWRVVEVTLDHNH-LLGCKLYKS 189
Query: 174 LPAYR---SIPIVDQDRIMLLSKAGCSISLIIRVLELEKGIDTCTLPFLERDIRNFIQSQ 230
+ R S P+ D I L R ++ G + L + +N S
Sbjct: 190 VKRKRKCVSSPVSDAKTIKLY-----------RACVVDNGSNVNPNSTLNKKFQNSTGSP 238
Query: 231 SSIG-KESDASNVLKLCKSLKDRDDSFKYDFTLDENNKLEHIIWAFGDSIRAYEAFGDVV 289
+ K D++ + ++ + +F Y +++ +L ++ WA S + FGDV+
Sbjct: 239 DLLNLKRGDSAAIYNYFCRMQLTNPNFFYLMDVNDEGQLRNVFWADAFSKVSCSYFGDVI 298
Query: 290 VFDTTYRINRYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFVNGKHPK 349
D++Y ++++PL + GV++HG + C L E + S+ W LK +L V + P+
Sbjct: 299 FIDSSYISGKFEIPLVTFTGVNHHGKTTLLSCGFLAGETMESYHWLLKVWL-SVMKRSPQ 357
Query: 350 TILTDQDVALKEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGS--RYNDFKYEFHK-VY 406
TI+TD+ L+ AI+ P + F + HI+ K+ LG Y+ + F K VY
Sbjct: 358 TIVTDRCKPLEAAISQVFPRSHQRFSLTHIMRKIPE----KLGGLHNYDAVRKAFTKAVY 413
Query: 407 HLECEDDFEHKWTLMIAQFGLGMDKHIDLLYSNRQFWALAYLKDFFFAGMTTTGRSESIN 466
+FE W M+ FG+ ++ + LY R WA YLKD FFAG+ E++
Sbjct: 414 ETLKVVEFEAAWGFMVHNFGVIENEWLRSLYEERAKWAPVYLKDTFFAGIAAAHPGETLK 473
Query: 467 SYIKRFLHVRTSLTDFVNQVGIAVNIRNQAGEEARMR-QKYHNPHIRTGFPIEEQAASML 525
+ +R++H +T L +F+++ +A+ +++ + + Q + ++T E Q + +
Sbjct: 474 PFFERYVHKQTPLKEFLDKYELALQKKHREETLSDIESQTLNTAELKTKCSFETQLSRIY 533
Query: 526 TPYAFKLLQHEIE-----LSTKYASTEEGENSYIVRFHTKVDGGR------IVKWIEEDK 574
T FK Q E+E ST + ++V+ + + R V +
Sbjct: 534 TRDMFKKFQIEVEEMYSCFSTTQVHVDGPFVIFLVKERVRGESSRREIRDFEVLYNRSVG 593
Query: 575 SINCSCKEFEFSGILCRHAIRVLVMKNYFHLPTKYLPFRWRQESSLVPTSSHIINCNDGS 634
+ C C F F G LCRHA+ VL +P +Y+ RWR++ + H +
Sbjct: 594 EVRCICSCFNFYGYLCRHALCVLNFNGVEEIPLRYILPRWRKDYKRL----HFADNGLTG 649
Query: 635 SFEFRSLVQW--------LQV--ESLKTKDREEVAIRELERVIRIITDMPEEQE 678
+ VQW LQV E + D +VA++ L+ + + + E+Q+
Sbjct: 650 FVDGTDRVQWFDQLYKNSLQVVEEGAVSLDHYKVAMQVLQESLDKVHSVEEKQD 703
>At4g19990 putative protein
Length = 672
Score = 177 bits (449), Expect = 2e-44
Identities = 164/685 (23%), Positives = 285/685 (40%), Gaps = 114/685 (16%)
Query: 31 NSSTCQEPQLEPLSLANNSADWIPYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKST 90
+S C QL+ N D EG+ F S+ +A++FY +A GF+ I K++
Sbjct: 2 SSGECSNVQLDDHRKNNLEID----EGREFESKEEAFEFYKEYANSVGFTT----IIKAS 53
Query: 91 KNQSENNPLGIYKREFVCHRAGITKQ-------SKVSEVESQRK-----RKTSRCNCGAK 138
+ +FVC R G K+ + + RK R +S+ +C A
Sbjct: 54 RRSRMTGK--FIDAKFVCTRYGSKKEDIDTGLGTDGFNIPQARKRGRINRSSSKTDCKAF 111
Query: 139 MLVTTRTIGFEEKWVVKYFHNHHNHQLLDDK--EVQFLPAYRSIPIVDQDRIMLLSKAGC 196
+ V R G +WVV+ HNH++ + ++ L R L K
Sbjct: 112 LHVKRRQDG---RWVVRSLVKEHNHEIFTGQADSLRELSGRRK-----------LEKLNG 157
Query: 197 SISLIIRVLELEKGIDTCTLPFLERDIRNFIQSQSSIGKESDASNVLKLCKSLKDRDDSF 256
+I ++ +LE G +ER + F QS
Sbjct: 158 AIVKEVKSRKLEDG-------DVERLLNFFTDMQS------------------------- 185
Query: 257 KYDFTLDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRINRYDMPLGIWVGVDNHGNS 316
L +I W Y F DVV DTT+ N Y +PL + GV++HG
Sbjct: 186 -----------LRNIFWVDAKGRFDYTCFSDVVSIDTTFIKNEYKLPLVAFTGVNHHGQF 234
Query: 317 IFFGC-VLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQDVALKEAIAMELPNTKHAFC 375
+ G +LL DE F W +++L ++G P+ ILT D LKEA+ P+++H F
Sbjct: 235 LLLGFGLLLTDESKSGFVWLFRAWLKAMHGCRPRVILTKHDQMLKEAVLEVFPSSRHCFY 294
Query: 376 IWHIVAKLSSWFS--FSLGSRYNDFKYEFHKVYHLECE-DDFEHKWTLMIAQFGLGMDKH 432
+W + ++ L + D E + + C+ +DFE W ++ +F + +
Sbjct: 295 MWDTLGQMPEKLGHVIRLEKKLVD---EINDAIYGSCQSEDFEKNWWEVVDRFHMRDNVW 351
Query: 433 IDLLYSNRQFWALAYLKDFFFAGMTTTGRSESINSYIKRFLHVRTSLTDFVNQVGIAVNI 492
+ LY +R++W Y+KD AGM T RS+S+NS + +++ +T+ F+ Q +
Sbjct: 352 LQSLYEDREYWVPVYMKDVSLAGMCTAQRSDSVNSGLDKYIQRKTTFKAFLEQYKKMIQE 411
Query: 493 RNQAGEEARMRQKYHNPHIRTGFPIEEQAASMLTPYAFKLLQHE----IELSTKYASTEE 548
R + E++ + Y P +++ P +Q A + T FK Q E + K S E+
Sbjct: 412 RYEEEEKSEIETLYKQPGLKSPSPFGKQMAEVYTREMFKKFQVEVLGGVACHPKKESEED 471
Query: 549 GENSYIVRFHT-KVDGGRIVKWIEEDKSINCSCKEFEFSGILCRHAIRVLVMKNYFHLPT 607
G N R + + +V W E + CSC+ FE G L +P+
Sbjct: 472 GVNKRTFRVQDYEQNRSFVVVWNSESSEVVCSCRLFELKGEL--------------SIPS 517
Query: 608 KYLPFRWRQESSLVPTSSHIINCNDGSSFEFRSLVQW--LQVESLKTKDREEVAIRELER 665
+Y+ RW +++ S + +D + E ++ L + SLK + ++
Sbjct: 518 QYVLKRWTKDA-----KSREVMESDQTDVESTKAQRYKDLCLRSLKLSEEASLSEESYNA 572
Query: 666 VIRIITDMPEEQEHAVDMEPNVPNS 690
V+ ++ + + E+ ++ N+ S
Sbjct: 573 VVNVLNEALRKWENKSNLIQNLEES 597
>At1g80010 hypothetical protein
Length = 696
Score = 173 bits (438), Expect = 4e-43
Identities = 157/615 (25%), Positives = 264/615 (42%), Gaps = 101/615 (16%)
Query: 54 PYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGI 113
P G F S DAY FY +AR+ GF+IR + ++ + + C+ G
Sbjct: 66 PTPGMEFESYDDAYSFYNSYARELGFAIRVKSSWTKRNSKEKRGAV------LCCNCQGF 119
Query: 114 TKQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWV--VKYFHNH-------HNHQ 164
++ +RK +R C A ++ R I F+ V VK HNH HN +
Sbjct: 120 KLLK-----DAHSRRKETRTGCQA--MIRLRLIHFDRWKVDQVKLDHNHSFDPQRAHNSK 172
Query: 165 LLDDKEVQFLPAYRSIPIVDQDRIMLLSKAGCSISLIIRVLELEKGIDTCTLPFLERDIR 224
PA ++ P + +R ++L + + T P L +
Sbjct: 173 SHKKSSSSASPATKTNPEPPPH-------------VQVRTIKLYRTLALDTPPALGTSLS 219
Query: 225 NFIQSQSSIGKESDASNVLKLCKSLKDRDD----------SFKYDFTLDENNKLEHIIWA 274
+ S S+ +S L+L + D +F Y L ++ L ++ W
Sbjct: 220 SGETSDLSLD-HFQSSRRLELRGGFRALQDFFFQIQLSSPNFLYLMDLADDGSLRNVFWI 278
Query: 275 FGDSIRAYEAFGDVVVFDTTYRINRYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSW 334
+ AY FGDV++FDTT N Y++PL +VG+++HG++I GC LL D+ ++ W
Sbjct: 279 DARARAAYSHFGDVLLFDTTCLSNAYELPLVAFVGINHHGDTILLGCGLLADQSFETYVW 338
Query: 335 ALKSFLYFVNGKHPKTILTDQDVALKEAIAMELPNTKHAFCIWHIVAKL---------SS 385
+++L + G+ P+ +T+Q A++ A++ P H + H++ + S
Sbjct: 339 LFRAWLTCMLGRPPQIFITEQCKAMRTAVSEVFPRAHHRLSLTHVLHNICQSVVQLQDSD 398
Query: 386 WFSFSLGSRYNDFKYEFHKVYHLECEDDFEHKWTLMIAQFGLGMDKHIDLLYSNRQFWAL 445
F +L N Y KV ++FE W MI +FG+ ++ I ++ +R+ WA
Sbjct: 399 LFPMAL----NRVVYGCLKV------EEFETAWEEMIIRFGMTNNETIRDMFQDRELWAP 448
Query: 446 AYLKDFFFAGMTTTGRSESINSYI-KRFLHVRTSLTDFVNQVGIAVNIRNQAGEEARMRQ 504
YLKD F AG T +I ++H TSL +F+ G E+ + +
Sbjct: 449 VYLKDTFLAGALTFPLGNVAAPFIFSGYVHENTSLREFLE------------GYESFLDK 496
Query: 505 KYHN------------PHIRTGFPIEEQAASMLTPYAFKLLQHEIE-LSTKYASTEEGEN 551
KY P ++T P E Q A + T F+ Q E+ +S+ + T+ N
Sbjct: 497 KYTREALCDSESLKLIPKLKTTHPYESQMAKVFTMEIFRRFQDEVSAMSSCFGVTQVHSN 556
Query: 552 ----SYIVRFHTKVDGGRIVKWIEEDKSIN-----CSCKEFEFSGILCRHAIRVLVMKNY 602
SY+V+ + D R + I E + C C F F+G CRH + +L
Sbjct: 557 GSASSYVVK-EREGDKVRDFEVIYETSAAAQVRCFCVCGGFSFNGYQCRHVLLLLSHNGL 615
Query: 603 FHLPTKYLPFRWRQE 617
+P +Y+ RWR++
Sbjct: 616 QEVPPQYILQRWRKD 630
>At4g38170 hypothetical protein
Length = 531
Score = 166 bits (420), Expect = 4e-41
Identities = 113/416 (27%), Positives = 193/416 (46%), Gaps = 14/416 (3%)
Query: 237 SDASNVLKLCKSLKDRDDSFKYDFTLDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYR 296
S +VL K + + F Y D N + WA Y FGD +VFDTTYR
Sbjct: 2 SRVEHVLNYLKRRQLENPGFLYAIEDDCGN----VFWADPTCRLNYTYFGDTLVFDTTYR 57
Query: 297 IN-RYDMPLGIWVGVDNHGNSIFFGCVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQ 355
RY +P + G ++HG + FGC L+ +E SF+W +++L ++ P +I +
Sbjct: 58 RGKRYQVPFAAFTGFNHHGQPVLFGCALILNESESSFAWLFQTWLQAMSAPPPPSITVEP 117
Query: 356 DVALKEAIAMELPNTKHAFCIWHIVAKLSSWFSFSLGSRYNDFKYEF-HKVYHLECEDDF 414
D ++ A++ T+ F I + + + + F+ EF + V E +F
Sbjct: 118 DRLIQVAVSRVFSQTRLRFSQPLIFEETEEKLAHVFQA-HPTFESEFINCVTETETAAEF 176
Query: 415 EHKWTLMIAQFGLGMDKHIDLLYSNRQFWALAYLKDFFFAGMTTTGRSESINSYIKRFLH 474
E W ++ ++ + + + +Y+ RQ W +++D F+ ++T S +NS+ + F+
Sbjct: 177 EASWDSIVRRYYMEDNDWLQSIYNARQQWVRVFIRDTFYGELSTNEGSSILNSFFQGFVD 236
Query: 475 VRTSLTDFVNQVGIAVNIRNQAGEEARMRQKYHNPHIRTGFPIEEQAASMLTPYAFKLLQ 534
T++ + Q A++ + +A P ++T P+E+QAAS+ T AF Q
Sbjct: 237 ASTTMQMLIKQYEKAIDSWREKELKADYEATNSTPVMKTPSPMEKQAASLYTRAAFIKFQ 296
Query: 535 HE----IELSTKYASTEEGENSYIVRFHTKVDGGRIVKWIEEDKSINCSCKEFEFSGILC 590
E + + S +Y V +V G V + + NCSC+ FE+SGI+C
Sbjct: 297 EEFVETLAIPANIISDSGTHTTYRVAKFGEVHKGHTVSFDSLEVKANCSCQMFEYSGIIC 356
Query: 591 RHAIRVLVMKNYFHLPTKYLPFRWRQESSLVPTSSH---IINCNDGSSFEFRSLVQ 643
RH + V KN LP++YL RW +E+ + T C + + F SL Q
Sbjct: 357 RHILAVFSAKNVLALPSRYLLRRWTKEAKIRGTEEQPEFSNGCQESLNLCFNSLRQ 412
>At3g59470 unknown protein
Length = 251
Score = 67.0 bits (162), Expect = 4e-11
Identities = 41/113 (36%), Positives = 61/113 (53%), Gaps = 11/113 (9%)
Query: 54 PYEGQIFNSEGDAYDFYCLFARKSGFSIRRDHIYKSTKNQSENNPLGIYKREFVCHRAGI 113
PY GQ F SE A+ FY +A K GF IR + K ++++ + +P+G R+ VC++ G
Sbjct: 70 PYVGQEFESEAAAHGFYNAYATKVGFVIR---VSKLSRSRHDGSPIG---RQLVCNKEGY 123
Query: 114 TKQSKVSEVESQRKRKTSRCNCGAKMLVTTRTIGFEEKWVVKYFHNHHNHQLL 166
SK +V R+R +R C A +L+ G KWV+ F HNH L+
Sbjct: 124 RLPSKRDKV--IRQRAETRVGCKAMILIRKENSG---KWVITKFVKEHNHSLM 171
>At4g09380 putative protein
Length = 960
Score = 65.1 bits (157), Expect = 1e-10
Identities = 78/338 (23%), Positives = 130/338 (38%), Gaps = 39/338 (11%)
Query: 262 LDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRINRYDMPLGIWVGVD--NHGNSIFF 319
L+ K +++ A G I + A V+V D T+ Y L I D +H + F
Sbjct: 384 LEGEKKFKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGMLVIATAQDPNHHHYPLAF 443
Query: 320 GCVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQDVALKEAIAMELPNTKHAFCIWHI 379
G ++ EK S+ W L++ + ++D+ ++K+A+ PN HA CIWH+
Sbjct: 444 G--IIDSEKDVSWIWFLENLKTVYSDVPGLVFISDRHQSIKKAVKTVYPNALHAACIWHL 501
Query: 380 VAKLSSWFSFSLGSRYNDFKYEFHKVYHLECEDDFEHKWTLMIAQFGLGMDKHIDLLYS- 438
+ F+ H E E +F H +L K D L
Sbjct: 502 CQNMRDRVKIDKDGAAVKFRDCAHAYTESEFEKEFGHFTSL--------WPKAADFLVKV 553
Query: 439 NRQFWALAYLKDFFFAGMTTTGRSESINSYIK--RFLHVRTSLTDFVNQVGIAVNIRNQA 496
+ W+ + K + + T+ +ESIN K R H+ + +++ N QA
Sbjct: 554 GFEKWSRCHFKGDKY-NIDTSNSAESINGVFKKARKYHLLPMIDVMISKFSEWFNEHRQA 612
Query: 497 GEEARMRQKYHNPHIRTGFPIEEQAASMLTPYAFKLLQHEIELSTKYASTEEGENSYIVR 556
PI Q + P +L +++ K E NSY
Sbjct: 613 SGSC---------------PITAQ----VVPTVENILHIRCQVAAKLTVFE--LNSYNQE 651
Query: 557 FHTKVDGGRIVKWIEEDKSINCSCKEFEFSGILCRHAI 594
++ V V ++ + K +CSCK F+ I C HA+
Sbjct: 652 YN--VIDLNSVSFLVDLKMKSCSCKRFDIDKIPCVHAM 687
>At4g28970 putative protein
Length = 914
Score = 63.5 bits (153), Expect = 4e-10
Identities = 78/338 (23%), Positives = 130/338 (38%), Gaps = 39/338 (11%)
Query: 262 LDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRINRYDMPLGIWVGVD--NHGNSIFF 319
L+ K +++ A G I + A V+V D T+ Y L I D +H + F
Sbjct: 384 LEGEKKFKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGMLVIATAQDPNHHHYPLAF 443
Query: 320 GCVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQDVALKEAIAMELPNTKHAFCIWHI 379
G ++ EK S+ W L+ + ++D+ ++K+A+ PN HA CIWH+
Sbjct: 444 G--IIDSEKDVSWIWFLEKLKTVYSDVPGLVFISDRHQSIKKAVKTVYPNALHAACIWHL 501
Query: 380 VAKLSSWFSFSLGSRYNDFKYEFHKVYHLECEDDFEHKWTLMIAQFGLGMDKHIDLLYS- 438
+ + F+ H E E +F H +L K D L
Sbjct: 502 CQNMRDRVTIDKDGAAVKFRDCAHAYTESEFEKEFGHFTSL--------WPKAADFLVKV 553
Query: 439 NRQFWALAYLKDFFFAGMTTTGRSESINSYIK--RFLHVRTSLTDFVNQVGIAVNIRNQA 496
+ W+ + K + + T+ +ESIN K R H+ + +++ N QA
Sbjct: 554 GFEKWSRCHFKGDKY-NIDTSNSAESINGVFKKARKYHLLPMIDVMISKFSEWFNEHRQA 612
Query: 497 GEEARMRQKYHNPHIRTGFPIEEQAASMLTPYAFKLLQHEIELSTKYASTEEGENSYIVR 556
PI Q + P +L +++ K E NSY
Sbjct: 613 SGSC---------------PITAQ----VVPTVENILHIRCQVAAKLTVFE--LNSYNQE 651
Query: 557 FHTKVDGGRIVKWIEEDKSINCSCKEFEFSGILCRHAI 594
++ V V ++ + K +CSCK F+ I C HA+
Sbjct: 652 YN--VIDLNSVSFLVDLKMKSCSCKCFDIDKIPCVHAM 687
>At3g47310 putative protein
Length = 735
Score = 59.3 bits (142), Expect = 8e-09
Identities = 75/338 (22%), Positives = 123/338 (36%), Gaps = 39/338 (11%)
Query: 262 LDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRINRYDMPLGIWVGVD--NHGNSIFF 319
L+ K +++ A G I + V+V D T+ Y L I D +H + F
Sbjct: 333 LEGEKKFKYLFIALGACIEGFRTMRKVIVVDATHLKTVYGGMLVIATAQDPNHHHYPLAF 392
Query: 320 GCVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQDVALKEAIAMELPNTKHAFCIWHI 379
G ++ E S+ W L+ + ++D+ ++K+ + PN HA CIWH+
Sbjct: 393 G--IIDSENDVSWIWFLEKLKTVYSDVPGLVFISDRHQSIKKVVKTVYPNALHAACIWHL 450
Query: 380 VAKLSSWFSFSLGSRYNDFKYEFHKVYHLECEDDFEHKWTLMIAQFGLGMDKHIDLLYS- 438
+ F+ H E E +F H +L K D L
Sbjct: 451 CQNMRDRVKIDKDGAAVKFRDCAHAYTESEFEKEFGHFTSL--------WPKAADFLVKV 502
Query: 439 NRQFWALAYLKDFFFAGMTTTGRSESINSYIK--RFLHVRTSLTDFVNQVGIAVNIRNQA 496
+ W+ + K + + T+ +ESIN K R H+ + +++ N Q
Sbjct: 503 GFEKWSRCHFKGDKY-NIDTSNSAESINGVFKKARKYHLLPMIDVMISKFSEWFNEHRQD 561
Query: 497 GEEARMRQKYHNPHIRTGFPIEEQAASMLTPYAFKLLQHEIELSTKYASTEEGENSYIVR 556
P P E + P A KL E+ NSY
Sbjct: 562 SSSC--------PITAQVVPTVENILHIRCPIAAKLTVFEL-------------NSYNQE 600
Query: 557 FHTKVDGGRIVKWIEEDKSINCSCKEFEFSGILCRHAI 594
++ V V ++ + K +CSCK F+ I C HA+
Sbjct: 601 YN--VIDLNSVSFLVDLKMKSCSCKCFDIDKIPCVHAM 636
>At5g27190 putative protein
Length = 779
Score = 58.9 bits (141), Expect = 1e-08
Identities = 74/336 (22%), Positives = 126/336 (37%), Gaps = 34/336 (10%)
Query: 262 LDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRINRYDMPLGIWVGVD-NHGNSIFFG 320
+D K +++ A G I ++ VV+ D T+ Y L D NH N I
Sbjct: 367 VDAQQKFKYLFVALGACIEGFKVMRKVVIVDATFLKTVYGGMLVFATAQDPNHHNYIIAS 426
Query: 321 CVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQDVALKEAIAMELPNTKHAFCIWHIV 380
V+ R E S+SW + ++D+ ++ ++I PN +H C+WH
Sbjct: 427 AVIDR-ENDASWSWFFNKLKTVIPDVPGLVFVSDRHQSIIKSIMQVFPNARHGHCVWH-- 483
Query: 381 AKLSSWFSFSLGSRYNDFKYEFHKVYHLECEDDFEHKWTLMIAQFGLGMDKHIDLLYSNR 440
LS + + + F H+ + +F ++ +F + ++D ++
Sbjct: 484 --LSQNVKVRVKTEKEEAAANFIACAHVYTQFEFTREYARFRRRFP-NVGPYLD-RWAPV 539
Query: 441 QFWALAYLKDFFFAGMTTTGRSESINSYIKRF-LHVRTSLTDFVNQVGIAVNIRNQAGEE 499
+ WA+ Y + + + T+ ES+NS +R + L D I I +
Sbjct: 540 ENWAMCYFEGDRY-NIDTSNACESLNSTFERARKYFLLPLLD-----AIIEKISEWFNKH 593
Query: 500 ARMRQKYHNPHIRTGFPIEEQAASMLTPYAFKLLQHEI-ELSTKYASTEEGENSYIVRFH 558
+ KY R P+ E L P A + E+ +Y+ EG SY V
Sbjct: 594 RKESVKYSK--TRGLVPVVENEIHSLCPIAKTMPVTELNSFERQYSVIGEGGISYYVDLL 651
Query: 559 TKVDGGRIVKWIEEDKSINCSCKEFEFSGILCRHAI 594
K CSC+ F+ C HAI
Sbjct: 652 RK----------------TCSCRRFDVDKYPCVHAI 671
>At1g36090 hypothetical protein
Length = 645
Score = 57.4 bits (137), Expect = 3e-08
Identities = 51/213 (23%), Positives = 86/213 (39%), Gaps = 14/213 (6%)
Query: 262 LDENNKLEHIIWAFGDSIRAYEAFGDVVVFDTTYRINRYDMPLGIWVGVD--NHGNSIFF 319
L+ K +++ A G I + A V+V D T+ Y L I D +H + F
Sbjct: 344 LEGEKKFKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGMLVIATAHDPNHHHYPLAF 403
Query: 320 GCVLLRDEKIPSFSWALKSFLYFVNGKHPKTILTDQDVALKEAIAMELPNTKHAFCIWHI 379
G ++ EK S+ W L+ + ++D+ ++K+A+ PN HA CIWH+
Sbjct: 404 G--IIDSEKDVSWIWFLEKLKTVYSDVPRLVFISDRHQSIKKAVKTVYPNALHAACIWHL 461
Query: 380 VAKLSSWFSFSLGSRYNDFKYEFHKVYHLECEDDFEHKWTLMIAQFGLGMDKHIDLLYS- 438
+ F+ H E E +F H +L K +D L
Sbjct: 462 CQNMRDRVKIDKDGAAVKFRDCAHAYTESEFEKEFGHFTSL--------WPKAVDFLVKV 513
Query: 439 NRQFWALAYLKDFFFAGMTTTGRSESINSYIKR 471
+ W+ + K + + T+ +ESIN K+
Sbjct: 514 GFEKWSRCHFKGDKY-NIDTSNSAESINGVFKK 545
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,184,704
Number of Sequences: 26719
Number of extensions: 822909
Number of successful extensions: 2250
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 2081
Number of HSP's gapped (non-prelim): 135
length of query: 757
length of database: 11,318,596
effective HSP length: 107
effective length of query: 650
effective length of database: 8,459,663
effective search space: 5498780950
effective search space used: 5498780950
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0079c.8


