

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0079b.3
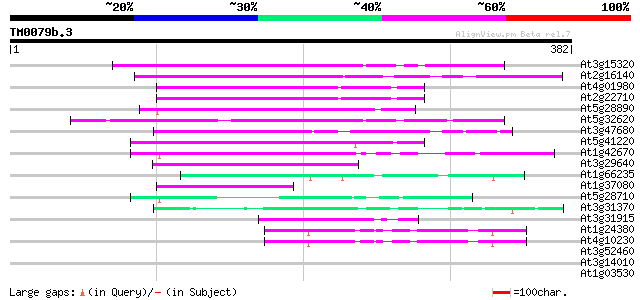
(382 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g15320 hypothetical protein 124 7e-29
At2g16140 pseudogene; similar to MURA transposase of maize Muta... 117 1e-26
At4g01980 hypothetical protein 116 2e-26
At2g22710 hypothetical protein 115 3e-26
At5g28890 putative protein 112 4e-25
At5g32620 putative protein 103 2e-22
At3g47680 unknown protein 103 2e-22
At5g41220 glutathione transferase-like 98 7e-21
At1g42670 unknown protein 98 7e-21
At3g29640 hypothetical protein 90 2e-18
At1g66235 putative protein 63 3e-10
At1g37080 hypothetical protein 60 2e-09
At5g28710 putative protein 58 8e-09
At3g31370 hypothetical protein 58 1e-08
At3g31915 hypothetical protein 50 3e-06
At1g24380 hypothetical protein 47 2e-05
At4g10230 hypothetical protein 46 4e-05
At3g52460 putative protein 39 0.006
At3g14010 unknown protein 37 0.018
At1g03530 unknown protein 37 0.018
>At3g15320 hypothetical protein
Length = 287
Score = 124 bits (312), Expect = 7e-29
Identities = 81/270 (30%), Positives = 127/270 (47%), Gaps = 13/270 (4%)
Query: 71 TQCEATPDDTQHEGLDDIDLEDEDQSSGK-KRTRWRVKDDLLLVQSWLNISKDPTVGTDQ 129
T+ A EG D EDE + R +W K+D++LV +WLN SKDP +G DQ
Sbjct: 9 TEVPAFSSQLSEEGSDLEGSEDEVKPKQSISRKKWTAKEDIVLVSAWLNTSKDPVIGNDQ 68
Query: 130 TAAKFWDRIRDQFDEYRDFD-TPPRTGKMLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHS 188
FW RI D P R K R+GK++K + F GCY TT SG S
Sbjct: 69 QGQSFWKRIAAYVAASPSLDGLPKREHAKCKHRWGKVNKSVTKFVGCYKTTTTHKTSGQS 128
Query: 189 EKDIMAEVHALFQVDHKKDFTHENVWRMVKDEPKWKGQSMKTNSRGQKKSGAGADGTSTD 248
E D+M + ++ D KK+FT ++ WR ++ + KW + + + K+ G DG ++
Sbjct: 129 EDDVMKLAYEIYFNDTKKNFTLDHAWRELRYDQKWCEATSRKGDKNAKRKKCG-DGNASS 187
Query: 249 PSASIDCDEYEATPPTTRPKGKKAEKRKAKTTDTASSTLSFAPHPDVLAMGKAK-MEMMA 307
++ D + PP G KA K KA+ S+T+ P + + +E
Sbjct: 188 QPIHVEDDSVMSRPP-----GVKAAKAKAR----KSATIKEGKKPATVKDDSGQSVEHFQ 238
Query: 308 NFREIRNRELDLQQADQQLKQSELQLRQEE 337
N E++ ++ D ++ + +Q E L + E
Sbjct: 239 NLWELKEKDWDRKEKQSKHQQLERLLSKTE 268
>At2g16140 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 311
Score = 117 bits (293), Expect = 1e-26
Identities = 80/292 (27%), Positives = 136/292 (46%), Gaps = 25/292 (8%)
Query: 86 DDIDLEDEDQSSGKKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEY 145
D LE+ ++R +W K+D++LV SWLN SKD +G +Q A FW RI +D
Sbjct: 42 DSPSLEEHVPKVKRERMKWSAKEDMVLVSSWLNTSKDAVIGNEQKANTFWSRIAAYYDAS 101
Query: 146 RDFD-TPPRTGKMLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEVHALFQVDH 204
+ R +K R+ K++ + F G Y + SG ++ D+++ H +F D+
Sbjct: 102 PQLNGLRKRMQGNIKQRWAKINDGVCKFVGSYEAASREKSSGQNDNDVISLAHEIFNNDY 161
Query: 205 KKDFTHENVWRMVKDEPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPT 264
F E+ WR+++ + KW Q+ S+ +K A TS PS ++
Sbjct: 162 GYKFPLEHAWRVLRHDQKWCSQA-SVMSKRRKCDKAAQPSTSQPPSHGVE-------EAM 213
Query: 265 TRPKGKKAEKRKAKTTDTASSTLSFAPHPDVLAMGKAKMEMMANFREIRNRELDLQQADQ 324
+RP G KA K KAK T T ++T V G A +E+ + + +++Q D
Sbjct: 214 SRPIGVKAAKAKAKKTVTKTTT--------VEDKGNAMLEIQSIW--------EIKQKDW 257
Query: 325 QLKQSELQLRQEELKFKKAENFRAYMDILNKNTSGMNDEELRTHNALRAFAL 376
+L+Q + + +E+ + K + ++ L + D E+ N L F L
Sbjct: 258 ELRQKDREQEKEDFEKKDRLSKTTLLESLIAKKEPLTDNEVTLKNKLIDFLL 309
>At4g01980 hypothetical protein
Length = 302
Score = 116 bits (290), Expect = 2e-26
Identities = 66/183 (36%), Positives = 93/183 (50%), Gaps = 5/183 (2%)
Query: 101 RTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFD-TPPRTGKMLK 159
R +W K+D++L+ +WLN SKDP VG +Q A FW RI D D P R K
Sbjct: 62 RKKWAAKEDIVLISAWLNTSKDPVVGNEQKAPAFWKRIASYVAANPDLDGVPKRASAQCK 121
Query: 160 CRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEVHALFQVDHKKDFTHENVWRMVKD 219
R+ K+++ + F GCY T SG +E D+M + LF+ D KK F+ ++ WR+++
Sbjct: 122 QRWAKMNELVMKFVGCYATTTNQKASGQTENDVMLFANELFENDMKKKFSLDHAWRLLRH 181
Query: 220 EPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKAKT 279
+ KW S +G K GT S ID ++ + RP G KA K KAK
Sbjct: 182 DQKWL-ISNAPKGKGIAKRRKVRVGTQAASSQPIDLEDDDV---MRRPPGVKAAKAKAKK 237
Query: 280 TDT 282
T T
Sbjct: 238 TPT 240
>At2g22710 hypothetical protein
Length = 300
Score = 115 bits (289), Expect = 3e-26
Identities = 66/183 (36%), Positives = 93/183 (50%), Gaps = 5/183 (2%)
Query: 101 RTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFD-TPPRTGKMLK 159
R +W K+D++L+ +WLN SKDP VG +Q A FW RI D D P R K
Sbjct: 60 RKKWAAKEDIVLISAWLNTSKDPVVGNEQKAPAFWKRIASYVAASPDLDGVPKRASAQCK 119
Query: 160 CRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEVHALFQVDHKKDFTHENVWRMVKD 219
R+ K+++ + F GCY T SG +E D+M + LF+ D KK F+ ++ WR+++
Sbjct: 120 QRWAKMNELVMKFVGCYATTTNQKASGQTENDVMLFANELFKNDMKKKFSLDHAWRLLRH 179
Query: 220 EPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKAKT 279
+ KW S +G K GT S ID ++ + RP G KA K KAK
Sbjct: 180 DQKWL-ISNAPKGKGIAKRRKVRVGTQAASSQPIDLEDDDV---MRRPPGVKAAKAKAKM 235
Query: 280 TDT 282
T T
Sbjct: 236 TPT 238
>At5g28890 putative protein
Length = 232
Score = 112 bits (279), Expect = 4e-25
Identities = 63/192 (32%), Positives = 98/192 (50%), Gaps = 7/192 (3%)
Query: 89 DLEDEDQSSGK---KRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEY 145
D + ED ++ K + +W K+D++L+ +WLN SKDP VG +Q A FW RI
Sbjct: 29 DEDHEDVNTPKVAIPKKKWSAKEDVILISAWLNTSKDPVVGNEQKAPAFWKRIATYVAAS 88
Query: 146 RDF-DTPPRTGKMLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEVHALFQVDH 204
D P R K R+ K+++ + F GCY T SG +E D+M + LF+ D
Sbjct: 89 PDLVGFPKRESAQCKQRWAKMNELVMKFVGCYATETNQKASGQTENDVMLFANELFENDM 148
Query: 205 KKDFTHENVWRMVKDEPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPT 264
KK F+ ++ WR+V+ E KW + R K+ G+ S+ P I+ ++Y+
Sbjct: 149 KKKFSLDHAWRLVRHEQKWIISNTPKEKRMSKRRKVGSQAKSSQP---INLEDYDVMARP 205
Query: 265 TRPKGKKAEKRK 276
R K KA+ +K
Sbjct: 206 LRVKVPKAKTKK 217
>At5g32620 putative protein
Length = 301
Score = 103 bits (256), Expect = 2e-22
Identities = 81/297 (27%), Positives = 124/297 (41%), Gaps = 20/297 (6%)
Query: 42 NPQYPMYPPQYQFQSQAPPTGSTGSKVSDTQCEATPDDTQHEGLDDIDLEDEDQSSGK-K 100
NP Y QSQ S T+ A EG + EDE +
Sbjct: 5 NPFYQSSGFMDLLQSQQEDMSQFAS--GSTEVPAFSSQLSEEGSELEGSEDEVKPKQSIS 62
Query: 101 RTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDTPPRTGKMLKC 160
R +W K+D++LV +WLN SKDP +G DQ A D P R K
Sbjct: 63 RKKWTAKEDIVLVSAWLNPSKDPVIGNDQQAYVAASPSLDGL--------PKREHAKCKQ 114
Query: 161 RFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEVHALFQVDHKKDFTHENVWRMVKDE 220
R+GK++K + F CY TT SG SE D+M + ++ D KK+FT ++ WR ++ +
Sbjct: 115 RWGKVNKSVTKFVACYKTTTTHKTSGQSEDDVMKLAYEIYFNDTKKNFTLDHAWRELRYD 174
Query: 221 PKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKAKTT 280
KW + + K+ G DG ++ ++ D + PP G KA K K + +
Sbjct: 175 QKWCEATSRKGDENAKRRKCG-DGNASSHPIHVEDDSIMSRPP-----GVKAAKAKGRKS 228
Query: 281 DTASSTLSFAPHPDVLAMGKAKMEMMANFREIRNRELDLQQADQQLKQSELQLRQEE 337
A D +E N E++ ++ D ++ + +Q E L + E
Sbjct: 229 AIVKEGKKPATVKDDSGQ---SVEHFQNLWELKEKDWDRKEKQSKHQQLERLLSKTE 282
>At3g47680 unknown protein
Length = 302
Score = 103 bits (256), Expect = 2e-22
Identities = 68/245 (27%), Positives = 122/245 (49%), Gaps = 19/245 (7%)
Query: 99 KKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFD-TPPRTGKM 157
++R +W +DL+LV +WLN SKD +G +Q FW RI + + R
Sbjct: 45 RERRKWSAGEDLVLVSAWLNTSKDAVIGNEQKGYAFWSRIAAYYGASPKLNGVEKRETGH 104
Query: 158 LKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEVHALFQVDHKKDFTHENVWRMV 217
+K R+ K+++ + F G Y T SG ++ D++A H ++ +H K FT E+ WR++
Sbjct: 105 IKQRWTKINEGVGKFVGSYEAATKQKSSGQNDDDVVALAHEIYNSEHGK-FTLEHAWRVL 163
Query: 218 KDEPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKA 277
+ E KW S K+ + +D +++ +EA +RP G KA K KA
Sbjct: 164 RFEQKWL-------SAPSTKATVMSKRRKSDKASTSQPQTHEAEEAMSRPIGVKAAKAKA 216
Query: 278 KTTDTASSTLSFAPHPDVLAMGKAKMEMMANFREIRNRELDLQQADQQLKQSELQLRQEE 337
K + ++T V G +E+ + + EI+ ++ +L+Q D++ ++ + + +QE
Sbjct: 217 KKAVSKTTT--------VEDKGNVMLEIQSIW-EIKQKDWELRQKDREQEKEDFE-KQER 266
Query: 338 LKFKK 342
L K
Sbjct: 267 LSRTK 271
>At5g41220 glutathione transferase-like
Length = 590
Score = 98.2 bits (243), Expect = 7e-21
Identities = 64/209 (30%), Positives = 96/209 (45%), Gaps = 11/209 (5%)
Query: 83 EGLDDIDLEDEDQSSGKKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQF 142
E DD +L K R +W +D +L+ +WLN SKDP V + A FW RI F
Sbjct: 252 EKSDDPNLVQNTTDRRKHRRKWSRAEDAILISAWLNTSKDPIVDNEHKACAFWKRIGAYF 311
Query: 143 DEYRDF-DTPPRTGKMLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEVHALFQ 201
+ + P R K R+ KL+ + F GCY++ SG SE D+ + ++
Sbjct: 312 NNSASLANLPKREPSHCKQRWSKLNDKVCKFVGCYDQALNQRSSGQSEDDVFQVAYQVYT 371
Query: 202 VDHKKDFTHENVWRMVKDEPKWKGQSMKTNSRG-------QKKSGAGADGTSTDP-SASI 253
++K +FT E+ WR ++ KW NS+G + +G +S++P S I
Sbjct: 372 NNYKSNFTLEHAWRELRHSKKWCSLYPFENSKGGGSSKRTKLNNGDRVYSSSSNPESVPI 431
Query: 254 DCDEYEATPPTTRPKGKKAEKRKAKTTDT 282
DE E P G K+ K+K K T
Sbjct: 432 ALDEEEQV--MDLPLGVKSSKQKEKKVAT 458
>At1g42670 unknown protein
Length = 579
Score = 98.2 bits (243), Expect = 7e-21
Identities = 80/300 (26%), Positives = 131/300 (43%), Gaps = 46/300 (15%)
Query: 83 EGLDDIDLEDEDQSSGKK----------RTRWRVKDDLLLVQSWLNISKDPTVGTDQTAA 132
E DD EDE +GKK R W +D++L+ WLN SKDP VG +Q A
Sbjct: 309 EWSDDDPSEDELPIAGKKGKKGKKAKKPRRNWSSTEDVVLISGWLNTSKDPVVGNEQKGA 368
Query: 133 KFWDRIRDQFDEYRDFDTPPRTGKM-LKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKD 191
FW+RI ++ + G + K R+ K++ + F G Y + SG ++ D
Sbjct: 369 AFWERIAVYYNSSPKLKGVEKRGHICCKQRWSKVNDAVNKFVGSYLAASKQQTSGQNDDD 428
Query: 192 IMAEVHALFQVDHKKDFTHENVWRMVKDEPKWKGQSMKTNSRGQKKSGAGADGTSTDPSA 251
+++ H +F D+ FT E+ WR + + KW QS ++ +K AD S
Sbjct: 429 VVSLAHQIFSKDYGCKFTCEHAWRERRYDQKWIAQSTHGKAKRRK---CEAD------SE 479
Query: 252 SIDCDEYEATPPTTRPKGKKAEKRKAKTTDTASSTLSFAPHPDVLAMGKAKMEMMANFRE 311
+ ++ EA RP G KA K AK A GKAK+ +
Sbjct: 480 PVGVEDKEA-----RPIGVKAAKAAAK------------------AKGKAKLSPVEGEET 516
Query: 312 IRNRELDLQQADQQLKQSELQLRQEELKFKKAENFRAYMDILNKNTSGMNDEELRTHNAL 371
+E+ Q+ ++K+ + +++ + K +N + + L T ++D E+ N L
Sbjct: 517 NALKEI---QSIWEIKEKDHAAKEKLIIIKDKKNRTKFFERLLGKTEPLSDIEIELKNKL 573
>At3g29640 hypothetical protein
Length = 171
Score = 89.7 bits (221), Expect = 2e-18
Identities = 47/142 (33%), Positives = 74/142 (52%), Gaps = 2/142 (1%)
Query: 98 GKKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFD-TPPRTGK 156
G + +W K+D++LV++WLN +KD +G DQ F +I D P R
Sbjct: 25 GLSQKKWFPKEDIVLVRAWLNTNKDHVIGNDQQCQSFLKQIASYVAISPQLDYLPKREHA 84
Query: 157 MLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEVHALFQVDHKKDFTHENVWRM 216
K R+ K++K + F GCY TT SG SE D+M + ++ D KK F E+ WR
Sbjct: 85 KCKQRWSKVNKSVTKFVGCYKTATTHKTSGQSEDDVMKLAYEIYYNDTKKKFNLEHTWRK 144
Query: 217 VKDEPKW-KGQSMKTNSRGQKK 237
++ + KW + ++K N ++K
Sbjct: 145 LRYDQKWCEAATIKGNKNAKRK 166
>At1g66235 putative protein
Length = 265
Score = 62.8 bits (151), Expect = 3e-10
Identities = 57/252 (22%), Positives = 100/252 (39%), Gaps = 29/252 (11%)
Query: 117 LNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDTPPRTGKMLKCRFGKLSKDIQFFTGCY 176
++IS+DP +G Q++ FWDR+ + F+ ++ R+ K L+CR + K + C
Sbjct: 1 MDISQDPIIGVYQSSDHFWDRVAESFENRKNPTWSKRSKKSLQCRLQTIEKASKKLHACI 60
Query: 177 NKVTTPWKSGHSEKDIMAEVHALFQVD--HKKDFTHENVWRMVKDEPKWKG--------- 225
SG S DI + + D K + ++VW ++++ K+K
Sbjct: 61 KLCENRRSSGASSDDIFNQAKEMLMQDKHFKSGWKFDHVWNIIRNFEKFKDGATQAKKVL 120
Query: 226 -----QSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKAKTT 280
++ +S Q SG+ + + D D D+ P RP K K K K
Sbjct: 121 NLCGLENPTLDSVSQASSGSFSFSLNLD-----DEDDIIGRSPYQRPIRVKKSKLKRKND 175
Query: 281 DTASSTLSFAPHPDVLAMGKAKMEMMANFREIRNRELDLQQADQQLKQ--SELQLRQEEL 338
+F K ME + R + L++Q L++ E ++ L
Sbjct: 176 QILDVIKTFEEG------NKQLMEQLKKTSAQRQQYLEMQSKSLALREQKEENKVLYRNL 229
Query: 339 KFKKAENFRAYM 350
+ N RAY+
Sbjct: 230 NSIEDPNLRAYL 241
>At1g37080 hypothetical protein
Length = 224
Score = 60.5 bits (145), Expect = 2e-09
Identities = 32/94 (34%), Positives = 49/94 (52%), Gaps = 1/94 (1%)
Query: 101 RTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDTP-PRTGKMLK 159
R +W K+D++L+ +WLN SKD VG +Q A FW R+ F + + R K
Sbjct: 68 RNKWTSKEDIVLISTWLNTSKDSVVGNEQRADAFWKRVVVYFASTLNVSSQIKREPSHSK 127
Query: 160 CRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIM 193
R+GK++K + F G + T SG +E +M
Sbjct: 128 QRWGKINKIVCMFGGSHEAANTQMASGMNEDVLM 161
>At5g28710 putative protein
Length = 264
Score = 58.2 bits (139), Expect = 8e-09
Identities = 55/240 (22%), Positives = 92/240 (37%), Gaps = 60/240 (25%)
Query: 83 EGLDDIDLEDEDQSSGKK-------RTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFW 135
E DD EDE +GKK R W +D++L+ WLN SKDP VG +Q A +
Sbjct: 47 EWSDDDPSEDEALIAGKKGKKAKKPRRNWSSIEDIVLICGWLNTSKDPMVGNEQKGAAYL 106
Query: 136 DRIRDQFDEYRDFDTPPRTGKMLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAE 195
+ Q SG ++ +++
Sbjct: 107 TASKQQ------------------------------------------TSGQNDDFVVSL 124
Query: 196 VHALFQVDHKKDFTHENVWRMVKDEPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDC 255
H +F D+ FT E+ WR ++ + KW QS ++ ++K A +D S+
Sbjct: 125 AHQIFSKDYGCKFTCEHAWRELRYDQKWIAQSTHGKAK-RRKCEADSD--------SVGV 175
Query: 256 DEYEATPPTTRPKGKKAEKRKAKTTDTASSTLSFAPHPDVLAMGKAKMEMMANFREIRNR 315
++ EA P + K KA K+ + ++ ++ A E + +E +NR
Sbjct: 176 EDKEARPISV--KAAKAAKQSPVEGEETNALKEIQSIWEIKDKDHAAKEKLIIIKEKKNR 233
>At3g31370 hypothetical protein
Length = 272
Score = 57.8 bits (138), Expect = 1e-08
Identities = 72/287 (25%), Positives = 103/287 (35%), Gaps = 84/287 (29%)
Query: 99 KKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDTPPRTGKML 158
K R +W + +D++L +WLN SKD VG
Sbjct: 62 KSRRKWGLTEDVVLSNAWLNTSKDH-VGC------------------------------- 89
Query: 159 KCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEVHALFQVDHKKDFTHENVWRMVK 218
C+F G Y T SG ++ D+M H F D+K T E+VWR V
Sbjct: 90 -CKF----------VGSYVATTKEKTSGENDNDVMNFAHHSFHTDYKTKLTLEHVWREVM 138
Query: 219 DEPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKAK 278
+ KW + Q+K S ++D + Y+ TRP KA K KAK
Sbjct: 139 FDQKWCSSNALNVPAKQRK-----QNESVGDHVTVDLEGYQ-----TRPIDVKAAKAKAK 188
Query: 279 TTDTASSTLSFAPHPDVLAMGKAKMEMMANFREIRNRELDLQQADQQLKQSELQLRQEEL 338
GKAK + + N D+Q +K+ L +++ L
Sbjct: 189 --------------------GKAKTRLTVE-GDANNTMTDMQYI-WGIKEKNLIFKEKPL 226
Query: 339 KFK--------KAENFRAYMDILNKNTSGMNDEELRTHNALRAFALS 377
K K N R +L KN S ++D EL + L LS
Sbjct: 227 NGKEKFLSEKNKLSNERLLESLLTKNES-VSDIELALKHKLITNMLS 272
>At3g31915 hypothetical protein
Length = 239
Score = 49.7 bits (117), Expect = 3e-06
Identities = 33/110 (30%), Positives = 53/110 (48%), Gaps = 12/110 (10%)
Query: 170 QFFTGCYNKVTTPWKSGHSEKDIMAEVHALFQVDHKKDFTHENVWRMVKDEPKW-KGQSM 228
Q G Y T SG ++ D++ H F D+K FT E+ WR V+ + KW ++
Sbjct: 61 QIDMGSYVAATKEKTSGQNDNDVLNFSHQSFYTDYKPKFTLEHAWREVRVDQKWCSSNAL 120
Query: 229 KTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKAK 278
++ +K++ + D + D ++ D+ TRP G KA K KAK
Sbjct: 121 NVPAKRRKQNESVGDHVTID----LEGDQ-------TRPIGVKATKGKAK 159
>At1g24380 hypothetical protein
Length = 273
Score = 46.6 bits (109), Expect = 2e-05
Identities = 48/189 (25%), Positives = 84/189 (44%), Gaps = 30/189 (15%)
Query: 174 GCYNKVTTPWKSGHSEKDIMAEVHALFQV--DHKKDFTHENVWRMVKDEPKWKGQSMKTN 231
GC N++ SG SE+DI+ + L +K+ F ++VW ++K K+ +MKT
Sbjct: 13 GCVNQIENKNPSGASEEDILNQAKMLLTQYEKYKRGFKFDHVWPILKGIEKFANDNMKTP 72
Query: 232 SRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKAKTTDTASSTLSFAP 291
+ Q G G D TS+ S SI+ T + P + D+ + S +
Sbjct: 73 AAFQ---GEGRDVTSSS-SFSIN------TESSPSPGMNSID----LNMDSEDANFSLSS 118
Query: 292 HPDVLAMGKAKMEMMANFREIRNRELDLQQADQQLK--------QSELQLRQEELKFKKA 343
P L K K + F+++ L+Q D+ +K ++E+Q ++ E+ K
Sbjct: 119 RPMGLKKAKRKQQSEEQFKQL------LEQNDKLIKAITKGTSERNEIQRQKIEVARMKE 172
Query: 344 ENFRAYMDI 352
EN + D+
Sbjct: 173 ENKILFADL 181
>At4g10230 hypothetical protein
Length = 273
Score = 45.8 bits (107), Expect = 4e-05
Identities = 48/189 (25%), Positives = 83/189 (43%), Gaps = 30/189 (15%)
Query: 174 GCYNKVTTPWKSGHSEKDIMAEVHALFQV--DHKKDFTHENVWRMVKDEPKWKGQSMKTN 231
GC N++ SG SE+DI+ + L +K+ F ++VW ++K K+ +MKT
Sbjct: 13 GCVNQIENKNPSGASEEDILNQAKMLLTQYEKYKRGFKFDHVWPILKGIEKFANDNMKTP 72
Query: 232 SRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKAKTTDTASSTLSFAP 291
Q G G D TS+ S SI+ T + P + D+ + S +
Sbjct: 73 PAFQ---GEGRDVTSSS-SFSIN------TESSPSPGMNSID----LNMDSEDANFSLSS 118
Query: 292 HPDVLAMGKAKMEMMANFREIRNRELDLQQADQQLK--------QSELQLRQEELKFKKA 343
P L K K + F+++ L+Q D+ +K ++E+Q ++ E+ K
Sbjct: 119 RPMGLKKAKRKQQSEEQFKQL------LEQNDKLIKAITKGTSERNEIQRQKIEVARMKE 172
Query: 344 ENFRAYMDI 352
EN + D+
Sbjct: 173 ENKILFADL 181
>At3g52460 putative protein
Length = 300
Score = 38.5 bits (88), Expect = 0.006
Identities = 25/68 (36%), Positives = 31/68 (44%), Gaps = 14/68 (20%)
Query: 3 PYPPQNQPHHPGGNFQNSQYQMFP----YTSQNQPLHPDENFTNPQYPMYPPQYQFQSQA 58
P PPQ+QP P +Q Q +P Y +QP P N+ N Y YP +QA
Sbjct: 30 PPPPQSQPPPP-----QTQQQTYPPVMGYPGYHQPPPPYPNYPNAPYQQYP-----YAQA 79
Query: 59 PPTGSTGS 66
PP GS
Sbjct: 80 PPASYYGS 87
Score = 33.1 bits (74), Expect = 0.26
Identities = 14/45 (31%), Positives = 22/45 (48%)
Query: 4 YPPQNQPHHPGGNFQNSQYQMFPYTSQNQPLHPDENFTNPQYPMY 48
YP +QP P N+ N+ YQ +PY + ++ Q P+Y
Sbjct: 53 YPGYHQPPPPYPNYPNAPYQQYPYAQAPPASYYGSSYPAQQNPVY 97
>At3g14010 unknown protein
Length = 595
Score = 37.0 bits (84), Expect = 0.018
Identities = 23/61 (37%), Positives = 28/61 (45%), Gaps = 11/61 (18%)
Query: 1 MSPYPPQNQP---------HHPGGNFQNSQYQMFPYTSQNQPLHPDENFTNPQYPMYPPQ 51
+ PYP NQP +HP G Q Q QM P Q Q + P + P Y M+PP
Sbjct: 525 LPPYPG-NQPQMMYHPQAYYHPNGQPQYPQQQMIP-GQQQQQMIPGQQHPRPVYYMHPPP 582
Query: 52 Y 52
Y
Sbjct: 583 Y 583
>At1g03530 unknown protein
Length = 801
Score = 37.0 bits (84), Expect = 0.018
Identities = 32/107 (29%), Positives = 47/107 (43%), Gaps = 19/107 (17%)
Query: 4 YPPQNQPHHPGGNFQNSQYQMFPYTSQNQPLHPDENFTNPQY-----PMYPPQYQFQSQA 58
+PPQ+Q H P N Q+QM P +Q ++ NPQY P P QFQ QA
Sbjct: 683 HPPQSQMHRPQSQM-NPQFQMPPQFQPHQ-----QSPMNPQYQMMHRPQSPANPQFQMQA 736
Query: 59 PPTGSTGSKVSDTQCEATPDDTQHEGLDDIDLEDEDQSSGKKRTRWR 105
+ + +Q +P D Q ++ + + SSG+ R R
Sbjct: 737 ----QSDVRPLQSQIPQSPSDLQ----SPMEPQSQGFSSGQSSERGR 775
Score = 30.8 bits (68), Expect = 1.3
Identities = 22/75 (29%), Positives = 27/75 (35%), Gaps = 1/75 (1%)
Query: 1 MSPYPPQNQPHHPGGNFQNSQYQMFPYTSQNQPLHPDENFTNPQYPMYPPQYQFQSQAPP 60
M +PP N P N QN YQ+ P +Q P N M+ PQ+ P
Sbjct: 568 MDGFPPNNAAWRPQSNQQNP-YQLPPIPNQMGMQIPFMAMQNQNQMMFQPQFNGGQMPMP 626
Query: 61 TGSTGSKVSDTQCEA 75
G G Q A
Sbjct: 627 GGPGGLNFFPGQASA 641
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.129 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,707,833
Number of Sequences: 26719
Number of extensions: 452605
Number of successful extensions: 1865
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 1711
Number of HSP's gapped (non-prelim): 184
length of query: 382
length of database: 11,318,596
effective HSP length: 101
effective length of query: 281
effective length of database: 8,619,977
effective search space: 2422213537
effective search space used: 2422213537
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0079b.3


