

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0078.10
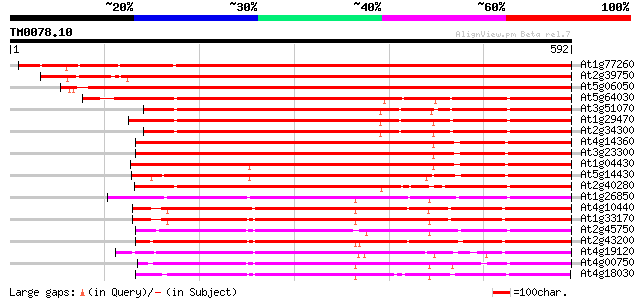
(592 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g77260 unknown protein 843 0.0
At2g39750 unknown protein 800 0.0
At5g06050 ankyrin-like protein 798 0.0
At5g64030 ankyrin-like protein 464 e-131
At3g51070 putative protein 464 e-131
At1g29470 unknown protein 457 e-129
At2g34300 unknown protein 451 e-127
At4g14360 450 e-127
At3g23300 ankyrin repeat protein 447 e-125
At1g04430 unknown protein 446 e-125
At5g14430 unknown protein 440 e-124
At2g40280 unknown protein 435 e-122
At1g26850 unknown protein 409 e-114
At4g10440 putative protein 402 e-112
At1g33170 unknown protein 402 e-112
At2g45750 hypothetical protein 395 e-110
At2g43200 hypothetical protein 394 e-109
At4g19120 ERD3 protein (ERD3) 393 e-109
At4g00750 unknown protein 377 e-104
At4g18030 unknown protein 376 e-104
>At1g77260 unknown protein
Length = 655
Score = 843 bits (2178), Expect = 0.0
Identities = 399/598 (66%), Positives = 477/598 (79%), Gaps = 20/598 (3%)
Query: 10 IDVLKASSSVKITALTLFSLTLLLFLFTRFSPTTNSP-LTFFLPSEQQPP---------- 58
+DV+K VK+ A S++ + FLF FS + + P L F + S
Sbjct: 8 LDVVKTPRLVKLIAFAFLSISTI-FLFNHFSDSFSYPSLPFPISSSSNVTEAIQTNITSV 66
Query: 59 ---LPPPPPPPRVKVFRLPPLPP-VERMGLVDENGVMTDNFTVGDSDLGSVVDLLSWKNE 114
P PPP PR+K+ PPLPP V R G+++ENG M+D+F +G D S+ +L S
Sbjct: 67 AAVAPSPPPRPRLKISP-PPLPPTVVRTGIINENGAMSDSFEIGGFDPDSIDELKSATGN 125
Query: 115 SRGVVEEEGAAAKVVVEKFKVCDSGMVDYVPCLDNAEAVARLNGSERGEKYERHCPEEGK 174
S V E+E +EK K+CD +DY+PCLDN E + RLN ++RGE YERHCP++
Sbjct: 126 S-SVEEKESPEVGFQIEKLKLCDKTKIDYIPCLDNEEEIKRLNNTDRGENYERHCPKQS- 183
Query: 175 GLNCLLPRPLGYRVPILWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQ 234
L+CL+P P GY+ PI WPQSRD++WF+NVPHTRLVEDKGGQNWI + DKFVFPGGGTQ
Sbjct: 184 -LDCLIPPPDGYKKPIQWPQSRDKIWFNNVPHTRLVEDKGGQNWIRREKDKFVFPGGGTQ 242
Query: 235 FIHGADKYLDQISAMVPDIAFGSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHEN 294
FIHGAD+YLDQIS M+PDI FGS TRVALDIGCGVASFGAFLMQRN TTLS+APKDVHEN
Sbjct: 243 FIHGADQYLDQISQMIPDITFGSRTRVALDIGCGVASFGAFLMQRNTTTLSVAPKDVHEN 302
Query: 295 QIQFALERGVPALAAVFATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGR 354
QIQFALERGVPA+ AVFAT RLL+PS +F+MIHCSRCRINWTRDDGILLLE NR+LRAG
Sbjct: 303 QIQFALERGVPAMVAVFATRRLLYPSQSFEMIHCSRCRINWTRDDGILLLEVNRMLRAGG 362
Query: 355 YFVWAAQPVYKHEESLQEQWKEMENLTSRLCWELVRKEGYIAIWRKPLNNSCYLSRDIPV 414
YFVWAAQPVYKHE++LQEQWKEM +LT+R+CWEL++KEGYIA+WRKPLNNSCY+SR+
Sbjct: 363 YFVWAAQPVYKHEDNLQEQWKEMLDLTNRICWELIKKEGYIAVWRKPLNNSCYVSREAGT 422
Query: 415 HPPLCESNDDPDDVWYVGLKACITQLPSDGYGVNVTTWPSRLHQPPDRLQSIKQDAIISR 474
PPLC +DDPDDVWYV +K CIT+LP +GYG NV+TWP+RLH PP+RLQSI+ DA ISR
Sbjct: 423 KPPLCRPDDDPDDVWYVDMKPCITRLPDNGYGANVSTWPARLHDPPERLQSIQMDAYISR 482
Query: 475 VELFRAESKYWNEIIDSYVRAYRWKEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNVVP 534
E+ +AES++W E+++SYVR +RWKE+ LRNV+DMRAGFGGFAAAL+DL +DCWVMN+VP
Sbjct: 483 KEIMKAESRFWLEVVESYVRVFRWKEFKLRNVLDMRAGFGGFAAALNDLGLDCWVMNIVP 542
Query: 535 VSGFNTLPVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCNISTIMLEM 592
VSGFNTLPV+YDRGL G MHDWCEPFDTYPRTYDL+HAA LFSVEKKRCNI+ IMLEM
Sbjct: 543 VSGFNTLPVIYDRGLQGAMHDWCEPFDTYPRTYDLIHAAFLFSVEKKRCNITNIMLEM 600
>At2g39750 unknown protein
Length = 694
Score = 800 bits (2067), Expect = 0.0
Identities = 374/576 (64%), Positives = 454/576 (77%), Gaps = 22/576 (3%)
Query: 33 LFLFTRFSPTTNSPLTFFLPSEQQPPL-------------PPPPPPPRVKVFRLPPLPPV 79
+F + PT ++ + QQPP PPPPPPP PP PV
Sbjct: 67 VFNLSAIIPTNHTQIEIPATIRQQPPSVVADTEKVKVEANPPPPPPPSPSP--PPPPGPV 124
Query: 80 ERMGLVDENGVMTDNFTVGDSDLGSVVDLLSWKNESRGVVEEEG---AAAKVVVEKFKVC 136
+ G+VD NGVM+D+F VG+ + +V D W N++ +VE + + A+V ++KF +C
Sbjct: 125 KSFGIVDANGVMSDDFEVGEVESDTVED---WGNQTE-IVEAKSDGDSKARVRIKKFGMC 180
Query: 137 DSGMVDYVPCLDNAEAVARLNGSERGEKYERHCPEEGKGLNCLLPRPLGYRVPILWPQSR 196
M +Y+PCLDN + + +L +ERGE++ERHCPE+GKGLNCL+P P GYR PI WP+SR
Sbjct: 181 PESMREYIPCLDNTDVIKKLKSTERGERFERHCPEKGKGLNCLVPPPKGYRQPIPWPKSR 240
Query: 197 DEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQISAMVPDIAFG 256
DEVWFSNVPHTRLVEDKGGQNWIS +KF FPGGGTQFIHGAD+YLDQ+S MV DI FG
Sbjct: 241 DEVWFSNVPHTRLVEDKGGQNWISRDKNKFKFPGGGTQFIHGADQYLDQMSKMVSDITFG 300
Query: 257 SNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALERGVPALAAVFATHRL 316
+ RVA+D+GCGVASFGA+L+ R+V T+S+APKDVHENQIQFALERGVPA+AA FAT RL
Sbjct: 301 KHIRVAMDVGCGVASFGAYLLSRDVMTMSVAPKDVHENQIQFALERGVPAMAAAFATRRL 360
Query: 317 LFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQPVYKHEESLQEQWKE 376
L+PS AFD+IHCSRCRINWTRDDGILLLE NR+LRAG YF WAAQPVYKHE +L+EQW E
Sbjct: 361 LYPSQAFDLIHCSRCRINWTRDDGILLLEINRMLRAGGYFAWAAQPVYKHEPALEEQWTE 420
Query: 377 MENLTSRLCWELVRKEGYIAIWRKPLNNSCYLSRDIPVHPPLCESNDDPDDVWYVGLKAC 436
M NLT LCW+LV+KEGY+AIW+KP NN CYLSR+ PPLC+ +DDPD+VWY LK C
Sbjct: 421 MLNLTISLCWKLVKKEGYVAIWQKPFNNDCYLSREAGTKPPLCDESDDPDNVWYTNLKPC 480
Query: 437 ITQLPSDGYGVNVTTWPSRLHQPPDRLQSIKQDAIISRVELFRAESKYWNEIIDSYVRAY 496
I+++P GYG NV WP+RLH PPDRLQ+IK D+ I+R ELF+AESKYWNEII YVRA
Sbjct: 481 ISRIPEKGYGGNVPLWPARLHTPPDRLQTIKFDSYIARKELFKAESKYWNEIIGGYVRAL 540
Query: 497 RWKEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNVVPVSGFNTLPVLYDRGLIGVMHDW 556
+WK+ LRNV+DMRAGFGGFAAAL+D ++DCWV++VVPVSG NTLPV+YDRGL+GVMHDW
Sbjct: 541 KWKKMKLRNVLDMRAGFGGFAAALNDHKLDCWVLSVVPVSGPNTLPVIYDRGLLGVMHDW 600
Query: 557 CEPFDTYPRTYDLLHAAGLFSVEKKRCNISTIMLEM 592
CEPFDTYPRTYD LHA+GLFS+E+KRC +STI+LEM
Sbjct: 601 CEPFDTYPRTYDFLHASGLFSIERKRCEMSTILLEM 636
>At5g06050 ankyrin-like protein
Length = 682
Score = 798 bits (2062), Expect = 0.0
Identities = 373/546 (68%), Positives = 444/546 (81%), Gaps = 18/546 (3%)
Query: 54 EQQPPLPP---PPPPP---RVKVFRLPPLPPVERMGLVDENGVMTDNFTVGDSDLGSVVD 107
E P LPP PPPPP +KVF G+V+ENG M+D F +GD D+ S
Sbjct: 74 ESHPILPPSLSPPPPPDSVELKVF-----------GIVNENGTMSDEFQIGDYDVESAET 122
Query: 108 LLSWKN-ESRGVVEEEGAAAKVVVEKFKVCDSGMVDYVPCLDNAEAVARLNGSERGEKYE 166
L + ES + + A+V V KF++C M +Y+PCLDN EA+ RLN + RGE++E
Sbjct: 123 LGNQTEFESSDDDDIKSTTARVSVRKFEICSENMTEYIPCLDNVEAIKRLNSTARGERFE 182
Query: 167 RHCPEEGKGLNCLLPRPLGYRVPILWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKF 226
R+CP +G GLNC +P P GYR PI WP+SRDEVWF+NVPHT+LVEDKGGQNWI +NDKF
Sbjct: 183 RNCPNDGMGLNCTVPIPQGYRSPIPWPRSRDEVWFNNVPHTKLVEDKGGQNWIYKENDKF 242
Query: 227 VFPGGGTQFIHGADKYLDQISAMVPDIAFGSNTRVALDIGCGVASFGAFLMQRNVTTLSI 286
FPGGGTQFIHGAD+YLDQIS M+PDI+FG++TRV LDIGCGVASFGA+LM RNV T+SI
Sbjct: 243 KFPGGGTQFIHGADQYLDQISQMIPDISFGNHTRVVLDIGCGVASFGAYLMSRNVLTMSI 302
Query: 287 APKDVHENQIQFALERGVPALAAVFATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEA 346
APKDVHENQIQFALERGVPA+ A F T RLL+PS AFD++HCSRCRINWTRDDGILLLE
Sbjct: 303 APKDVHENQIQFALERGVPAMVAAFTTRRLLYPSQAFDLVHCSRCRINWTRDDGILLLEV 362
Query: 347 NRLLRAGRYFVWAAQPVYKHEESLQEQWKEMENLTSRLCWELVRKEGYIAIWRKPLNNSC 406
NR+LRAG YFVWAAQPVYKHE++L+EQW+EM NLT+RLCW LV+KEGYIAIW+KP+NN+C
Sbjct: 363 NRMLRAGGYFVWAAQPVYKHEKALEEQWEEMLNLTTRLCWVLVKKEGYIAIWQKPVNNTC 422
Query: 407 YLSRDIPVHPPLCESNDDPDDVWYVGLKACITQLPSDGYGVNVTTWPSRLHQPPDRLQSI 466
YLSR V PPLC S DDPD+VWYV LKACIT++ +GYG N+ WP+RL PPDRLQ+I
Sbjct: 423 YLSRGAGVSPPLCNSEDDPDNVWYVDLKACITRIEENGYGANLAPWPARLLTPPDRLQTI 482
Query: 467 KQDAIISRVELFRAESKYWNEIIDSYVRAYRWKEYNLRNVMDMRAGFGGFAAALHDLQID 526
+ D+ I+R ELF AESKYW EII +YV A WK+ LRNV+DMRAGFGGFAAAL +L++D
Sbjct: 483 QIDSYIARKELFVAESKYWKEIISNYVNALHWKQIGLRNVLDMRAGFGGFAAALAELKVD 542
Query: 527 CWVMNVVPVSGFNTLPVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCNIS 586
CWV+NV+PVSG NTLPV+YDRGL+GVMHDWCEPFDTYPRTYDLLHAAGLFS+E+KRCN++
Sbjct: 543 CWVLNVIPVSGPNTLPVIYDRGLLGVMHDWCEPFDTYPRTYDLLHAAGLFSIERKRCNMT 602
Query: 587 TIMLEM 592
T+MLEM
Sbjct: 603 TMMLEM 608
>At5g64030 ankyrin-like protein
Length = 829
Score = 464 bits (1193), Expect = e-131
Identities = 238/529 (44%), Positives = 332/529 (61%), Gaps = 32/529 (6%)
Query: 77 PPVERMGLVDENGVMTDNFTVGDSDLGSVVDLLSWKNESRGVVEEEGAAAKVVVEKFKVC 136
PP ++ L++E +F+ + ES+ E + + + K+ +C
Sbjct: 255 PPGAQLELLNETTAQNGSFS-------------TQATESKNEKEAQKGSGDKLDYKWALC 301
Query: 137 DSGM-VDYVPCLDNAEAVARLNGSERGEKYERHCPEEGKGLNCLLPRPLGYRVPILWPQS 195
++ DY+PCLDN +A+ L ++ E ERHCP+ CL+P P GY+ PI WP+S
Sbjct: 302 NTTAGPDYIPCLDNVQAIRSLPSTKHYEHRERHCPDSPP--TCLVPLPDGYKRPIEWPKS 359
Query: 196 RDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQISAMVPDIAF 255
R+++W++NVPHT+L E KG QNW+ + + FPGGGTQF HGA Y+D I VP IA+
Sbjct: 360 REKIWYTNVPHTKLAEYKGHQNWVKVTGEYLTFPGGGTQFKHGALHYIDFIQESVPAIAW 419
Query: 256 GSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALERGVPALAAVFATHR 315
G +RV LD+GCGVASFG FL R+V T+S+APKD HE Q+QFALERG+PA++AV T R
Sbjct: 420 GKRSRVVLDVGCGVASFGGFLFDRDVITMSLAPKDEHEAQVQFALERGIPAISAVMGTTR 479
Query: 316 LLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQPVYKHEESLQEQWK 375
L FP FD++HC+RCR+ W + G LLLE NR+LR G +FVW+A PVY+ + E WK
Sbjct: 480 LPFPGRVFDIVHCARCRVPWHIEGGKLLLELNRVLRPGGFFVWSATPVYQKKTEDVEIWK 539
Query: 376 EMENLTSRLCWELV--RKEGY----IAIWRKPLNNSCYLSRDIPVHPPLCESNDDPDDVW 429
M L ++CWELV K+ +A +RKP +N CY +R PV PP+C +DDP+ W
Sbjct: 540 AMSELIKKMCWELVSINKDTINGVGVATYRKPTSNECYKNRSEPV-PPICADSDDPNASW 598
Query: 430 YVGLKACITQLPSDGYGVN---VTTWPSRLHQPPDRLQSIKQDAIISRV--ELFRAESKY 484
V L+AC+ P D WP+RL + P L S Q + + E F A+ ++
Sbjct: 599 KVPLQACMHTAPEDKTQRGSQWPEQWPARLEKAPFWLSS-SQTGVYGKAAPEDFSADYEH 657
Query: 485 WNEII-DSYVRAYRWKEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNVVPVSGFNTLPV 543
W ++ SY+ ++RNVMDMRA +GGFAAAL DL++ WVMNVVP+ +TL +
Sbjct: 658 WKRVVTKSYLNGLGINWASVRNVMDMRAVYGGFAAALRDLKV--WVMNVVPIDSPDTLAI 715
Query: 544 LYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCNISTIMLEM 592
+Y+RGL G+ HDWCE F TYPR+YDLLHA LFS K+RCN++ ++ E+
Sbjct: 716 IYERGLFGIYHDWCESFSTYPRSYDLLHADHLFSKLKQRCNLTAVIAEV 764
>At3g51070 putative protein
Length = 895
Score = 464 bits (1193), Expect = e-131
Identities = 234/466 (50%), Positives = 307/466 (65%), Gaps = 24/466 (5%)
Query: 142 DYVPCLDNAEAVARLNGSERGEKYERHCPEEGKGLNCLLPRPLGYRVPILWPQSRDEVWF 201
DY+PCLDN EA+ +L E ERHCPE+ CL+P P GY+ I WP+SRD++W+
Sbjct: 381 DYIPCLDNEEAIMKLRSRRHFEHRERHCPEDPP--TCLVPLPEGYKEAIKWPESRDKIWY 438
Query: 202 SNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQISAMVPDIAFGSNTRV 261
NVPHT+L E KG QNW+ + + FPGGGTQFIHGA Y+D + + +IA+G TRV
Sbjct: 439 HNVPHTKLAEVKGHQNWVKVTGEFLTFPGGGTQFIHGALHYIDFLQQSLKNIAWGKRTRV 498
Query: 262 ALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALERGVPALAAVFATHRLLFPSH 321
LD+GCGVASFG FL +R+V +S+APKD HE Q+QFALER +PA++AV + RL FPS
Sbjct: 499 ILDVGCGVASFGGFLFERDVIAMSLAPKDEHEAQVQFALERKIPAISAVMGSKRLPFPSR 558
Query: 322 AFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQPVYKHEESLQEQWKEMENLT 381
FD+IHC+RCR+ W + G+LLLE NR+LR G YFVW+A PVY+ E + WKEM LT
Sbjct: 559 VFDLIHCARCRVPWHNEGGMLLLELNRMLRPGGYFVWSATPVYQKLEEDVQIWKEMSALT 618
Query: 382 SRLCWELV-----RKEGY-IAIWRKPLNNSCYLSRDIPVHPPLCESNDDPDDVWYVGLKA 435
LCWELV + G AI++KP N CY R PPLC++NDD + WYV L+A
Sbjct: 619 KSLCWELVTINKDKLNGIGAAIYQKPATNECYEKRK-HNKPPLCKNNDDANAAWYVPLQA 677
Query: 436 CITQLPSD------GYGVNVTTWPSRLHQPPDRLQSIKQDAIISR--VELFRAESKYWNE 487
C+ ++P++ + VN WP RL PP L S Q I + F + ++W
Sbjct: 678 CMHKVPTNVVERGSKWPVN---WPRRLQTPPYWLNS-SQMGIYGKPAPRDFTTDYEHWKH 733
Query: 488 IIDS-YVRAYRWKEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNVVPVSGFNTLPVLYD 546
++ Y+ N+RNVMDMRA +GGFAAAL DLQ+ WVMNVV ++ +TLP++Y+
Sbjct: 734 VVSKVYMNEIGISWSNVRNVMDMRAVYGGFAAALKDLQV--WVMNVVNINSPDTLPIIYE 791
Query: 547 RGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCNISTIMLEM 592
RGL G+ HDWCE F TYPR+YDLLHA LFS + RCN+ +M E+
Sbjct: 792 RGLFGIYHDWCESFSTYPRSYDLLHADHLFSKLRTRCNLVPVMAEV 837
>At1g29470 unknown protein
Length = 768
Score = 457 bits (1177), Expect = e-129
Identities = 234/480 (48%), Positives = 311/480 (64%), Gaps = 20/480 (4%)
Query: 126 AKVVVEKFKVCD-SGMVDYVPCLDNAEAVARLNGSERGEKYERHCPEEGKGLNCLLPRPL 184
A+V K+KVC+ + DY+PCLDN +A+ +L+ ++ E ERHCPEE CL+ P
Sbjct: 231 AQVSSIKWKVCNVTAGPDYIPCLDNWQAIRKLHSTKHYEHRERHCPEESP--RCLVSLPE 288
Query: 185 GYRVPILWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLD 244
GY+ I WP+SR+++W++N+PHT+L E KG QNW+ + + FPGGGTQF +GA Y+D
Sbjct: 289 GYKRSIKWPKSREKIWYTNIPHTKLAEVKGHQNWVKMSGEYLTFPGGGTQFKNGALHYID 348
Query: 245 QISAMVPDIAFGSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALERGV 304
+ PDIA+G+ TRV LD+GCGVASFG +L R+V LS APKD HE Q+QFALERG+
Sbjct: 349 FLQESYPDIAWGNRTRVILDVGCGVASFGGYLFDRDVLALSFAPKDEHEAQVQFALERGI 408
Query: 305 PALAAVFATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQPVY 364
PA++ V T RL FP FD+IHC+RCR+ W + G LLLE NR LR G +FVW+A PVY
Sbjct: 409 PAMSNVMGTKRLPFPGSVFDLIHCARCRVPWHIEGGKLLLELNRALRPGGFFVWSATPVY 468
Query: 365 KHEESLQEQWKEMENLTSRLCWELVR------KEGYIAIWRKPLNNSCYLSRDIPVHPPL 418
+ E WK M LT +CWEL+ E AI++KP++N CY R PPL
Sbjct: 469 RKTEEDVGIWKAMSKLTKAMCWELMTIKKDELNEVGAAIYQKPMSNKCYNERS-QNEPPL 527
Query: 419 CESNDDPDDVWYVGLKACITQLPSDGY---GVNVTTWPSRLHQPPDRLQSIKQDAIISR- 474
C+ +DD + W V L+ACI ++ D V +WP R+ P L S Q+ + +
Sbjct: 528 CKDSDDQNAAWNVPLEACIHKVTEDSSKRGAVWPESWPERVETVPQWLDS--QEGVYGKP 585
Query: 475 -VELFRAESKYWNEIID-SYVRAYRWKEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNV 532
E F A+ + W I+ SY+ +RNVMDMRA +GGFAAAL DL++ WVMNV
Sbjct: 586 AQEDFTADHERWKTIVSKSYLNGMGIDWSYVRNVMDMRAVYGGFAAALKDLKL--WVMNV 643
Query: 533 VPVSGFNTLPVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCNISTIMLEM 592
VP+ +TLP++Y+RGL G+ HDWCE F TYPRTYDLLHA LFS KKRCN+ +M E+
Sbjct: 644 VPIDSPDTLPIIYERGLFGIYHDWCESFSTYPRTYDLLHADHLFSSLKKRCNLVGVMAEV 703
>At2g34300 unknown protein
Length = 770
Score = 451 bits (1160), Expect = e-127
Identities = 228/463 (49%), Positives = 302/463 (64%), Gaps = 19/463 (4%)
Query: 142 DYVPCLDNAEAVARLNGSERGEKYERHCPEEGKGLNCLLPRPLGYRVPILWPQSRDEVWF 201
DY+PCLDN +A+ +L+ + E ERHCPEE +CL+ P GY+ I WP+SR+++W+
Sbjct: 250 DYIPCLDNWQAIKKLHTTMHYEHRERHCPEESP--HCLVSLPDGYKRSIKWPKSREKIWY 307
Query: 202 SNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQISAMVPDIAFGSNTRV 261
+NVPHT+L E KG QNW+ + + FPGGGTQF +GA Y+D I P IA+G+ TRV
Sbjct: 308 NNVPHTKLAEIKGHQNWVKMSGEHLTFPGGGTQFKNGALHYIDFIQQSHPAIAWGNRTRV 367
Query: 262 ALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALERGVPALAAVFATHRLLFPSH 321
LD+GCGVASFG +L +R+V LS APKD HE Q+QFALERG+PA+ V T RL FP
Sbjct: 368 ILDVGCGVASFGGYLFERDVLALSFAPKDEHEAQVQFALERGIPAMLNVMGTKRLPFPGS 427
Query: 322 AFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQPVYKHEESLQEQWKEMENLT 381
FD+IHC+RCR+ W + G LLLE NR LR G +FVW+A PVY+ E WK M LT
Sbjct: 428 VFDLIHCARCRVPWHIEGGKLLLELNRALRPGGFFVWSATPVYRKNEEDSGIWKAMSELT 487
Query: 382 SRLCWELVR------KEGYIAIWRKPLNNSCYLSRDIPVHPPLCESNDDPDDVWYVGLKA 435
+CW+LV E AI++KP +N CY R PPLC+ +DD + W V L+A
Sbjct: 488 KAMCWKLVTIKKDKLNEVGAAIYQKPTSNKCYNKRP-QNEPPLCKDSDDQNAAWNVPLEA 546
Query: 436 CITQLPSDGY---GVNVTTWPSRLHQPPDRLQSIKQDAIISR--VELFRAESKYWNEIID 490
C+ ++ D V WP R+ P+ L S Q+ + + E F A+ + W I+
Sbjct: 547 CMHKVTEDSSKRGAVWPNMWPERVETAPEWLDS--QEGVYGKPAPEDFTADQEKWKTIVS 604
Query: 491 -SYVRAYRWKEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNVVPVSGFNTLPVLYDRGL 549
+Y+ N+RNVMDMRA +GGFAAAL DL++ WVMNVVPV +TLP++Y+RGL
Sbjct: 605 KAYLNDMGIDWSNVRNVMDMRAVYGGFAAALKDLKL--WVMNVVPVDAPDTLPIIYERGL 662
Query: 550 IGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCNISTIMLEM 592
G+ HDWCE F+TYPRTYDLLHA LFS +KRCN+ ++M E+
Sbjct: 663 FGIYHDWCESFNTYPRTYDLLHADHLFSTLRKRCNLVSVMAEI 705
>At4g14360
Length = 608
Score = 450 bits (1158), Expect = e-127
Identities = 223/472 (47%), Positives = 297/472 (62%), Gaps = 19/472 (4%)
Query: 133 FKVCDSGMVDYVPCLD-NAEAVARLN-GSERGEKYERHCPEEGKGLNCLLPRPLGYRVPI 190
F VCD + +PCLD N RL E YERHCP + NCL+P P GY+VPI
Sbjct: 76 FPVCDDRHSELIPCLDRNLIYQMRLKLDLSLMEHYERHCPPPERRFNCLIPPPNGYKVPI 135
Query: 191 LWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQISAMV 250
WP+SRDEVW N+PHT L +K QNW+ +K DK FPGGGT F +GADKY+ ++ M+
Sbjct: 136 KWPKSRDEVWKVNIPHTHLAHEKSDQNWMVVKGDKINFPGGGTHFHYGADKYIASMANML 195
Query: 251 --PDIAF--GSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALERGVPA 306
P+ G R D+GCGVASFG +L+ ++ T+S+AP DVH+NQIQFALERG+PA
Sbjct: 196 NYPNNVLNNGGRLRTVFDVGCGVASFGGYLLSSDILTMSLAPNDVHQNQIQFALERGIPA 255
Query: 307 LAAVFATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQPVYKH 366
V T RL +PS +F++ HCSRCRI+W + DGILLLE +R+LR G YF +++ Y
Sbjct: 256 SLGVLGTKRLPYPSRSFELSHCSRCRIDWLQRDGILLLELDRVLRPGGYFAYSSPEAYAQ 315
Query: 367 EESLQEQWKEMENLTSRLCWELVRKEGYIAIWRKPLNNSCYLSRDIPVHPPLCESNDDPD 426
+E W+EM L R+CW++ K IW+KPL N CYL R+ PPLC S++DPD
Sbjct: 316 DEEDLRIWREMSALVERMCWKIAAKRNQTVIWQKPLTNDCYLEREPGTQPPLCRSDNDPD 375
Query: 427 DVWYVGLKACITQLPSDGY---GVNVTTWPSRLHQPPDRLQSIKQDAIISRVELFRAESK 483
VW V ++ACIT + G + WP+RL PP RL +F +++
Sbjct: 376 AVWGVNMEACITSYSDHDHKTKGSGLAPWPARLTSPPPRLADFGYS-----TGMFEKDTE 430
Query: 484 YWNEIIDSY--VRAYRWKEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNVVPVSGFNTL 541
W + +D+Y + + R + +RN+MDM+A G FAAAL + D WVMNVVP G NTL
Sbjct: 431 LWRQRVDTYWDLLSPRIESDTVRNIMDMKASMGSFAAALKEK--DVWVMNVVPEDGPNTL 488
Query: 542 PVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFS-VEKKRCNISTIMLEM 592
++YDRGL+G +H WCE F TYPRTYDLLHA + S ++KK C+ ++LEM
Sbjct: 489 KLIYDRGLMGAVHSWCEAFSTYPRTYDLLHAWDIISDIKKKGCSEVDLLLEM 540
>At3g23300 ankyrin repeat protein
Length = 611
Score = 447 bits (1149), Expect = e-125
Identities = 220/472 (46%), Positives = 297/472 (62%), Gaps = 19/472 (4%)
Query: 133 FKVCDSGMVDYVPCLD-NAEAVARLN-GSERGEKYERHCPEEGKGLNCLLPRPLGYRVPI 190
F VCD + +PCLD N RL E YERHCP + NCL+P P GY++PI
Sbjct: 79 FPVCDDRHSELIPCLDRNLIYQMRLKLDLSLMEHYERHCPPPERRFNCLIPPPPGYKIPI 138
Query: 191 LWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQISAMV 250
WP+SRDEVW N+PHT L +K QNW+ +K +K FPGGGT F +GADKY+ ++ M+
Sbjct: 139 KWPKSRDEVWKVNIPHTHLAHEKSDQNWMVVKGEKINFPGGGTHFHYGADKYIASMANML 198
Query: 251 --PDIAF--GSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALERGVPA 306
P+ G R LD+GCGVASFG +L+ + T+S+AP DVH+NQIQFALERG+PA
Sbjct: 199 NFPNNVLNNGGRLRTFLDVGCGVASFGGYLLASEIMTMSLAPNDVHQNQIQFALERGIPA 258
Query: 307 LAAVFATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQPVYKH 366
V T RL +PS +F++ HCSRCRI+W + DGILLLE +R+LR G YF +++ Y
Sbjct: 259 YLGVLGTKRLPYPSRSFELAHCSRCRIDWLQRDGILLLELDRVLRPGGYFAYSSPEAYAQ 318
Query: 367 EESLQEQWKEMENLTSRLCWELVRKEGYIAIWRKPLNNSCYLSRDIPVHPPLCESNDDPD 426
+E W+EM L R+CW + K IW+KPL N CYL R+ PPLC S+ DPD
Sbjct: 319 DEEDLRIWREMSALVGRMCWTIAAKRNQTVIWQKPLTNDCYLGREPGTQPPLCNSDSDPD 378
Query: 427 DVWYVGLKACITQLPSDGY---GVNVTTWPSRLHQPPDRLQSIKQDAIISRVELFRAESK 483
V+ V ++ACITQ + G + WP+RL PP RL ++F +++
Sbjct: 379 AVYGVNMEACITQYSDHDHKTKGSGLAPWPARLTSPPPRLADFGYS-----TDIFEKDTE 433
Query: 484 YWNEIIDSY--VRAYRWKEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNVVPVSGFNTL 541
W + +D+Y + + + + +RN+MDM+A G FAAAL + D WVMNVVP G NTL
Sbjct: 434 TWRQRVDTYWDLLSPKIQSDTVRNIMDMKASMGSFAAALKEK--DVWVMNVVPEDGPNTL 491
Query: 542 PVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFS-VEKKRCNISTIMLEM 592
++YDRGL+G +H WCE F TYPRTYDLLHA + S ++K+ C+ ++LEM
Sbjct: 492 KLIYDRGLMGAVHSWCEAFSTYPRTYDLLHAWDIISDIKKRGCSAEDLLLEM 543
>At1g04430 unknown protein
Length = 623
Score = 446 bits (1147), Expect = e-125
Identities = 224/477 (46%), Positives = 299/477 (61%), Gaps = 19/477 (3%)
Query: 128 VVVEKFKVCDSGMVDYVPCLD-NAEAVARLN-GSERGEKYERHCPEEGKGLNCLLPRPLG 185
VV + F VCD + +PCLD N RL E YERHCP + NCL+P P G
Sbjct: 79 VVAKSFPVCDDRHSEIIPCLDRNFIYQMRLKLDLSLMEHYERHCPPPERRFNCLIPPPSG 138
Query: 186 YRVPILWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQ 245
Y+VPI WP+SRDEVW +N+PHT L ++K QNW+ K +K FPGGGT F +GADKY+
Sbjct: 139 YKVPIKWPKSRDEVWKANIPHTHLAKEKSDQNWMVEKGEKISFPGGGTHFHYGADKYIAS 198
Query: 246 ISAMVP---DIAFGSNT-RVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALE 301
I+ M+ D+ R LD+GCGVASFGA+L+ ++ T+S+AP DVH+NQIQFALE
Sbjct: 199 IANMLNFSNDVLNDEGRLRTVLDVGCGVASFGAYLLASDIMTMSLAPNDVHQNQIQFALE 258
Query: 302 RGVPALAAVFATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQ 361
RG+PA V T RL +PS +F+ HCSRCRI+W + DG+LLLE +R+LR G YF +++
Sbjct: 259 RGIPAYLGVLGTKRLPYPSRSFEFAHCSRCRIDWLQRDGLLLLELDRVLRPGGYFAYSSP 318
Query: 362 PVYKHEESLQEQWKEMENLTSRLCWELVRKEGYIAIWRKPLNNSCYLSRDIPVHPPLCES 421
Y +E + WKEM L R+CW + K +W+KPL+N CYL R+ PPLC S
Sbjct: 319 EAYAQDEENLKIWKEMSALVERMCWRIAVKRNQTVVWQKPLSNDCYLEREPGTQPPLCRS 378
Query: 422 NDDPDDVWYVGLKACITQLPSDGY---GVNVTTWPSRLHQPPDRLQSIKQDAIISRVELF 478
+ DPD V V ++ACIT + G + WP+RL P RL ++F
Sbjct: 379 DADPDAVAGVSMEACITPYSKHDHKTKGSGLAPWPARLTSSPPRLADFGYS-----TDMF 433
Query: 479 RAESKYWNEIIDSY--VRAYRWKEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNVVPVS 536
+++ W + +DSY + + + K +RN+MDM+A G FAAAL D D WVMNVV
Sbjct: 434 EKDTELWKQQVDSYWNLMSSKVKSNTVRNIMDMKAHMGSFAAALKDK--DVWVMNVVSPD 491
Query: 537 GFNTLPVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFS-VEKKRCNISTIMLEM 592
G NTL ++YDRGLIG H+WCE F TYPRTYDLLHA +FS ++ K C+ +++EM
Sbjct: 492 GPNTLKLIYDRGLIGTNHNWCEAFSTYPRTYDLLHAWSIFSDIKSKGCSAEDLLIEM 548
>At5g14430 unknown protein
Length = 612
Score = 440 bits (1132), Expect = e-124
Identities = 218/478 (45%), Positives = 305/478 (63%), Gaps = 23/478 (4%)
Query: 129 VVEKFKVCDSGMVDYVPCLD---NAEAVARLNGSERGEKYERHCPEEGKGLNCLLPRPLG 185
V + +CDS + +PCLD + + +LN S E YE HCP + NCL+P P+G
Sbjct: 76 VPKSVPICDSRHSELIPCLDRNLHYQLKLKLNLSLM-EHYEHHCPPSERRFNCLVPPPVG 134
Query: 186 YRVPILWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQ 245
Y++P+ WP SRDEVW +N+PHT L ++K QNW+ + DK FPGGGT F +GADKY+
Sbjct: 135 YKIPLRWPVSRDEVWKANIPHTHLAQEKSDQNWMVVNGDKINFPGGGTHFHNGADKYIVS 194
Query: 246 ISAMVP----DIAFGSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALE 301
++ M+ + G + R LD+GCGVASFGA+L+ ++ +S+AP DVH+NQIQFALE
Sbjct: 195 LAQMLKFPGDKLNNGGSIRNVLDVGCGVASFGAYLLSHDIIAMSLAPNDVHQNQIQFALE 254
Query: 302 RGVPALAAVFATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQ 361
RG+P+ V T RL +PS +F++ HCSRCRI+W + DGILLLE +RLLR G YFV+++
Sbjct: 255 RGIPSTLGVLGTKRLPYPSRSFELAHCSRCRIDWLQRDGILLLELDRLLRPGGYFVYSSP 314
Query: 362 PVYKHEESLQEQWKEMENLTSRLCWELVRKEGYIAIWRKPLNNSCYLSRDIPVHPPLCES 421
Y H+ ++ M +L R+CW++V K IW KP++NSCYL RD V PPLC S
Sbjct: 315 EAYAHDPENRKIGNAMHDLFKRMCWKVVAKRDQSVIWGKPISNSCYLKRDPGVLPPLCPS 374
Query: 422 NDDPDDVWYVGLKACIT----QLPSDGYGVNVTTWPSRLHQPPDRLQSIKQDAIISRVEL 477
DDPD W V +KACI+ ++ + + + WP RL PP RL+ I E
Sbjct: 375 GDDPDATWNVSMKACISPYSVRMHKERWS-GLVPWPRRLTAPPPRLEEIGVTP-----EQ 428
Query: 478 FRAESKYWNEIIDSYVRAYR--WKEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNVVPV 535
FR +++ W + Y + + ++ ++RNVMDM + GGFAAAL+D D WVMNV+PV
Sbjct: 429 FREDTETWRLRVIEYWKLLKPMVQKNSIRNVMDMSSNLGGFAAALNDK--DVWVMNVMPV 486
Query: 536 SGFNTLPVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKR-CNISTIMLEM 592
+ ++YDRGLIG HDWCE FDTYPRT+DL+HA F+ + R C+ +++EM
Sbjct: 487 QSSPRMKIIYDRGLIGATHDWCEAFDTYPRTFDLIHAWNTFTETQARGCSFEDLLIEM 544
>At2g40280 unknown protein
Length = 589
Score = 435 bits (1119), Expect = e-122
Identities = 223/468 (47%), Positives = 298/468 (63%), Gaps = 20/468 (4%)
Query: 132 KFKVCDSG-MVDYVPCLDNAEAVARLNGSERGEKYERHCPEEGKGLNCLLPRPLGYRVPI 190
K+ +C VDY+PCLDN A+ +L E ERHCPE CLLP P Y+ P+
Sbjct: 79 KWDLCKGAESVDYIPCLDNYAAIKQLKSRRHMEHRERHCPEPSP--KCLLPLPDNYKPPV 136
Query: 191 LWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQISAMV 250
WP+SRD +W+ NVPH +LVE K QNW+ + + VFPGGGTQF G Y++ I +
Sbjct: 137 PWPKSRDMIWYDNVPHPKLVEYKKEQNWVKKEGEFLVFPGGGTQFKFGVTHYVEFIEKAL 196
Query: 251 PDIAFGSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALERGVPALAAV 310
P I +G N RV LD+GCGVASFG L+ ++V T+S APKD HE QIQFALERG+PA +V
Sbjct: 197 PSIKWGKNIRVVLDVGCGVASFGGSLLDKDVITMSFAPKDEHEAQIQFALERGIPATLSV 256
Query: 311 FATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQPVYKHEESL 370
T +L FPS+AFD+IHC+RCR++W D G LLE NR+LR G +F+W+A PVY+ +
Sbjct: 257 IGTQQLTFPSNAFDLIHCARCRVHWDADGGKPLLELNRVLRPGGFFIWSATPVYRDNDRD 316
Query: 371 QEQWKEMENLTSRLCWELVRK----EGY-IAIWRKPLNNSCYLSRDIPVHPPLCESNDDP 425
W EM +LT +CW++V K G + I++KP + SCY R PPLC+ +
Sbjct: 317 SRIWNEMVSLTKSICWKVVTKTVDSSGIGLVIYQKPTSESCYNKRSTQ-DPPLCDKK-EA 374
Query: 426 DDVWYVGLKACITQLPSDGYGVNVTTWPSRLHQPPDRLQSIKQDAIISRVELFRAESKYW 485
+ WYV L C+++LPS NV +WP P RL S+K +I + E + +++ W
Sbjct: 375 NGSWYVPLAKCLSKLPSG----NVQSWPELW---PKRLVSVKPQSISVKAETLKKDTEKW 427
Query: 486 N-EIIDSYVRAYRWKEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNVVPVSGFNTLPVL 544
+ + D Y++ +RNVMDM AGFGGFAAAL +L + WVMNVVPV +TL V+
Sbjct: 428 SASVSDVYLKHLAVNWSTVRNVMDMNAGFGGFAAALINLPL--WVMNVVPVDKPDTLSVV 485
Query: 545 YDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCNISTIMLEM 592
YDRGLIGV HDWCE +TYPRTYDLLH++ L +RC I ++ E+
Sbjct: 486 YDRGLIGVYHDWCESVNTYPRTYDLLHSSFLLGDLTQRCEIVQVVAEI 533
>At1g26850 unknown protein
Length = 616
Score = 409 bits (1052), Expect = e-114
Identities = 218/505 (43%), Positives = 301/505 (59%), Gaps = 25/505 (4%)
Query: 104 SVVDLLSWKNESRGVVEEEGAAAKVVVEKFKVCDSGMVDYVPCLDNAEAVARLNGSERGE 163
++V L+++ G GA+ V+ F+ CD DY PC D A+ S
Sbjct: 57 NIVPSLNFETHHAGESSLVGASEAAKVKAFEPCDGRYTDYTPCQDQRRAMTFPRDSMIYR 116
Query: 164 KYERHCPEEGKGLNCLLPRPLGYRVPILWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKN 223
ERHC E + L+CL+P P GY P WP+SRD V ++N P+ L +K QNWI +
Sbjct: 117 --ERHCAPENEKLHCLIPAPKGYVTPFSWPKSRDYVPYANAPYKALTVEKAIQNWIQYEG 174
Query: 224 DKFVFPGGGTQFIHGADKYLDQISAMVPDIAFGSNTRVALDIGCGVASFGAFLMQRNVTT 283
D F FPGGGTQF GADKY+DQ+++++P R ALD GCGVAS+GA+L RNV
Sbjct: 175 DVFRFPGGGTQFPQGADKYIDQLASVIP--MENGTVRTALDTGCGVASWGAYLWSRNVRA 232
Query: 284 LSIAPKDVHENQIQFALERGVPALAAVFATHRLLFPSHAFDMIHCSRCRINWTRDDGILL 343
+S AP+D HE Q+QFALERGVPA+ V T +L +P+ AFDM HCSRC I W +DG+ L
Sbjct: 233 MSFAPRDSHEAQVQFALERGVPAVIGVLGTIKLPYPTRAFDMAHCSRCLIPWGANDGMYL 292
Query: 344 LEANRLLRAGRYFVWAAQPV---------YKHEESLQEQWKEMENLTSRLCWELVRKEGY 394
+E +R+LR G Y++ + P+ + +E LQE+ +++E LCWE + G
Sbjct: 293 MEVDRVLRPGGYWILSGPPINWKVNYKAWQRPKEDLQEEQRKIEEAAKLLCWEKKYEHGE 352
Query: 395 IAIWRKPLNNSCYLSRDIPVHPPLCESNDDPDDVWYVGLKACITQLP-----SDGYGVNV 449
IAIW+K +N+ SR C++ DD DDVWY ++ACIT P + G +
Sbjct: 353 IAIWQKRVNDEACRSRQDDPRANFCKT-DDTDDVWYKKMEACITPYPETSSSDEVAGGEL 411
Query: 450 TTWPSRLHQPPDRLQSIKQDAIISRVELFRAESKYWNEIIDSYVRAYRWKEY-NLRNVMD 508
+P RL+ P R+ S + V+ + +++ W + + +Y R + RN+MD
Sbjct: 412 QAFPDRLNAVPPRISSGSISGV--TVDAYEDDNRQWKKHVKAYKRINSLLDTGRYRNIMD 469
Query: 509 MRAGFGGFAAALHDLQIDCWVMNVVP-VSGFNTLPVLYDRGLIGVMHDWCEPFDTYPRTY 567
M AGFGGFAAAL ++ WVMNVVP ++ N L V+Y+RGLIG+ HDWCE F TYPRTY
Sbjct: 470 MNAGFGGFAAALESQKL--WVMNVVPTIAEKNRLGVVYERGLIGIYHDWCEAFSTYPRTY 527
Query: 568 DLLHAAGLFSVEKKRCNISTIMLEM 592
DL+HA LFS+ K +CN I+LEM
Sbjct: 528 DLIHANHLFSLYKNKCNADDILLEM 552
>At4g10440 putative protein
Length = 633
Score = 402 bits (1032), Expect = e-112
Identities = 205/488 (42%), Positives = 294/488 (60%), Gaps = 42/488 (8%)
Query: 130 VEKFKVCDSGMVDYVPCLDNAEAVARLNGSERGEKY--------ERHCPEEGKGLNCLLP 181
++ F+ C+ + +Y PC D +RG ++ ERHCP + + L CL+P
Sbjct: 90 IKYFEPCELSLSEYTPCEDR----------QRGRRFDRNMMKYRERHCPVKDELLYCLIP 139
Query: 182 RPLGYRVPILWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADK 241
P Y++P WPQSRD W+ N+PH L +K QNWI ++ D+F FPGGGT F GAD
Sbjct: 140 PPPNYKIPFKWPQSRDYAWYDNIPHKELSVEKAVQNWIQVEGDRFRFPGGGTMFPRGADA 199
Query: 242 YLDQISAMVPDIAFGSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALE 301
Y+D I+ ++P G R A+D GCGVASFGA+L++R++ +S AP+D HE Q+QFALE
Sbjct: 200 YIDDIARLIPLTDGG--IRTAIDTGCGVASFGAYLLKRDIMAVSFAPRDTHEAQVQFALE 257
Query: 302 RGVPALAAVFATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQ 361
RGVPA+ + + RL +P+ AFD+ HCSRC I W ++DG+ L+E +R+LR G Y++ +
Sbjct: 258 RGVPAIIGIMGSRRLPYPARAFDLAHCSRCLIPWFKNDGLYLMEVDRVLRPGGYWILSGP 317
Query: 362 PV---------YKHEESLQEQWKEMENLTSRLCWELVRKEGYIAIWRKPLNN-SCYLSRD 411
P+ + EE L+++ +E++ LCW+ V ++G ++IW+KPLN+ C +
Sbjct: 318 PINWKQYWRGWERTEEDLKKEQDSIEDVAKSLCWKKVTEKGDLSIWQKPLNHIECKKLKQ 377
Query: 412 IPVHPPLCESNDDPDDVWYVGLKACITQL-----PSDGYGVNVTTWPSRLHQPPDRLQSI 466
PP+C S+D+ D WY L+ CIT L P D G + WP R P R+ I
Sbjct: 378 NNKSPPIC-SSDNADSAWYKDLETCITPLPETNNPDDSAGGALEDWPDRAFAVPPRI--I 434
Query: 467 KQDAIISRVELFRAESKYWNEIIDSYVRAY-RWKEYNLRNVMDMRAGFGGFAAALHDLQI 525
+ E FR +++ W E I Y + RN+MDM A GGFAA++ L+
Sbjct: 435 RGTIPEMNAEKFREDNEVWKERIAHYKKIVPELSHGRFRNIMDMNAFLGGFAASM--LKY 492
Query: 526 DCWVMNVVPVSG-FNTLPVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCN 584
WVMNVVPV TL V+Y+RGLIG DWCE F TYPRTYD++HA GLFS+ + RC+
Sbjct: 493 PSWVMNVVPVDAEKQTLGVIYERGLIGTYQDWCEGFSTYPRTYDMIHAGGLFSLYEHRCD 552
Query: 585 ISTIMLEM 592
++ I+LEM
Sbjct: 553 LTLILLEM 560
>At1g33170 unknown protein
Length = 709
Score = 402 bits (1032), Expect = e-112
Identities = 205/488 (42%), Positives = 296/488 (60%), Gaps = 41/488 (8%)
Query: 130 VEKFKVCDSGMVDYVPCLDNAEAVARLNGSERGEKY--------ERHCPEEGKGLNCLLP 181
V+ F+ CD + +Y PC D ERG ++ ERHCP + + L CL+P
Sbjct: 107 VKYFEPCDMSLSEYTPCEDR----------ERGRRFDRNMMKYRERHCPSKDELLYCLIP 156
Query: 182 RPLGYRVPILWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADK 241
P Y++P WPQSRD W+ N+PH L +K QNWI ++ ++F FPGGGT F GAD
Sbjct: 157 PPPNYKIPFKWPQSRDYAWYDNIPHKELSIEKAIQNWIQVEGERFRFPGGGTMFPRGADA 216
Query: 242 YLDQISAMVPDIAFGSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALE 301
Y+D I+ ++P + G+ R A+D GCGVASFGA+L++R++ +S AP+D HE Q+QFALE
Sbjct: 217 YIDDIARLIP-LTDGA-IRTAIDTGCGVASFGAYLLKRDIVAMSFAPRDTHEAQVQFALE 274
Query: 302 RGVPALAAVFATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQ 361
RGVPA+ + + RL +P+ AFD+ HCSRC I W ++DG+ L E +R+LR G Y++ +
Sbjct: 275 RGVPAIIGIMGSRRLPYPARAFDLAHCSRCLIPWFQNDGLYLTEVDRVLRPGGYWILSGP 334
Query: 362 PV---------YKHEESLQEQWKEMENLTSRLCWELVRKEGYIAIWRKPLNN-SCYLSRD 411
P+ + +E L+++ +E+ LCW+ V ++G ++IW+KP+N+ C +
Sbjct: 335 PINWKKYWKGWERSQEDLKQEQDSIEDAARSLCWKKVTEKGDLSIWQKPINHVECNKLKR 394
Query: 412 IPVHPPLCESNDDPDDVWYVGLKACITQLP----SDGY-GVNVTTWPSRLHQPPDRLQSI 466
+ PPLC +D PD WY L++C+T LP SD + G + WP+R P R+ I
Sbjct: 395 VHKTPPLCSKSDLPDFAWYKDLESCVTPLPEANSSDEFAGGALEDWPNRAFAVPPRI--I 452
Query: 467 KQDAIISRVELFRAESKYWNEIIDSYVRAY-RWKEYNLRNVMDMRAGFGGFAAALHDLQI 525
E FR +++ W E I Y + RN+MDM A GGFAAA+ ++
Sbjct: 453 GGTIPDINAEKFREDNEVWKERISYYKQIMPELSRGRFRNIMDMNAYLGGFAAAM--MKY 510
Query: 526 DCWVMNVVPVSG-FNTLPVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCN 584
WVMNVVPV TL V+++RG IG DWCE F TYPRTYDL+HA GLFS+ + RC+
Sbjct: 511 PSWVMNVVPVDAEKQTLGVIFERGFIGTYQDWCEGFSTYPRTYDLIHAGGLFSIYENRCD 570
Query: 585 ISTIMLEM 592
++ I+LEM
Sbjct: 571 VTLILLEM 578
>At2g45750 hypothetical protein
Length = 631
Score = 395 bits (1016), Expect = e-110
Identities = 208/486 (42%), Positives = 289/486 (58%), Gaps = 39/486 (8%)
Query: 133 FKVCDSGMVDYVPCLDNAEAVARLNGSERGEKYERHCPEEGKGLNCLLPRPLGYRVPILW 192
F C + + ++ PC D ++ ER E +RHCPE + L C +P P GY+ P W
Sbjct: 87 FPSCAAALSEHTPCEDAKRSLKF--SRERLEYRQRHCPEREEILKCRIPAPYGYKTPFRW 144
Query: 193 PQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQISAMVPD 252
P SRD WF+NVPHT L +K QNW+ +ND+F FPGGGT F GAD Y+D I ++ D
Sbjct: 145 PASRDVAWFANVPHTELTVEKKNQNWVRYENDRFWFPGGGTMFPRGADAYIDDIGRLI-D 203
Query: 253 IAFGSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALERGVPALAAVFA 312
++ GS R A+D GCGVASFGA+L+ RN+TT+S AP+D HE Q+QFALERGVPA+ + A
Sbjct: 204 LSDGS-IRTAIDTGCGVASFGAYLLSRNITTMSFAPRDTHEAQVQFALERGVPAMIGIMA 262
Query: 313 THRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQPVYKHEESLQE 372
T RL +PS AFD+ HCSRC I W ++DG L+E +R+LR G Y++ + P+ + Q+
Sbjct: 263 TIRLPYPSRAFDLAHCSRCLIPWGQNDGAYLMEVDRVLRPGGYWILSGPPI-----NWQK 317
Query: 373 QWK--------------EMENLTSRLCWELVRKEGYIAIWRKPLNN-SCYLSRDIPVHPP 417
+WK ++E + LCW+ V + +AIW+KP N+ C +R++ +P
Sbjct: 318 RWKGWERTMDDLNAEQTQIEQVARSLCWKKVVQRDDLAIWQKPFNHIDCKKTREVLKNPE 377
Query: 418 LCESNDDPDDVWYVGLKACITQLP--------SDGYGVNVTTWPSRLHQPPDRLQSIKQD 469
C + DPD WY + +C+T LP G V WP+RL+ P R+ +
Sbjct: 378 FCRHDQDPDMAWYTKMDSCLTPLPEVDDAEDLKTVAGGKVEKWPARLNAIPPRVNKGALE 437
Query: 470 AIISRVELFRAESKYWNEIIDSYVRA-YRWKEY-NLRNVMDMRAGFGGFAAALHDLQIDC 527
I E F +K W + + Y + Y+ E RN++DM A GGFAAAL D +
Sbjct: 438 EITP--EAFLENTKLWKQRVSYYKKLDYQLGETGRYRNLVDMNAYLGGFAAALADDPV-- 493
Query: 528 WVMNVVPVSG-FNTLPVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCNIS 586
WVMNVVPV NTL V+Y+RGLIG +WCE TYPRTYD +HA +F++ + +C
Sbjct: 494 WVMNVVPVEAKLNTLGVIYERGLIGTYQNWCEAMSTYPRTYDFIHADSVFTLYQGQCEPE 553
Query: 587 TIMLEM 592
I+LEM
Sbjct: 554 EILLEM 559
>At2g43200 hypothetical protein
Length = 611
Score = 394 bits (1011), Expect = e-109
Identities = 214/476 (44%), Positives = 293/476 (60%), Gaps = 27/476 (5%)
Query: 133 FKVCDSGMVDYVPCLDNAEAVARLNGSERGEKYERHCPEEGKG-LNCLLPRPLGYRVPIL 191
F +C +Y+PC D + AR ER + ERHCP+ + CL+P+P GY+ P
Sbjct: 91 FPLCPKNFTNYLPCHD--PSTARQYSIERHYRRERHCPDIAQEKFRCLVPKPTGYKTPFP 148
Query: 192 WPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQISAMVP 251
WP+SR WF NVP RL E K QNW+ ++ D+FVFPGGGT F G Y+D I +++P
Sbjct: 149 WPESRKYAWFRNVPFKRLAELKKTQNWVRLEGDRFVFPGGGTSFPGGVKDYVDVILSVLP 208
Query: 252 DIAFGSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALERGVPALAAVF 311
+A GS R LDIGCGVASFGAFL+ + T+SIAP+D+HE Q+QFALERG+PA+ V
Sbjct: 209 -LASGS-IRTVLDIGCGVASFGAFLLNYKILTMSIAPRDIHEAQVQFALERGLPAMLGVL 266
Query: 312 ATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQPV-----YKH 366
+T++L +PS +FDM+HCSRC +NWT DG+ L+E +R+LR Y+V + PV +K+
Sbjct: 267 STYKLPYPSRSFDMVHCSRCLVNWTSYDGLYLMEVDRVLRPEGYWVLSGPPVASRVKFKN 326
Query: 367 E----ESLQEQWKEMENLTSRLCWELVRKEGYIAIWRKPLNN-SCYLSRDIPVHPPLCES 421
+ + LQ Q +++ ++ RLCWE + + + IWRKP N+ C P LC S
Sbjct: 327 QKRDSKELQNQMEKLNDVFRRLCWEKIAESYPVVIWRKPSNHLQCRKRLKALKFPGLC-S 385
Query: 422 NDDPDDVWYVGLKACITQLP--SDGYGVNVTTWPSRL-HQPPDRLQSIKQDAIISRVELF 478
+ DPD WY ++ CIT LP +D + WP RL H P + SI+ I F
Sbjct: 386 SSDPDAAWYKEMEPCITPLPDVNDTNKTVLKNWPERLNHVPRMKTGSIQGTTIAG----F 441
Query: 479 RAESKYWNEIIDSYVRAYRW-KEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNVVPVS- 536
+A++ W + Y +++ RNV+DM AG GGFAAAL ++ WVMNVVP
Sbjct: 442 KADTNLWQRRVLYYDTKFKFLSNGKYRNVIDMNAGLGGFAAAL--IKYPMWVMNVVPFDL 499
Query: 537 GFNTLPVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCNISTIMLEM 592
NTL V+YDRGLIG +WCE TYPRTYDL+HA G+FS+ +C+I I+LEM
Sbjct: 500 KPNTLGVVYDRGLIGTYMNWCEALSTYPRTYDLIHANGVFSLYLDKCDIVDILLEM 555
>At4g19120 ERD3 protein (ERD3)
Length = 600
Score = 393 bits (1010), Expect = e-109
Identities = 208/503 (41%), Positives = 298/503 (58%), Gaps = 40/503 (7%)
Query: 112 KNESRGVVEEEGAAAKVVVEKFKVCDSGMVDYVPCLDNAEAVARLNGSERGEKYERHCPE 171
K ES + ++ K V F C S DY PC D + + G+ R ERHCP
Sbjct: 51 KAESSSLDVDDSLQVKSV--SFSECSSDYQDYTPCTDPRKW--KKYGTHRLTFMERHCPP 106
Query: 172 EGKGLNCLLPRPLGYRVPILWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGG 231
CL+P P GY+ PI WP+S+DE W+ NVP+ + + K QNW+ + +KF+FPGG
Sbjct: 107 VFDRKQCLVPPPDGYKPPIRWPKSKDECWYRNVPYDWINKQKSNQNWLRKEGEKFIFPGG 166
Query: 232 GTQFIHGADKYLDQISAMVPDIAFGSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDV 291
GT F HG Y+D + ++P++ G+ R A+D GCGVAS+G L+ R + T+S+AP+D
Sbjct: 167 GTMFPHGVSAYVDLMQDLIPEMKDGT-IRTAIDTGCGVASWGGDLLDRGILTVSLAPRDN 225
Query: 292 HENQIQFALERGVPALAAVFATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLR 351
HE Q+QFALERG+PA+ + +T RL FPS++FDM HCSRC I WT G+ LLE +R+LR
Sbjct: 226 HEAQVQFALERGIPAILGIISTQRLPFPSNSFDMAHCSRCLIPWTEFGGVYLLEVHRILR 285
Query: 352 AGRYFVWAAQPV-YKH-----EESLQEQ---WKEMENLTSRLCWELVRKEGYIAIWRKPL 402
G ++V + PV Y++ + +++EQ +++++ L S +C+++ K+ IA+W+K
Sbjct: 286 PGGFWVLSGPPVNYENRWKGWDTTIEEQRSNYEKLQELLSSMCFKMYAKKDDIAVWQKSP 345
Query: 403 NNSCY--LSRDIPVHPPLCESNDDPDDVWYVGLKACITQLPSDGYG----VNVTTWPSRL 456
+N CY LS D +PP C+ + +PD WY L+ C+ +PS + WP RL
Sbjct: 346 DNLCYNKLSNDPDAYPPKCDDSLEPDSAWYTPLRPCVV-VPSPKLKKTDLESTPKWPERL 404
Query: 457 HQPPDRLQSIKQDAIISRVELFRAESKYWNEIIDSYVRAYRWKEY-------NLRNVMDM 509
H P+R+ + +F+ + W RA +K+ +RNVMDM
Sbjct: 405 HTTPERISDVPG----GNGNVFKHDDSKWK------TRAKHYKKLLPAIGSDKIRNVMDM 454
Query: 510 RAGFGGFAAALHDLQIDCWVMNVVPVSGFNTLPVLYDRGLIGVMHDWCEPFDTYPRTYDL 569
+GG AAAL + WVMNVV NTLPV++DRGLIG HDWCE F TYPRTYDL
Sbjct: 455 NTAYGGLAAAL--VNDPLWVMNVVSSYAANTLPVVFDRGLIGTYHDWCEAFSTYPRTYDL 512
Query: 570 LHAAGLFSVEKKRCNISTIMLEM 592
LH GLF+ E +RC++ +MLEM
Sbjct: 513 LHVDGLFTSESQRCDMKYVMLEM 535
>At4g00750 unknown protein
Length = 633
Score = 377 bits (967), Expect = e-104
Identities = 204/482 (42%), Positives = 283/482 (58%), Gaps = 39/482 (8%)
Query: 136 CDSGMVDYVPCLDNAEAVAR-LN-GSERGEKYERHCPEEGKGLNCLLPRPLGYRVPILWP 193
C +Y PC E V R LN ER ERHCPE+ + + C +P P GY +P WP
Sbjct: 99 CGVEFSEYTPC----EFVNRSLNFPRERLIYRERHCPEKHEIVRCRIPAPYGYSLPFRWP 154
Query: 194 QSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQISAMVPDI 253
+SRD WF+NVPHT L +K QNW+ + D+F+FPGGGT F GAD Y+D+I ++ ++
Sbjct: 155 ESRDVAWFANVPHTELTVEKKNQNWVRYEKDRFLFPGGGTMFPRGADAYIDEIGRLI-NL 213
Query: 254 AFGSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALERGVPALAAVFAT 313
GS R A+D GCGVASFGA+LM RN+ T+S AP+D HE Q+QFALERGVPA+ V A+
Sbjct: 214 KDGS-IRTAIDTGCGVASFGAYLMSRNIVTMSFAPRDTHEAQVQFALERGVPAIIGVLAS 272
Query: 314 HRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQPV---------Y 364
RL FP+ AFD+ HCSRC I W + +G L+E +R+LR G Y++ + P+
Sbjct: 273 IRLPFPARAFDIAHCSRCLIPWGQYNGTYLIEVDRVLRPGGYWILSGPPINWQRHWKGWE 332
Query: 365 KHEESLQEQWKEMENLTSRLCWELVRKEGYIAIWRKPLNN-SCYLSRDIPVHPPLCESND 423
+ + L + ++E + LCW + + +A+W+KP N+ C +R PP C
Sbjct: 333 RTRDDLNSEQSQIERVARSLCWRKLVQREDLAVWQKPTNHVHCKRNRIALGRPPFCH-RT 391
Query: 424 DPDDVWYVGLKACITQLP-------SDGYGVNVTTWPSRLHQPPDRLQS-----IKQDAI 471
P+ WY L+ C+T LP + G + WP RL+ P R++S I +D
Sbjct: 392 LPNQGWYTKLETCLTPLPEVTGSEIKEVAGGQLARWPERLNALPPRIKSGSLEGITEDEF 451
Query: 472 ISRVELFRAESKYWNEIIDSYVRAYRWKEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMN 531
+S E ++ Y+ + R+ RN +DM A GGFA+AL D + WVMN
Sbjct: 452 VSNTEKWQRRVSYYKKYDQQLAETGRY-----RNFLDMNAHLGGFASALVDDPV--WVMN 504
Query: 532 VVPV-SGFNTLPVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCNISTIML 590
VVPV + NTL V+Y+RGLIG +WCE TYPRTYD +HA +FS+ K RC++ I+L
Sbjct: 505 VVPVEASVNTLGVIYERGLIGTYQNWCEAMSTYPRTYDFIHADSVFSLYKDRCDMEDILL 564
Query: 591 EM 592
EM
Sbjct: 565 EM 566
>At4g18030 unknown protein
Length = 621
Score = 376 bits (965), Expect = e-104
Identities = 208/479 (43%), Positives = 284/479 (58%), Gaps = 37/479 (7%)
Query: 133 FKVCDSGMVDYVPCL--DNAEAVARLNGSERGEKYERHCPEEGKGLNCLLPRPLGYRVPI 190
FK CD + DY PC D A R N R ERHCP + + L CL+P P GY P
Sbjct: 85 FKPCDVKLKDYTPCQEQDRAMKFPRENMIYR----ERHCPPDNEKLRCLVPAPKGYMTPF 140
Query: 191 LWPQSRDEVWFSNVPHTRLVEDKGGQNWISIKNDKFVFPGGGTQFIHGADKYLDQISAMV 250
WP+SRD V ++N P L +K GQNW+ + + F FPGGGT F GAD Y++++++++
Sbjct: 141 PWPKSRDYVHYANAPFKSLTVEKAGQNWVQFQGNVFKFPGGGTMFPQGADAYIEELASVI 200
Query: 251 PDIAFGSNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVHENQIQFALERGVPALAAV 310
P I GS R ALD GCGVAS+GA++++RNV T+S AP+D HE Q+QFALERGVPA+ AV
Sbjct: 201 P-IKDGS-VRTALDTGCGVASWGAYMLKRNVLTMSFAPRDNHEAQVQFALERGVPAIIAV 258
Query: 311 FATHRLLFPSHAFDMIHCSRCRINWTRDDGILLLEANRLLRAGRYFVWAAQPV------- 363
+ L +P+ AFDM CSRC I WT ++G L+E +R+LR G Y+V + P+
Sbjct: 259 LGSILLPYPARAFDMAQCSRCLIPWTANEGTYLMEVDRVLRPGGYWVLSGPPINWKTWHK 318
Query: 364 --YKHEESLQEQWKEMENLTSRLCWELVRKEGYIAIWRKPLNN-SCYLSRDIPVHPPLCE 420
+ + L + K +E + LCWE ++G IAI+RK +N+ SC R PV C+
Sbjct: 319 TWNRTKAELNAEQKRIEGIAESLCWEKKYEKGDIAIFRKKINDRSC--DRSTPV--DTCK 374
Query: 421 SNDDPDDVWYVGLKACITQLP-----SDGYGVNVTTWPSRLHQPPDRLQSIKQDAIISRV 475
D DDVWY ++ C+T P + G + +P RL P + +I+ V
Sbjct: 375 -RKDTDDVWYKEIETCVTPFPKVSNEEEVAGGKLKKFPERLFAVPPSISK----GLINGV 429
Query: 476 --ELFRAESKYWNEIIDSYVRAYRW-KEYNLRNVMDMRAGFGGFAAALHDLQIDCWVMNV 532
E ++ + W + + Y R R RNVMDM AG GGFAAAL + WVMNV
Sbjct: 430 DEESYQEDINLWKKRVTGYKRINRLIGSTRYRNVMDMNAGLGGFAAALESPK--SWVMNV 487
Query: 533 VPVSGFNTLPVLYDRGLIGVMHDWCEPFDTYPRTYDLLHAAGLFSVEKKRCNISTIMLE 591
+P NTL V+Y+RGLIG+ HDWCE F TYPRTYD +HA+G+FS+ + C + I+LE
Sbjct: 488 IPTINKNTLSVVYERGLIGIYHDWCEGFSTYPRTYDFIHASGVFSLYQHSCKLEDILLE 546
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,085,656
Number of Sequences: 26719
Number of extensions: 744123
Number of successful extensions: 6824
Number of sequences better than 10.0: 204
Number of HSP's better than 10.0 without gapping: 139
Number of HSP's successfully gapped in prelim test: 69
Number of HSP's that attempted gapping in prelim test: 3715
Number of HSP's gapped (non-prelim): 1352
length of query: 592
length of database: 11,318,596
effective HSP length: 105
effective length of query: 487
effective length of database: 8,513,101
effective search space: 4145880187
effective search space used: 4145880187
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0078.10


