

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0076a.9
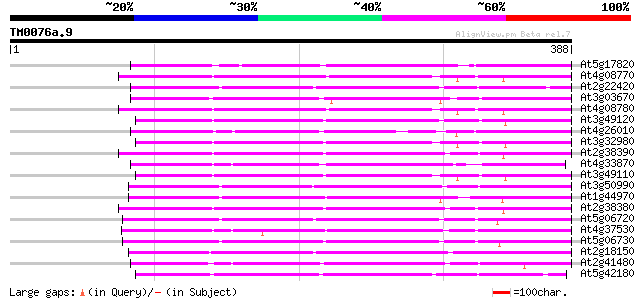
(388 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g17820 peroxidase ATP13a 214 6e-56
At4g08770 peroxidase C2 precursor like protein 213 1e-55
At2g22420 putative peroxidase 211 7e-55
At3g03670 putative peroxidase 207 1e-53
At4g08780 peroxidase C2 precursor like protein 206 2e-53
At3g49120 peroxidase 204 5e-53
At4g26010 peroxidase like protein 204 7e-53
At3g32980 peroxidase, putative 204 7e-53
At2g38390 peroxidase 201 6e-52
At4g33870 putative peroxidase 197 6e-51
At3g49110 peroxidase 197 6e-51
At3g50990 peroxidase-like protein 197 8e-51
At1g44970 peroxidase like protein 197 8e-51
At2g38380 peroxidase like protein 197 1e-50
At5g06720 peroxidase (emb|CAA68212.1) 195 4e-50
At4g37530 peroxidase - like protein 195 4e-50
At5g06730 peroxidase 192 3e-49
At2g18150 putative peroxidase 191 4e-49
At2g41480 peroxidase like protein 191 6e-49
At5g42180 peroxidase (emb|CAA66960.1) 190 1e-48
>At5g17820 peroxidase ATP13a
Length = 313
Score = 214 bits (545), Expect = 6e-56
Identities = 121/306 (39%), Positives = 175/306 (56%), Gaps = 17/306 (5%)
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
LR GFYS SCP AE I+ + + P A +LR+ FHDCFV GCDAS+L+DST
Sbjct: 24 LRVGFYSQSCPQAETIVRNLVRQRFGVTPTVTAALLRMHFHDCFVKGCDASLLIDST--- 80
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
N EK++ NG + + DL+D IK++LE CP VSCAD + + +++ALAG P
Sbjct: 81 -NSEKTAGPNGSVREF-DLIDRIKAQLEAACPSTVSCADIVTLATRDSVALAGGPSYSIP 138
Query: 204 GGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIF 263
GRRD S D LP P S V LF KG + + V LLGAH++G +C +F
Sbjct: 139 TGRRDGRVSNNL---DVTLPGPTISVSGAVSLFTNKGMNTFDAVALLGAHTVGQGNCGLF 195
Query: 264 MDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRY-RNEPVNFDNTPTVMDNLFYRDL 322
DR+ +F+ T +PDP++ P + LR C N T ++ P+ FDN F++
Sbjct: 196 SDRITSFQGTGRPDPSMDPALVTSLRNTCRNSATAALDQSSPLRFDNQ-------FFKQ- 247
Query: 323 VDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVRK 382
+ K + +L D L +DP+T V + A++ A F ++F M K+ +++VLTG +GE+R+
Sbjct: 248 IRKRRGVLQVDQRLASDPQTRGIVARYANNNAFFKRQFVRAMVKMGAVDVLTGRNGEIRR 307
Query: 383 ICRATN 388
CR N
Sbjct: 308 NCRRFN 313
>At4g08770 peroxidase C2 precursor like protein
Length = 346
Score = 213 bits (542), Expect = 1e-55
Identities = 124/320 (38%), Positives = 177/320 (54%), Gaps = 15/320 (4%)
Query: 76 QPNTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASI 135
Q + + L FY +CP I +R++P+ A+ILRL FHDCFV GCDASI
Sbjct: 16 QVSLSHAQLSPSFYDKTCPQVFDIATTTIVNALRSDPRIAASILRLHFHDCFVNGCDASI 75
Query: 136 LLDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALA 195
LLD+T EK +F N +G D++D +K+ +E+ CP VSCAD + + E++ LA
Sbjct: 76 LLDNT-TSFRTEKDAFGNANSARGFDVIDKMKAAVEKACPKTVSCADLLAIAAQESVVLA 134
Query: 196 GMPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFS-PEEMVILLGAHS 254
G P + GRRDSL + +D NLP P ++ +++ + F+ G ++V L G H+
Sbjct: 135 GGPSWRVPNGRRDSLRGFMDLAND-NLPAPFFTLNQLKDRFKNVGLDRASDLVALSGGHT 193
Query: 255 IGAAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD---NTP 311
G C MDR+YNF NT PDP L +L+ LR+ C PR N+ V D TP
Sbjct: 194 FGKNQCQFIMDRLYNFSNTGLPDPTLDKSYLSTLRKQC-----PRNGNQSVLVDFDLRTP 248
Query: 312 TVMDNLFYRDLVDKGKSLLLTDAHLVTDP---RTAPTVGQMADDQALFHKRFAEVMTKLT 368
T+ DN +Y +L + K L+ +D L + P T P V + AD Q F FA+ M +++
Sbjct: 249 TLFDNKYYVNL-KENKGLIQSDQELFSSPDASDTLPLVREYADGQGKFFDAFAKAMIRMS 307
Query: 369 SLNVLTGNDGEVRKICRATN 388
SL+ LTG GE+R CR N
Sbjct: 308 SLSPLTGKQGEIRLNCRVVN 327
>At2g22420 putative peroxidase
Length = 329
Score = 211 bits (536), Expect = 7e-55
Identities = 124/305 (40%), Positives = 177/305 (57%), Gaps = 8/305 (2%)
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
LR FYS++CP AE I+ + + +++A+++R QFHDCFV GCDAS+LLD TPN
Sbjct: 23 LRPRFYSETCPEAESIVRREMKKAMIKEARSVASVMRFQFHDCFVNGCDASLLLDDTPNM 82
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
EK S N L+ ++VDDIK LE+ CP VSCAD ++ + +A+AL G P +
Sbjct: 83 LG-EKLSLSNIDSLRSFEVVDDIKEALEKACPATVSCADIVIMAARDAVALTGGPDWEVK 141
Query: 204 GGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIF 263
GR+DSL + + D+ +P P +A +++LF+R S ++MV L G+HSIG C
Sbjct: 142 LGRKDSL-TASQQDSDDIMPSPRANATFLIDLFERFNLSVKDMVALSGSHSIGQGRCFSI 200
Query: 264 MDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNLFYRDLV 323
M R+YN + KPDPAL P + +L ++C G N + D TP V DN +++DLV
Sbjct: 201 MFRLYNQSGSGKPDPALEPSYRKKLDKLCPLGGD---ENVTGDLDATPQVFDNQYFKDLV 257
Query: 324 DKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVRKI 383
G+ L +D L T+ T V ++DQ F + FAE M KL L +G GE+R
Sbjct: 258 -SGRGFLNSDQTLYTNLVTREYVKMFSEDQDEFFRAFAEGMVKLGDLQ--SGRPGEIRFN 314
Query: 384 CRATN 388
CR N
Sbjct: 315 CRVVN 319
>At3g03670 putative peroxidase
Length = 321
Score = 207 bits (526), Expect = 1e-53
Identities = 120/309 (38%), Positives = 176/309 (56%), Gaps = 14/309 (4%)
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L+ FYS+SCPNAE I+ + + +P A + R+ FHDCFV GCDAS+L+D P
Sbjct: 23 LKFKFYSESCPNAETIVENLVRQQFARDPSITAALTRMHFHDCFVQGCDASLLID--PTT 80
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
+ + + ++G +L+D+IK+ LE +CP VSC+D + + +A+ L G P
Sbjct: 81 SQLSEKNAGPNFSVRGFELIDEIKTALEAQCPSTVSCSDIVTLATRDAVFLGGGPSYVVP 140
Query: 204 GGRRDSLYSLATVVDDNN--LPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCD 261
GRRD S +D N LP P S + M+ F KG + + V LLGAH++G A C
Sbjct: 141 TGRRDGFVSNP---EDANEILPPPFISVEGMLSFFGNKGMNVFDSVALLGAHTVGIASCG 197
Query: 262 IFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPG--TPRYRNEPVNFDNTPTVMDNLFY 319
F+DRV NF+ T PDP++ P LR C+ PG ++ PV TP DNLF+
Sbjct: 198 NFVDRVTNFQGTGLPDPSMDPTLAGRLRNTCAVPGGFAALDQSMPV----TPVSFDNLFF 253
Query: 320 RDLVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGE 379
+ ++ K +LL D + +DP T+ V Q A + LF ++FA M K+ +++VLTG+ GE
Sbjct: 254 GQIRER-KGILLIDQLIASDPATSGVVLQYASNNELFKRQFAIAMVKMGAVDVLTGSAGE 312
Query: 380 VRKICRATN 388
+R CRA N
Sbjct: 313 IRTNCRAFN 321
>At4g08780 peroxidase C2 precursor like protein
Length = 346
Score = 206 bits (524), Expect = 2e-53
Identities = 121/320 (37%), Positives = 174/320 (53%), Gaps = 15/320 (4%)
Query: 76 QPNTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASI 135
Q + + L FY +CP I+ + +R++P+ A+ILRL FHDCFV GCDASI
Sbjct: 16 QVSLSHAQLSPSFYDKTCPQVFDIVTNTIVNALRSDPRIAASILRLHFHDCFVNGCDASI 75
Query: 136 LLDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALA 195
LLD+T EK +F N +G D++D +K+ +E+ CP VSCAD + + E++ LA
Sbjct: 76 LLDNT-TSFRTEKDAFGNANSARGFDVIDKMKAAIEKACPRTVSCADMLAIAAKESIVLA 134
Query: 196 GMPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFS-PEEMVILLGAHS 254
G P GRRDSL + +D NLP P+ + ++ + F+ G ++V L G H+
Sbjct: 135 GGPSWMVPNGRRDSLRGFMDLAND-NLPGPSSTLKQLKDRFKNVGLDRSSDLVALSGGHT 193
Query: 255 IGAAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD---NTP 311
G + C MDR+YNF T PDP L +L LR+ C PR N+ V D TP
Sbjct: 194 FGKSQCQFIMDRLYNFGETGLPDPTLDKSYLATLRKQC-----PRNGNQSVLVDFDLRTP 248
Query: 312 TVMDNLFYRDLVDKGKSLLLTDAHLVTDP---RTAPTVGQMADDQALFHKRFAEVMTKLT 368
T+ DN +Y +L + K L+ +D L + P T P V AD Q F F + + +++
Sbjct: 249 TLFDNKYYVNL-KENKGLIQSDQELFSSPDAADTLPLVRAYADGQGTFFDAFVKAIIRMS 307
Query: 369 SLNVLTGNDGEVRKICRATN 388
SL+ LTG GE+R CR N
Sbjct: 308 SLSPLTGKQGEIRLNCRVVN 327
>At3g49120 peroxidase
Length = 353
Score = 204 bits (520), Expect = 5e-53
Identities = 117/306 (38%), Positives = 168/306 (54%), Gaps = 11/306 (3%)
Query: 88 FYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNGDNVE 147
FY SCPN I+ + +R++P+ A+ILRL FHDCFV GCDASILLD+T E
Sbjct: 36 FYDRSCPNVTNIVRETIVNELRSDPRIAASILRLHFHDCFVNGCDASILLDNT-TSFRTE 94
Query: 148 KSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPLGGRR 207
K +F N +G ++D +K+ +E CP VSCAD + + +++ LAG P + GRR
Sbjct: 95 KDAFGNANSARGFPVIDRMKAAVERACPRTVSCADMLTIAAQQSVTLAGGPSWRVPLGRR 154
Query: 208 DSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFS-PEEMVILLGAHSIGAAHCDIFMDR 266
DSL + + + NLP P ++ ++ F+ G P ++V L G H+ G C +DR
Sbjct: 155 DSLQAFLELA-NANLPAPFFTLPQLKASFRNVGLDRPSDLVALSGGHTFGKNQCQFILDR 213
Query: 267 VYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD-NTPTVMDNLFYRDLVDK 325
+YNF NT PDP L +L LR +C G R+ V+FD TPTV DN +Y +L ++
Sbjct: 214 LYNFSNTGLPDPTLNTTYLQTLRGLCPLNGN---RSALVDFDLRTPTVFDNKYYVNLKER 270
Query: 326 GKSLLLTDAHLVTDPR---TAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVRK 382
K L+ +D L + P T P V AD F F E M ++ ++ TG G++R
Sbjct: 271 -KGLIQSDQELFSSPNATDTIPLVRAYADGTQTFFNAFVEAMNRMGNITPTTGTQGQIRL 329
Query: 383 ICRATN 388
CR N
Sbjct: 330 NCRVVN 335
>At4g26010 peroxidase like protein
Length = 310
Score = 204 bits (519), Expect = 7e-53
Identities = 123/308 (39%), Positives = 171/308 (54%), Gaps = 23/308 (7%)
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
LR GFYS SCP AE I+A A R++ A LR+QFHDCFV GCDAS+L+D P G
Sbjct: 22 LRTGFYSRSCPRAESIVASVVANRFRSDKSITAAFLRMQFHDCFVRGCDASLLIDPRP-G 80
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
EKS+ N ++G +++D+ K +LE CP VSCAD + + +++ALAG PR
Sbjct: 81 RPSEKSTGPNA-SVRGYEIIDEAKRQLEAACPRTVSCADIVTLATRDSVALAGGPRFSVP 139
Query: 204 GGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILL-GAHSIGAAHCDI 262
GRRD L S +D NLP P ++LF +G + +MV L+ G HS+G AHC +
Sbjct: 140 TGRRDGLRSNP---NDVNLPGPTIPVSASIQLFAAQGMNTNDMVTLIGGGHSVGVAHCSL 196
Query: 263 FMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNF--DNTPTVMDNLFYR 320
F DR+ D A+ P + LR+ CS+P N+P F T +DN Y
Sbjct: 197 FQDRL--------SDRAMEPSLKSSLRRKCSSP------NDPTTFLDQKTSFTVDNAIYG 242
Query: 321 DLVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEV 380
+ + + + +L D +L D T+ V A LF KRFAE + K+ ++ VLTG GE+
Sbjct: 243 E-IRRQRGILRIDQNLGLDRSTSGIVSGYASSNTLFRKRFAEALVKMGTIKVLTGRSGEI 301
Query: 381 RKICRATN 388
R+ CR N
Sbjct: 302 RRNCRVFN 309
>At3g32980 peroxidase, putative
Length = 352
Score = 204 bits (519), Expect = 7e-53
Identities = 120/308 (38%), Positives = 168/308 (53%), Gaps = 15/308 (4%)
Query: 88 FYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNGDNVE 147
FY ++CP+ I+ D +R++P+ A+ILRL FHDCFV GCDASILLD+T E
Sbjct: 35 FYDNTCPSVFTIVRDTIVNELRSDPRIAASILRLHFHDCFVNGCDASILLDNT-TSFRTE 93
Query: 148 KSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPLGGRR 207
K + N +G ++D +K+ +E CP VSCAD + + +A+ LAG P + GRR
Sbjct: 94 KDAAPNANSARGFPVIDRMKAAVETACPRTVSCADILTIAAQQAVNLAGGPSWRVPLGRR 153
Query: 208 DSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFS-PEEMVILLGAHSIGAAHCDIFMDR 266
DSL + + + NLP P ++ ++ FQ G P ++V L G H+ G C MDR
Sbjct: 154 DSLQAFFALA-NTNLPAPFFTLPQLKASFQNVGLDRPSDLVALSGGHTFGKNQCQFIMDR 212
Query: 267 VYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD---NTPTVMDNLFYRDLV 323
+YNF NT PDP L +L LR C PR N+ V D TPTV DN +Y +L
Sbjct: 213 LYNFSNTGLPDPTLNTTYLQTLRGQC-----PRNGNQTVLVDFDLRTPTVFDNKYYVNLK 267
Query: 324 DKGKSLLLTDAHLVTDPR---TAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEV 380
+ K L+ TD L + P T P V + AD F F E M ++ ++ LTG G++
Sbjct: 268 EL-KGLIQTDQELFSSPNATDTIPLVREYADGTQKFFNAFVEAMNRMGNITPLTGTQGQI 326
Query: 381 RKICRATN 388
R+ CR N
Sbjct: 327 RQNCRVVN 334
>At2g38390 peroxidase
Length = 349
Score = 201 bits (511), Expect = 6e-52
Identities = 120/317 (37%), Positives = 175/317 (54%), Gaps = 10/317 (3%)
Query: 76 QPNTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASI 135
Q + N LR FY +CP II D +RT+P+ A++LRL FHDCFV GCDASI
Sbjct: 23 QASNSNAQLRPDFYFRTCPPIFNIIGDTIVNELRTDPRIAASLLRLHFHDCFVRGCDASI 82
Query: 136 LLDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALA 195
LLD++ EK + N ++G D++D +K+ +E CP VSCAD + S ++ L+
Sbjct: 83 LLDNS-TSFRTEKDAAPNKNSVRGFDVIDRMKAAIERACPRTVSCADIITIASQISVLLS 141
Query: 196 GMPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFS-PEEMVILLGAHS 254
G P GRRDS+ + + + LP P + ++ F G + P ++V L G H+
Sbjct: 142 GGPWWPVPLGRRDSVEAFFALA-NTALPSPFSTLTQLKTAFADVGLNRPSDLVALSGGHT 200
Query: 255 IGAAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDN-TPTV 313
G A C R+YNF TN+PDP+L P +L ELR++C G VNFD+ TPT
Sbjct: 201 FGKAQCQFVTPRLYNFNGTNRPDPSLNPTYLVELRRLCPQNGNGTVL---VNFDSVTPTT 257
Query: 314 MDNLFYRDLVDKGKSLLLTDAHLVTDP--RTAPTVGQMADDQALFHKRFAEVMTKLTSLN 371
D +Y +L++ GK L+ +D L + P T P V Q + + +F F + M ++ +L
Sbjct: 258 FDRQYYTNLLN-GKGLIQSDQVLFSTPGADTIPLVNQYSSNTFVFFGAFVDAMIRMGNLK 316
Query: 372 VLTGNDGEVRKICRATN 388
LTG GE+R+ CR N
Sbjct: 317 PLTGTQGEIRQNCRVVN 333
>At4g33870 putative peroxidase
Length = 358
Score = 197 bits (502), Expect = 6e-51
Identities = 119/301 (39%), Positives = 160/301 (52%), Gaps = 17/301 (5%)
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L +Y +SCP AEKIIA A +I P I+RL FHDCF+ GCDAS+LLD+
Sbjct: 68 LHYDYYRESCPTAEKIIAKAIRDIYNVTPSVAPPIIRLLFHDCFIEGCDASVLLDAD-EA 126
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
EK + N + LKG D++D +KS+LE CPG+VSCAD +V + EA+ + P
Sbjct: 127 HTSEKDASPN-LSLKGFDVIDAVKSELENVCPGVVSCADLLVLAAREAVLVVNFPSLTLS 185
Query: 204 GGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIF 263
G + A ++ LP P+ + +++ F +GF+ E V L GAHSIG HC F
Sbjct: 186 SGFAAAYRDFA----EHELPAPDATLSVILQRFSFRGFNERETVSLFGAHSIGITHCTFF 241
Query: 264 MDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNLFYRDLV 323
+R+YNF T KPDP L P FL EL+ C + + P + P +M N
Sbjct: 242 KNRLYNFSATGKPDPELNPGFLQELKTKCPFSVSTSSPSAPPDI-GLPPLMQN------- 293
Query: 324 DKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVRKI 383
K L+ +D L+ T V A D LF + FA M KL+S NVLTG G+VR
Sbjct: 294 ---KGLMSSDQQLMGSEVTEMWVRAYASDPLLFRREFAMSMMKLSSYNVLTGPLGQVRTS 350
Query: 384 C 384
C
Sbjct: 351 C 351
>At3g49110 peroxidase
Length = 354
Score = 197 bits (502), Expect = 6e-51
Identities = 117/308 (37%), Positives = 163/308 (51%), Gaps = 15/308 (4%)
Query: 88 FYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNGDNVE 147
FY SCP I+ D +R++P+ +ILRL FHDCFV GCDASILLD+T E
Sbjct: 37 FYDTSCPTVTNIVRDTIVNELRSDPRIAGSILRLHFHDCFVNGCDASILLDNT-TSFRTE 95
Query: 148 KSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPLGGRR 207
K + N +G ++D +K+ +E CP VSCAD + + +++ LAG P K GRR
Sbjct: 96 KDALGNANSARGFPVIDRMKAAVERACPRTVSCADMLTIAAQQSVTLAGGPSWKVPLGRR 155
Query: 208 DSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFS-PEEMVILLGAHSIGAAHCDIFMDR 266
DSL + + + NLP P ++ ++ F+ G P ++V L GAH+ G C MDR
Sbjct: 156 DSLQAFLDLA-NANLPAPFFTLPQLKANFKNVGLDRPSDLVALSGAHTFGKNQCRFIMDR 214
Query: 267 VYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD---NTPTVMDNLFYRDLV 323
+YNF NT PDP L +L LR C PR N+ V D TP V DN +Y +L
Sbjct: 215 LYNFSNTGLPDPTLNTTYLQTLRGQC-----PRNGNQSVLVDFDLRTPLVFDNKYYVNLK 269
Query: 324 DKGKSLLLTDAHLVTDPR---TAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEV 380
++ K L+ +D L + P T P V AD F F E M ++ ++ TG G++
Sbjct: 270 EQ-KGLIQSDQELFSSPNATDTIPLVRAYADGTQTFFNAFVEAMNRMGNITPTTGTQGQI 328
Query: 381 RKICRATN 388
R CR N
Sbjct: 329 RLNCRVVN 336
>At3g50990 peroxidase-like protein
Length = 336
Score = 197 bits (501), Expect = 8e-51
Identities = 115/308 (37%), Positives = 176/308 (56%), Gaps = 8/308 (2%)
Query: 83 NLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPN 142
+L FY +SCPNA+ I+ A +P+ A+ILRL FHDCFV GCDAS+LLDS+
Sbjct: 32 SLSPQFYENSCPNAQAIVQSYVANAYFNDPRMAASILRLHFHDCFVNGCDASVLLDSSGT 91
Query: 143 GDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKP 202
++ EK S N +G +++D+IKS LE ECP VSCAD + ++ +++ + G P +
Sbjct: 92 MES-EKRSNANRDSARGFEVIDEIKSALENECPETVSCADLLALVARDSIVICGGPSWEV 150
Query: 203 LGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDI 262
GRRD+ + + N+P P + ++ +F +G ++V LLG+H+IG + C
Sbjct: 151 YLGRRDA-REASLIGSMENIPSPESTLQTILTMFNFQGLDLTDLVALLGSHTIGNSRCIG 209
Query: 263 FMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDN-TPTVMDNLFYRD 321
F R+YN N PD L + + L+Q C G + N D TPT DN +Y++
Sbjct: 210 FRQRLYNHTGNNDPDQTLNQDYASMLQQGCPISGNDQ---NLFNLDYVTPTKFDNYYYKN 266
Query: 322 LVDKGKSLLLTDAHLVTDP-RTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEV 380
LV+ + LL +D L T T V A+++ F ++FA+ M K+ +++ LTG DGE+
Sbjct: 267 LVN-FRGLLSSDEILFTQSIETMEMVKYYAENEGAFFEQFAKSMVKMGNISPLTGTDGEI 325
Query: 381 RKICRATN 388
R+ICR N
Sbjct: 326 RRICRRVN 333
>At1g44970 peroxidase like protein
Length = 346
Score = 197 bits (501), Expect = 8e-51
Identities = 118/313 (37%), Positives = 171/313 (53%), Gaps = 17/313 (5%)
Query: 83 NLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPN 142
NL FY SCP A++I+ + + P+ A++LRL FHDCFV GCDASILLD +
Sbjct: 44 NLYPQFYQFSCPQADEIVMTVLEKAIAKEPRMAASLLRLHFHDCFVQGCDASILLDDSAT 103
Query: 143 GDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKP 202
+ EK++ N ++G ++D+IK+KLE+ CP VSCAD + + + L+G P +
Sbjct: 104 IRS-EKNAGPNKNSVRGFQVIDEIKAKLEQACPQTVSCADILALAARGSTILSGGPSWEL 162
Query: 203 LGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDI 262
GRRDS + + N+P PN + ++ +FQRKG + E++V L G H+IG A C
Sbjct: 163 PLGRRDSRTASLNGAN-TNIPAPNSTIQNLLTMFQRKGLNEEDLVSLSGGHTIGVARCTT 221
Query: 263 FMDRVYNFKNTNKPDPALRPPFLNELRQICSNPG-----TPRYRNEPVNFDNTPTVMDNL 317
F R+YN N+PD L + LR IC G +P P FDNT
Sbjct: 222 FKQRLYNQNGNNQPDETLERSYYYGLRSICPPTGGDNNISPLDLASPARFDNT------- 274
Query: 318 FYRDLVDKGKSLLLTDAHLVTD--PRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTG 375
Y L+ GK LL +D L+T +T V A+D+ LF ++FA+ M + ++ LTG
Sbjct: 275 -YFKLLLWGKGLLTSDEVLLTGNVGKTGALVKAYAEDERLFFQQFAKSMVNMGNIQPLTG 333
Query: 376 NDGEVRKICRATN 388
+GE+RK C N
Sbjct: 334 FNGEIRKSCHVIN 346
>At2g38380 peroxidase like protein
Length = 349
Score = 197 bits (500), Expect = 1e-50
Identities = 118/317 (37%), Positives = 173/317 (54%), Gaps = 10/317 (3%)
Query: 76 QPNTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASI 135
Q + N LR FY +CP II + + ++T+P+ A++LRL FHDCFV GCDASI
Sbjct: 23 QASNSNAQLRPDFYFGTCPFVFDIIGNIIVDELQTDPRIAASLLRLHFHDCFVRGCDASI 82
Query: 136 LLDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALA 195
LLD++ EK + N +G +++D +K LE CPG VSCAD + S ++ L+
Sbjct: 83 LLDNS-TSFRTEKDAAPNANSARGFNVIDRMKVALERACPGRVSCADILTIASQISVLLS 141
Query: 196 GMPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFS-PEEMVILLGAHS 254
G P GRRDS+ + + + LP P ++ ++ F G + ++V L G H+
Sbjct: 142 GGPWWPVPLGRRDSVEAFFALA-NTALPSPFFNLTQLKTAFADVGLNRTSDLVALSGGHT 200
Query: 255 IGAAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD-NTPTV 313
G A C R+YNF TN PDP+L P +L ELR++C G VNFD TP
Sbjct: 201 FGRAQCQFVTPRLYNFNGTNSPDPSLNPTYLVELRRLCPQNGNGTVL---VNFDVVTPDA 257
Query: 314 MDNLFYRDLVDKGKSLLLTDAHLVTDP--RTAPTVGQMADDQALFHKRFAEVMTKLTSLN 371
D+ +Y +L GK L+ +D L + P T P V Q + D ++F + F + M ++ +L
Sbjct: 258 FDSQYYTNL-RNGKGLIQSDQELFSTPGADTIPLVNQYSSDMSVFFRAFIDAMIRMGNLR 316
Query: 372 VLTGNDGEVRKICRATN 388
LTG GE+R+ CR N
Sbjct: 317 PLTGTQGEIRQNCRVVN 333
>At5g06720 peroxidase (emb|CAA68212.1)
Length = 335
Score = 195 bits (495), Expect = 4e-50
Identities = 114/313 (36%), Positives = 176/313 (55%), Gaps = 9/313 (2%)
Query: 79 TINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLD 138
T + L FYS +CPNA I+ + ++++ + A+++RL FHDCFV GCDASILLD
Sbjct: 27 TSSAQLNATFYSGTCPNASAIVRSTIQQALQSDTRIGASLIRLHFHDCFVNGCDASILLD 86
Query: 139 STPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMP 198
T + + EK++ N +G ++VD+IK+ LE CPG+VSC+D + S +++LAG P
Sbjct: 87 DTGSIQS-EKNAGPNVNSARGFNVVDNIKTALENACPGVVSCSDVLALASEASVSLAGGP 145
Query: 199 RQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAA 258
L GRRDSL + ++++P P S + F G + ++V L GAH+ G A
Sbjct: 146 SWTVLLGRRDSL-TANLAGANSSIPSPIESLSNITFKFSAVGLNTNDLVALSGAHTFGRA 204
Query: 259 HCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD-NTPTVMDNL 317
C +F +R++NF T PDP L L+ L+Q+C G+ + N D +TP DN
Sbjct: 205 RCGVFNNRLFNFSGTGNPDPTLNSTLLSTLQQLCPQNGS---ASTITNLDLSTPDAFDNN 261
Query: 318 FYRDLVDKGKSLLLTDAHL--VTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTG 375
++ +L LL +D L T T V A +Q LF + FA+ M + +++ LTG
Sbjct: 262 YFANL-QSNDGLLQSDQELFSTTGSSTIAIVTSFASNQTLFFQAFAQSMINMGNISPLTG 320
Query: 376 NDGEVRKICRATN 388
++GE+R C+ N
Sbjct: 321 SNGEIRLDCKKVN 333
>At4g37530 peroxidase - like protein
Length = 329
Score = 195 bits (495), Expect = 4e-50
Identities = 119/316 (37%), Positives = 173/316 (54%), Gaps = 12/316 (3%)
Query: 78 NTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILL 137
N + LR FY+ +CPN E+I+ +A + ++ + LRL FHDCFV GCDAS+++
Sbjct: 21 NLSSAQLRGDFYAGTCPNVEQIVRNAVQKKIQQTFTTIPATLRLYFHDCFVNGCDASVMI 80
Query: 138 DSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEE----CPGIVSCADTMVFLSFEAMA 193
ST N + EK N + L G IK+K + C VSCAD + + + +
Sbjct: 81 AST-NTNKAEKDHEDN-LSLAGDGFDTVIKAKEAVDAVPNCRNKVSCADILTMATRDVVN 138
Query: 194 LAGMPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAH 253
LAG P+ GRRD L S A+ V LP P + +++ LF G SP +M+ L GAH
Sbjct: 139 LAGGPQYAVELGRRDGLSSSASSVT-GKLPKPTFDLNQLNALFAENGLSPNDMIALSGAH 197
Query: 254 SIGAAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD-NTPT 312
++G AHC +R+YNF TN DP + ++ EL+ C PR +N D NTP
Sbjct: 198 TLGFAHCTKVFNRLYNFNKTNNVDPTINKDYVTELKASCPQNIDPRV---AINMDPNTPR 254
Query: 313 VMDNLFYRDLVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNV 372
DN++Y++L +GK L +D L TD R+ PTV A++ LF++ F M KL + V
Sbjct: 255 QFDNVYYKNL-QQGKGLFTSDQVLFTDSRSKPTVDLWANNGQLFNQAFISSMIKLGRVGV 313
Query: 373 LTGNDGEVRKICRATN 388
TG++G +R+ C A N
Sbjct: 314 KTGSNGNIRRDCGAFN 329
>At5g06730 peroxidase
Length = 358
Score = 192 bits (487), Expect = 3e-49
Identities = 111/313 (35%), Positives = 170/313 (53%), Gaps = 9/313 (2%)
Query: 79 TINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLD 138
T + L FYS +CPNA I+ + ++++ + +++RL FHDCFV GCD S+LLD
Sbjct: 28 TSSAQLNATFYSGTCPNASAIVRSTIQQALQSDARIGGSLIRLHFHDCFVNGCDGSLLLD 87
Query: 139 STPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMP 198
T + + EK++ N +G ++VD IK+ LE CPGIVSC+D + S +++LAG P
Sbjct: 88 DTSSIQS-EKNAPANANSTRGFNVVDSIKTALENACPGIVSCSDILALASEASVSLAGGP 146
Query: 199 RQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAA 258
L GRRD L + + +++LP P + + F G ++V L GAH+ G
Sbjct: 147 SWTVLLGRRDGLTANLSGA-NSSLPSPFEGLNNITSKFVAVGLKTTDVVSLSGAHTFGRG 205
Query: 259 HCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD-NTPTVMDNL 317
C F +R++NF T PDP L L+ L+Q+C G+ N D +TP DN
Sbjct: 206 QCVTFNNRLFNFNGTGNPDPTLNSTLLSSLQQLCPQNGS---NTGITNLDLSTPDAFDNN 262
Query: 318 FYRDLVDKGKSLLLTDAHLV--TDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTG 375
++ +L LL +D L T T P V A +Q LF + F + M K+ +++ LTG
Sbjct: 263 YFTNL-QSNNGLLQSDQELFSNTGSATVPIVNSFASNQTLFFEAFVQSMIKMGNISPLTG 321
Query: 376 NDGEVRKICRATN 388
+ GE+R+ C+ N
Sbjct: 322 SSGEIRQDCKVVN 334
>At2g18150 putative peroxidase
Length = 338
Score = 191 bits (486), Expect = 4e-49
Identities = 112/307 (36%), Positives = 168/307 (54%), Gaps = 6/307 (1%)
Query: 83 NLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPN 142
NL GFY SCP AE+I+ A+ V + A+++RL FHDCFV GCD S+LLD T
Sbjct: 35 NLFPGFYRSSCPRAEEIVRSVVAKAVARETRMAASLMRLHFHDCFVQGCDGSLLLD-TSG 93
Query: 143 GDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKP 202
EK+S N +G ++VD+IK+ LE ECP VSCAD + + ++ L G P
Sbjct: 94 SIVTEKNSNPNSRSARGFEVVDEIKAALENECPNTVSCADALTLAARDSSVLTGGPSWMV 153
Query: 203 LGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDI 262
GRRDS S + +NN+P PN + + +V F +G ++V L G+H+IG + C
Sbjct: 154 PLGRRDST-SASLSGSNNNIPAPNNTFNTIVTRFNNQGLDLTDVVALSGSHTIGFSRCTS 212
Query: 263 FMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD-NTPTVMDNLFYRD 321
F R+YN PD L + LRQ C G + +E D N+ DN ++++
Sbjct: 213 FRQRLYNQSGNGSPDRTLEQSYAANLRQRCPRSGGDQNLSE---LDINSAGRFDNSYFKN 269
Query: 322 LVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVR 381
L++ L + ++ ++ V + A+DQ F ++FAE M K+ +++ LTG+ GE+R
Sbjct: 270 LIENMGLLNSDEVLFSSNEQSRELVKKYAEDQEEFFEQFAESMIKMGNISPLTGSSGEIR 329
Query: 382 KICRATN 388
K CR N
Sbjct: 330 KNCRKIN 336
>At2g41480 peroxidase like protein
Length = 328
Score = 191 bits (485), Expect = 6e-49
Identities = 115/311 (36%), Positives = 170/311 (53%), Gaps = 17/311 (5%)
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L+ G+YS SCP AE I+ ++P +LRL FHDCFV GCD S+L+ G
Sbjct: 29 LKNGYYSTSCPKAESIVRSTVESHFDSDPTISPGLLRLHFHDCFVQGCDGSVLI----KG 84
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
+ E+++ N + L+G +++DD K++LE CPG+VSCAD + + +++ L+ P +
Sbjct: 85 KSAEQAALPN-LGLRGLEVIDDAKARLEAVCPGVVSCADILALAARDSVDLSDGPSWRVP 143
Query: 204 GGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIF 263
GR+D SLAT + +NLP P S + FQ KG ++V LLGAH+IG C F
Sbjct: 144 TGRKDGRISLAT--EASNLPSPLDSVAVQKQKFQDKGLDTHDLVTLLGAHTIGQTDCLFF 201
Query: 264 MDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD-NTPTVMDNLFYRDL 322
R+YNF T DP + P FL +L+ +C G R V D +P+ D F+++L
Sbjct: 202 RYRLYNFTVTGNSDPTISPSFLTQLKTLCPPNGDGSKR---VALDIGSPSKFDESFFKNL 258
Query: 323 VDKGKSLLLTDAHLVTDPRTAPTVGQMADDQA-----LFHKRFAEVMTKLTSLNVLTGND 377
D G ++L +D L +D T V + A F F + M K++S++V T D
Sbjct: 259 RD-GNAILESDQRLWSDAETNAVVKKYASRLRGLLGFRFDYEFGKAMIKMSSIDVKTDVD 317
Query: 378 GEVRKICRATN 388
GEVRK+C N
Sbjct: 318 GEVRKVCSKVN 328
>At5g42180 peroxidase (emb|CAA66960.1)
Length = 317
Score = 190 bits (483), Expect = 1e-48
Identities = 107/298 (35%), Positives = 163/298 (53%), Gaps = 10/298 (3%)
Query: 88 FYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNGDNVE 147
+Y +CP A+ I+ +A + + + A +LR+ FHDCFV GCD S+LLDS G N
Sbjct: 27 YYDHTCPQADHIVTNAVKKAMSNDQTVPAALLRMHFHDCFVRGCDGSVLLDS--KGKNKA 84
Query: 148 KSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPLGGRR 207
+ I L ++D+ K LEE+CPGIVSCAD + + +A+AL+G P GR+
Sbjct: 85 EKDGPPNISLHAFYVIDNAKKALEEQCPGIVSCADILSLAARDAVALSGGPTWAVPKGRK 144
Query: 208 DSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIFMDRV 267
D S A ++ LP P ++ ++ + F ++G S ++V L G H++G AHC F +R+
Sbjct: 145 DGRISKA--IETRQLPAPTFNISQLRQNFGQRGLSMHDLVALSGGHTLGFAHCSSFQNRL 202
Query: 268 YNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNLFYRDLVDKGK 327
+ F + DP L P F L +C P +N N D T T DN++Y+ L+ +GK
Sbjct: 203 HKFNTQKEVDPTLNPSFAARLEGVC--PAHNTVKNAGSNMDGTVTSFDNIYYKMLI-QGK 259
Query: 328 SLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVRKICR 385
SL +D L+ P T V + A+ F + F + M K++S ++GN EVR CR
Sbjct: 260 SLFSSDESLLAVPSTKKLVAKYANSNEEFERAFVKSMIKMSS---ISGNGNEVRLNCR 314
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.139 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,638,778
Number of Sequences: 26719
Number of extensions: 457540
Number of successful extensions: 1646
Number of sequences better than 10.0: 90
Number of HSP's better than 10.0 without gapping: 83
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1320
Number of HSP's gapped (non-prelim): 100
length of query: 388
length of database: 11,318,596
effective HSP length: 101
effective length of query: 287
effective length of database: 8,619,977
effective search space: 2473933399
effective search space used: 2473933399
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0076a.9


