

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0076a.12
(430 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
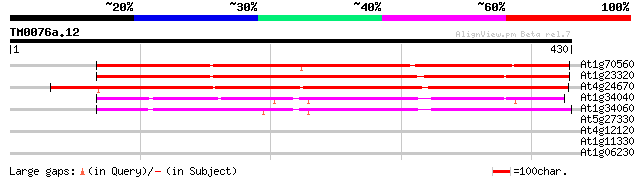
At1g70560 alliinase like protein 377 e-105
At1g23320 putative alliinase 365 e-101
At4g24670 putative alliin lyase 344 5e-95
At1g34040 putative protein 254 8e-68
At1g34060 unknown protein 253 1e-67
At5g27330 glutamic acid-rich protein 28 7.5
At4g12120 unknown protein 28 7.5
At1g11330 receptor-like protein kinase, putative 28 9.8
At1g06230 Ring3-like bromodomain protein 28 9.8
>At1g70560 alliinase like protein
Length = 391
Score = 377 bits (968), Expect = e-105
Identities = 183/365 (50%), Positives = 254/365 (69%), Gaps = 7/365 (1%)
Query: 67 LNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVV 126
+NLD GDP + +YW+K + C V I+GC+LMSY SD +N+CW++ PEL+DAI+ LH VV
Sbjct: 25 VNLDHGDPTAYEEYWRKMGDRCTVTIRGCDLMSYFSDMTNLCWFLEPELEDAIKDLHGVV 84
Query: 127 RNAVTKDKYIVIGTGTTQLFMAALFALSPSMTTSHPINVVSATPYYSEYKDQLDLFCSER 186
NA T+D+YIV+GTG+TQL AA+ ALS S+ S P++VV+A P+YS Y ++ S
Sbjct: 85 GNAATEDRYIVVGTGSTQLCQAAVHALS-SLARSQPVSVVAAAPFYSTYVEETTYVRSGM 143
Query: 187 FQWGGDAGEYDKNETYIEVVNYPNNPDGTIKEPMVK--SIVEGKLVHDLAYYWPHYTPIT 244
++W GDA +DK YIE+V PNNPDGTI+E +V E K++HD AYYWPHYTPIT
Sbjct: 144 YKWEGDAWGFDKKGPYIELVTSPNNPDGTIRETVVNRPDDDEAKVIHDFAYYWPHYTPIT 203
Query: 245 HQANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIM 304
+ +HD+MLFT SK TGHAGSRIGWA+VKD EVAKKMV ++ ++SIGV+KESQ R AKI+
Sbjct: 204 RRQDHDIMLFTFSKITGHAGSRIGWALVKDKEVAKKMVEYIIVNSIGVSKESQVRTAKIL 263
Query: 305 GVICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKAYCNFANES 364
V+ + +S + FF+Y + +++ RWEKLR ++ FT+ KYP+A+CN+ +S
Sbjct: 264 NVL---KETCKSESESENFFKYGREMMKNRWEKLREVVKESDAFTLPKYPEAFCNYFGKS 320
Query: 365 FEALPAFVWLKCEEGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIKR 424
E+ PAF WL +E + L + K+ R G R G+ KHVR+SM+ +D F ++R
Sbjct: 321 LESYPAFAWLGTKEETDLVSE-LRRHKVMSRAGERCGSDKKHVRVSMLSREDVFNVFLER 379
Query: 425 LSNAK 429
L+N K
Sbjct: 380 LANMK 384
>At1g23320 putative alliinase
Length = 388
Score = 365 bits (937), Expect = e-101
Identities = 178/363 (49%), Positives = 246/363 (67%), Gaps = 7/363 (1%)
Query: 67 LNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVV 126
+NLD+GDP F +YW KK + C VVI +LMSY SDT NVCW++ PEL+ AI+ LH +
Sbjct: 26 INLDQGDPTAFQEYWMKKKDRCTVVIPAWDLMSYFSDTKNVCWFLEPELEKAIKALHGAI 85
Query: 127 RNAVTKDKYIVIGTGTTQLFMAALFALSPSMTTSHPINVVSATPYYSEYKDQLDLFCSER 186
NA T+++YIV+GTG++QL AALFALS S++ P+++V+A PYYS Y ++ S
Sbjct: 86 GNAATEERYIVVGTGSSQLCQAALFALS-SLSEVKPVSIVAAVPYYSTYVEEASYLQSTL 144
Query: 187 FQWGGDAGEYDKNETYIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPITHQ 246
++W GDA +DK YIE+V PNNPDG ++EP+V GK++HDLAYYWPHYTPIT +
Sbjct: 145 YKWEGDARTFDKKGPYIELVTSPNNPDGIMREPVVNRREGGKVIHDLAYYWPHYTPITRR 204
Query: 247 ANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMGV 306
+HD+MLFT SK TGHAGSRIGWA+VKDIEVAKKMV ++ ++SIGV+KESQTRA I+
Sbjct: 205 QDHDLMLFTFSKITGHAGSRIGWALVKDIEVAKKMVHYLTINSIGVSKESQTRATTILNE 264
Query: 307 ICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKAYCNFANESFE 366
+ + + FFEY ++ RWE+LR +E FT+ YP+ +CNF ++
Sbjct: 265 LTKTCR-----TQSESFFEYGYEKMKSRWERLREVVESGDAFTLPNYPQDFCNFFGKTLS 319
Query: 367 ALPAFVWLKCEEGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIKRLS 426
PAF WL +E + + L + K+ RGG R G ++VR+SM+ DD+F ++RL+
Sbjct: 320 TSPAFAWLGYKEE-RDLGSLLKEKKVLTRGGDRCGCNKRYVRVSMLSRDDDFDVSLQRLA 378
Query: 427 NAK 429
K
Sbjct: 379 TIK 381
>At4g24670 putative alliin lyase
Length = 440
Score = 344 bits (883), Expect = 5e-95
Identities = 182/409 (44%), Positives = 256/409 (62%), Gaps = 18/409 (4%)
Query: 32 RARGFFQEHTLYPSMVVAKAASPINKNGFTSPSST-----------LNLDKGDPLVFGKY 80
R RG + T Y S+ + + + +S S +NL GDP V+ +Y
Sbjct: 33 RERGDSWDRTAYVSIWPVVSTTASESSSLSSASCNYSKIEEDDDRIINLKFGDPTVYERY 92
Query: 81 WKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVVRNAVTKDKYIVIGT 140
W++ E +VI G + +SY SD +N+CW++ PEL I R+H VV NAVT+D++IV+GT
Sbjct: 93 WQENGEVTTMVIPGWQSLSYFSDENNLCWFLEPELAKEIVRVHKVVGNAVTQDRFIVVGT 152
Query: 141 GTTQLFMAALFALSPSMTTSHPINVVSATPYYSEYKDQLDLFCSERFQWGGDAGEYDKNE 200
G+TQL+ AAL+ALSP S PINVVSATPYYS Y D S ++WGGDA Y ++
Sbjct: 153 GSTQLYQAALYALSPH-DDSGPINVVSATPYYSTYPLITDCLKSGLYRWGGDAKTYKEDG 211
Query: 201 TYIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPITHQANHDVMLFTLSKCT 260
YIE+V PNNPDG ++E +V S EG L+HDLAYYWP YTPIT A+HDVMLFT SK T
Sbjct: 212 PYIELVTSPNNPDGFLRESVVNS-TEGILIHDLAYYWPQYTPITSPADHDVMLFTASKST 270
Query: 261 GHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMGVICDGYQKFESIEPD 320
GHAG RIGWA+VKD E A+KM+ ++++++IGV+K+SQ R AK++ V+ D
Sbjct: 271 GHAGIRIGWALVKDRETARKMIEYIELNTIGVSKDSQLRVAKVLKVVSDSCGNVTG---- 326
Query: 321 QLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKAYCNFANESFEALPAFVWLKCEEGI 380
+ FF++ + + ERW+ L+++ + K F+V + CNF FE PAF W KCEEGI
Sbjct: 327 KSFFDHSYDAMYERWKLLKQAAKDTKRFSVPDFVSQRCNFFGRVFEPQPAFAWFKCEEGI 386
Query: 381 ENAENYL-GKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIKRLSNA 428
+ E +L + KI + G FG +VR+SM+ D F + R++++
Sbjct: 387 VDCEKFLREEKKILTKSGKYFGDELSNVRISMLDRDTNFNIFLHRITSS 435
>At1g34040 putative protein
Length = 457
Score = 254 bits (648), Expect = 8e-68
Identities = 142/371 (38%), Positives = 222/371 (59%), Gaps = 29/371 (7%)
Query: 67 LNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVV 126
++ + GDPL +W +K+EE VV G MSY + + +M EL+ I +LH+VV
Sbjct: 90 VDANSGDPLFLEPFWIRKAEESAVVESGWHRMSYTFNGYGL--FMSAELEKIIRKLHNVV 147
Query: 127 RNAVTKDKYIVIGTGTTQLFMAALFALSPSMTTSHPINVVSATPYYSEYKDQLDLFCSER 186
NAVT +++I+ G G TQL A++ ALS + + S P +V++ PYY+ YK Q D F S
Sbjct: 148 GNAVTDNRFIIFGAGATQLLAASVHALSQTNSLS-PSRLVTSVPYYNLYKQQADFFNSTN 206
Query: 187 FQWGGDAGEYDKNET------YIEVVNYPNNPDGTIKEPMVKSIVEG---KLVHDLAYYW 237
++ GDA + ++E IE+V PNNPDG +K +++++G K +HD AYYW
Sbjct: 207 LKFEGDASAWKRSERNDDIKQVIEIVTSPNNPDGKLK----RAVLDGPNVKYIHDYAYYW 262
Query: 238 PHYTPITHQANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQ 297
P+++PIT QA+ D+ LF+LSK TGHAGSR GWA+VK+ V +KM +++ +SS+GV++++Q
Sbjct: 263 PYFSPITRQADEDLSLFSLSKTTGHAGSRFGWALVKEKTVYEKMKIYISLSSMGVSRDTQ 322
Query: 298 TRAAKIMGVIC-DGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKA 356
RA +++ V+ DG + F + L++RWE L + + F++
Sbjct: 323 LRALQLLKVVIGDGGNE---------IFRFGYGTLKKRWEILNKIFSMSTRFSLETIKPE 373
Query: 357 YCNFANESFEALPAFVWLKCEEGIENAENY--LGKLKIRGRGGVRFGAGSKHVRLSMIGT 414
YCN+ + E P++ W+KCE E+ + Y KI GR G FG+ + VRLS+I +
Sbjct: 374 YCNYFKKVREFTPSYAWVKCERP-EDTDCYEIFKAAKITGRNGEMFGSDERFVRLSLIRS 432
Query: 415 DDEFTELIKRL 425
D+F +LI L
Sbjct: 433 QDDFDQLIAML 443
>At1g34060 unknown protein
Length = 463
Score = 253 bits (647), Expect = 1e-67
Identities = 137/376 (36%), Positives = 217/376 (57%), Gaps = 28/376 (7%)
Query: 67 LNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVV 126
++ + GDPL +W +++E +++ G MSY+ + Y+ EL+ I +LH VV
Sbjct: 91 VDANSGDPLFLEPFWMRQAERSAILVSGWHRMSYIYEDGT---YVSRELEKVIRKLHSVV 147
Query: 127 RNAVTKDKYIVIGTGTTQLFMAALFALS-PSMTTSHPINVVSATPYYSEYKDQLDLFCSE 185
NAVT +++++ G+GTTQL AA+ ALS + + S P ++++ PYY+ YKDQ + F S
Sbjct: 148 GNAVTDNRFVIFGSGTTQLLAAAVHALSLTNSSVSSPARLLTSIPYYAMYKDQAEFFDSA 207
Query: 186 RFQWGGDA------GEYDKNETYIEVVNYPNNPDGTIKEPMVKSIVEG---KLVHDLAYY 236
++ G+A G D IEVV PNNPDG +K +++++G K +HD AYY
Sbjct: 208 HLKFEGNASAWKQSGRNDNITQVIEVVTSPNNPDGKLK----RAVLDGPNVKTLHDYAYY 263
Query: 237 WPHYTPITHQANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKES 296
WPH++PITH + D+ LF+LSK TGHAGSR GW +VKD + +KM F++++S+GV+KE+
Sbjct: 264 WPHFSPITHPVDEDLSLFSLSKTTGHAGSRFGWGLVKDKAIYEKMDRFIRLTSMGVSKET 323
Query: 297 QTRAAKIMGVIC-DGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPK 355
Q +++ V+ DG + F + +++RWE L + + F++
Sbjct: 324 QLHVLQLLKVVVGDGGNE---------IFSFGYGTVKKRWETLNKIFSMSTRFSLQTIKP 374
Query: 356 AYCNFANESFEALPAFVWLKCEEGIE-NAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGT 414
YCN+ + E P++ W+KCE + N KI GR G FG+ + VRLS+I +
Sbjct: 375 EYCNYFKKVREFTPSYAWVKCERPEDTNCYEIFRAAKITGRNGNVFGSEERFVRLSLIRS 434
Query: 415 DDEFTELIKRLSNAKY 430
D+F +LI L Y
Sbjct: 435 QDDFDQLIAMLKKLVY 450
>At5g27330 glutamic acid-rich protein
Length = 628
Score = 28.5 bits (62), Expect = 7.5
Identities = 14/47 (29%), Positives = 25/47 (52%)
Query: 111 MLPELKDAIERLHHVVRNAVTKDKYIVIGTGTTQLFMAALFALSPSM 157
M+ E+K + ++ H V +A K + + + T L MAA A + S+
Sbjct: 581 MVEEMKKELAKMKHSVEDAHKKKSFWTLVSSVTSLLMAASVAYAASL 627
>At4g12120 unknown protein
Length = 662
Score = 28.5 bits (62), Expect = 7.5
Identities = 24/88 (27%), Positives = 37/88 (41%), Gaps = 11/88 (12%)
Query: 96 ELMSYLSDTSNVCWYMLPE--LKDAIERLHHVVRNAVTKDKYIVIGTGTTQLFMAALFAL 153
E L D + W L + + DA ERLH + N V+K+K + +
Sbjct: 302 EKKEVLLDEEDSIWVELRDAHIADASERLHEKMTNFVSKNKAAQLKHSSKDF-------- 353
Query: 154 SPSMTTSHPINVVSATPYYSEYKDQLDL 181
+++ +V A P YSE D+L L
Sbjct: 354 -GDLSSKDLQKMVHALPQYSEQIDKLSL 380
>At1g11330 receptor-like protein kinase, putative
Length = 840
Score = 28.1 bits (61), Expect = 9.8
Identities = 15/44 (34%), Positives = 25/44 (56%), Gaps = 2/44 (4%)
Query: 73 DPLVFGKYWKKKSEECMVVIKGC--ELMSYLSDTSNVCWYMLPE 114
DP VF K ++K+ E+C+ + C E+ + + SNV W + E
Sbjct: 753 DPAVFDKCFEKEIEKCVHIGLLCVQEVANDRPNVSNVIWMLTTE 796
>At1g06230 Ring3-like bromodomain protein
Length = 766
Score = 28.1 bits (61), Expect = 9.8
Identities = 12/25 (48%), Positives = 17/25 (68%)
Query: 55 INKNGFTSPSSTLNLDKGDPLVFGK 79
++KN +PS TL+L+ GD LV K
Sbjct: 179 VDKNPIEAPSQTLSLEDGDTLVVDK 203
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,775,940
Number of Sequences: 26719
Number of extensions: 422907
Number of successful extensions: 998
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 967
Number of HSP's gapped (non-prelim): 10
length of query: 430
length of database: 11,318,596
effective HSP length: 102
effective length of query: 328
effective length of database: 8,593,258
effective search space: 2818588624
effective search space used: 2818588624
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0076a.12


