

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0076a.11
(428 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
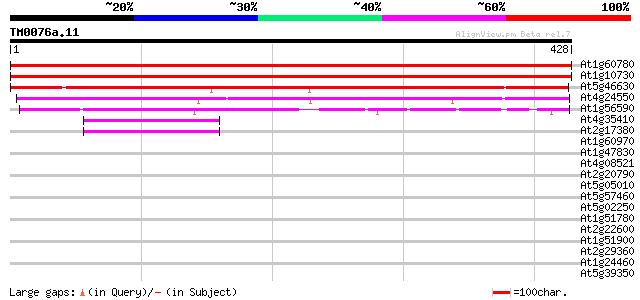
Score E
Sequences producing significant alignments: (bits) Value
At1g60780 putative Clathrin Coat Assembly protein 795 0.0
At1g10730 putative clathrin-coat assembly protein 760 0.0
At5g46630 AP47/50p (gb|AAB88283.1) 363 e-100
At4g24550 clathrin coat assembly like protein 256 1e-68
At1g56590 clathrin coat assembly like protein 151 8e-37
At4g35410 clathrin assembly protein AP19 homolog 46 3e-05
At2g17380 clathrin assembly protein AP19, small subunit 44 1e-04
At1g60970 coatomer zeta subunit like protein 34 0.14
At1g47830 clathrin coat assembly protein AP17, putative 34 0.18
At4g08521 putative coatomer protein 33 0.30
At2g20790 unknown protein 33 0.39
At5g05010 coatomer delta subunit (delta-coat protein) (delta-COP) 32 0.67
At5g57460 unknown protein 32 0.88
At5g02250 ribonuclease II-like protein 31 1.1
At1g51780 hypothetical protein 30 3.3
At2g22600 putative RNA-binding protein 29 4.4
At1g51900 Hypothetical protein 29 4.4
At2g29360 putative tropinone reductase 28 7.4
At1g24460 unknown protein 28 7.4
At5g39350 putative protein 28 9.7
>At1g60780 putative Clathrin Coat Assembly protein
Length = 428
Score = 795 bits (2054), Expect = 0.0
Identities = 389/428 (90%), Positives = 414/428 (95%)
Query: 1 MAGAASALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISY 60
MAGAASALFLLDIKGRVL+WRDYRGDV+A AERFFTKLIEK+ D+QS DPV +DNG++Y
Sbjct: 1 MAGAASALFLLDIKGRVLVWRDYRGDVSAAQAERFFTKLIEKEGDSQSNDPVAYDNGVTY 60
Query: 61 MFVQHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEI 120
MFVQHSNVYLMIA+RQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDE+
Sbjct: 61 MFVQHSNVYLMIASRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEM 120
Query: 121 MDFGYPQYTEAKILSEFIKTDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFLDVVES 180
MDFGYPQYTEA+ILSEFIKTDAYRMEVTQRPPMAVTNAVSWRSEGI YKKNEVFLDV+E+
Sbjct: 121 MDFGYPQYTEARILSEFIKTDAYRMEVTQRPPMAVTNAVSWRSEGIQYKKNEVFLDVIEN 180
Query: 181 VNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTTKGKAIDLEDIK 240
VNILVNSNGQI+RSDVVGALKMRTYL+GMPECKLGLNDRVLLEAQGR TKGKAIDLEDIK
Sbjct: 181 VNILVNSNGQIVRSDVVGALKMRTYLTGMPECKLGLNDRVLLEAQGRATKGKAIDLEDIK 240
Query: 241 FHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATVEKHSKSRIEIMVKA 300
FHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEA +E HS+SR+E+++KA
Sbjct: 241 FHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEAQIESHSRSRVEMLIKA 300
Query: 301 RSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRSFPGGKEYMLR 360
RSQFKERSTATNVEIELPVP DA NP VRTS+GSA+YAPEKDAL+WKI+SFPG KEYMLR
Sbjct: 301 RSQFKERSTATNVEIELPVPTDASNPTVRTSLGSASYAPEKDALVWKIKSFPGNKEYMLR 360
Query: 361 AEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQALPWVRYITMA 420
AEF LPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQALPWVRYITMA
Sbjct: 361 AEFHLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQALPWVRYITMA 420
Query: 421 GEYELRLI 428
GEYELRL+
Sbjct: 421 GEYELRLV 428
>At1g10730 putative clathrin-coat assembly protein
Length = 428
Score = 760 bits (1962), Expect = 0.0
Identities = 372/428 (86%), Positives = 406/428 (93%)
Query: 1 MAGAASALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISY 60
MAGAASALFLLDIKGRVL+WRDYRGDVTA AERFFTKLIE + D+QS DPV +DNG++Y
Sbjct: 1 MAGAASALFLLDIKGRVLVWRDYRGDVTAAQAERFFTKLIETEGDSQSNDPVAYDNGVTY 60
Query: 61 MFVQHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEI 120
MFVQHSN+YLMIA+RQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDE+
Sbjct: 61 MFVQHSNIYLMIASRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEM 120
Query: 121 MDFGYPQYTEAKILSEFIKTDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFLDVVES 180
MDFGYPQ+TEA+ILSEFIKTDAYRMEVTQRPPMAVTN+VSWRSEG+ +KKNEVFLDV+ES
Sbjct: 121 MDFGYPQFTEARILSEFIKTDAYRMEVTQRPPMAVTNSVSWRSEGLKFKKNEVFLDVIES 180
Query: 181 VNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTTKGKAIDLEDIK 240
VNILVNSNGQI+RSDVVGALKMRTYLSGMPECKLGLNDR+LLEAQGR KGKAIDLEDIK
Sbjct: 181 VNILVNSNGQIVRSDVVGALKMRTYLSGMPECKLGLNDRILLEAQGRAIKGKAIDLEDIK 240
Query: 241 FHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATVEKHSKSRIEIMVKA 300
FHQCVRLARFENDRTISFIPPDG+FDLMTYRLSTQVKPLIWVEA +E+HS+SR+E++VKA
Sbjct: 241 FHQCVRLARFENDRTISFIPPDGSFDLMTYRLSTQVKPLIWVEAHIERHSRSRVEMLVKA 300
Query: 301 RSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRSFPGGKEYMLR 360
RSQFK+RS AT+VEIELPVP DA NP+VRTS+GSAAYAPEKDAL+WKI+ F G KE+ L+
Sbjct: 301 RSQFKDRSYATSVEIELPVPTDAYNPDVRTSLGSAAYAPEKDALVWKIQYFYGNKEHTLK 360
Query: 361 AEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQALPWVRYITMA 420
A+F LPSI AEEATPERKAPIRVKFEIP F VSGIQVRYLKIIEKSGYQA PWVRYITMA
Sbjct: 361 ADFHLPSIAAEEATPERKAPIRVKFEIPKFIVSGIQVRYLKIIEKSGYQAHPWVRYITMA 420
Query: 421 GEYELRLI 428
GEYELRL+
Sbjct: 421 GEYELRLM 428
>At5g46630 AP47/50p (gb|AAB88283.1)
Length = 438
Score = 363 bits (932), Expect = e-100
Identities = 192/440 (43%), Positives = 276/440 (62%), Gaps = 17/440 (3%)
Query: 1 MAGAASALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISY 60
M AASA++ L+++G VLI R YR DV + F T ++ Q PV G S+
Sbjct: 1 MPVAASAIYFLNLRGDVLINRTYRDDVGGNMVDAFRTHIM--QTKELGNCPVRQIGGCSF 58
Query: 61 MFVQHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFE-ELEEESLRDNFVVVYELLDE 119
++++ SNVY++I N N A F+ V +FK YF +E+++R+NFV++YELLDE
Sbjct: 59 VYMRISNVYIVIVVSSNANVACGFKFVVEAVALFKSYFGGAFDEDAIRNNFVLIYELLDE 118
Query: 120 IMDFGYPQYTEAKILSEFIKTDAYRMEVTQRPP--------MAVTNAVSWRSEGISYKKN 171
IMDFGYPQ +IL +I + R + +P + VT AV WR EG++YKKN
Sbjct: 119 IMDFGYPQNLSPEILKLYITQEGVRSPFSSKPKDKPVPNATLQVTGAVGWRREGLAYKKN 178
Query: 172 EVFLDVVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGR---- 227
EVFLD+VESVN+L++S G ++R DV G + M+ +LSGMP+ KLGLND++ LE +
Sbjct: 179 EVFLDIVESVNLLMSSKGNVLRCDVTGKVLMKCFLSGMPDLKLGLNDKIGLEKESEMKSR 238
Query: 228 -TTKGKAIDLEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATV 286
GK I+L+D+ FHQCV L RF +++T+SF+PPDG F+LM YR++ V V T+
Sbjct: 239 PAKSGKTIELDDVTFHQCVNLTRFNSEKTVSFVPPDGEFELMKYRITEGVNLPFRVLPTI 298
Query: 287 EKHSKSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIW 346
++ ++R+E+ VK +S F + A V +++PVP N + + G A Y P D L+W
Sbjct: 299 KELGRTRMEVNVKVKSVFGAKMFALGVVVKIPVPKQTAKTNFQVTTGRAKYNPSIDCLVW 358
Query: 347 KIRSFPGGKEYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKS 406
KIR FPG E L AE L S E+ + R PI+++F++P FT SG++VR+LK+ EKS
Sbjct: 359 KIRKFPGQTESTLSAEIELISTMGEKKSWTR-PPIQMEFQVPMFTASGLRVRFLKVWEKS 417
Query: 407 GYQALPWVRYITMAGEYELR 426
GY + WVRYIT AG YE+R
Sbjct: 418 GYNTVEWVRYITKAGSYEIR 437
>At4g24550 clathrin coat assembly like protein
Length = 451
Score = 256 bits (655), Expect = 1e-68
Identities = 143/450 (31%), Positives = 241/450 (52%), Gaps = 30/450 (6%)
Query: 6 SALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHD-NGISYMFVQ 64
S F+L +G +++RDYR +V E FF K+ + D ++ P + + +G++Y V+
Sbjct: 4 SQFFVLSQRGDNIVFRDYRAEVPKGSTETFFRKVKFWKEDGNAEAPPIFNVDGVNYFHVK 63
Query: 65 HSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFG 124
+Y + TR N + + +L L R+ V K Y L E+S R NFV+VYELLDE++DFG
Sbjct: 64 VVGLYFVATTRVNVSPSLVLELLQRIARVIKDYLGVLNEDSFRKNFVLVYELLDEVIDFG 123
Query: 125 YPQYTEAKILSEFIKTDA----------------YRMEVTQRPPMAVTNAVSWRSEGISY 168
Y Q T ++L +I + + + P AVT +V G
Sbjct: 124 YVQTTSTEVLKSYIFNEPIVVSPARLQPIDPAAIFTQGAKRMPGTAVTKSVVANDPG-GR 182
Query: 169 KKNEVFLDVVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRT 228
++ E+F+D++E +++ +S+G I+ S++ G ++M++YLSG PE +L LN+ + + GR+
Sbjct: 183 RREEIFVDIIEKISVTFSSSGYILTSEIDGTIQMKSYLSGNPEIRLALNEDLNIGRGGRS 242
Query: 229 ------TKGKAIDLEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWV 282
+ G + L+D FH+ VRL F++DRT+S +PPDG F +M YR++ + KP V
Sbjct: 243 VYDYRSSSGSGVILDDCNFHESVRLDSFDSDRTLSLVPPDGEFPVMNYRMTQEFKPPFHV 302
Query: 283 EATVEKHSKSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAA----YA 338
+E+ + + E+++K R++F A + +++P+P + G+A +
Sbjct: 303 NTLIEEAGRLKAEVIIKIRAEFPSDIIANTITVQMPLPNYTSRASFELEPGAAGQRTDFK 362
Query: 339 PEKDALIWKIRSFPGGKEYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVR 398
L W ++ GG E+ LRA+ T E P+ + F IP + VS +QV+
Sbjct: 363 ESNKMLEWNLKKIVGGGEHTLRAKLTFSQEFHGNITKE-AGPVSMTFTIPMYNVSKLQVK 421
Query: 399 YLKIIEK-SGYQALPWVRYITMAGEYELRL 427
YL+I +K S Y WVRY+T A Y R+
Sbjct: 422 YLQIAKKSSSYNPYRWVRYVTQANSYVARI 451
>At1g56590 clathrin coat assembly like protein
Length = 480
Score = 151 bits (381), Expect = 8e-37
Identities = 119/443 (26%), Positives = 201/443 (44%), Gaps = 53/443 (11%)
Query: 8 LFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMF-VQHS 66
+FL+ G V++ + G +F Q D+ PV+ + Y+F +
Sbjct: 68 IFLISDSGEVMLEKQLTGHRVDRSICAWFWDQYISQGDSFKALPVIA-SPTHYLFQIVRD 126
Query: 67 NVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFGYP 126
+ + ++ + FL RV DV Y L E+ ++DNF++VYELLDE++D G+P
Sbjct: 127 GITFLACSQVEMPPLMAIEFLCRVADVLSEYLGGLNEDLIKDNFIIVYELLDEMIDNGFP 186
Query: 127 QYTEAKILSEFIK-----------TDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFL 175
TE IL E I V+ P + V WR Y NEV++
Sbjct: 187 LTTEPSILKEMIAPPNLVSKMLSVVTGNASNVSDTLPSGAGSCVPWRPTDPKYSSNEVYV 246
Query: 176 DVVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTTKGKAID 235
D+VE ++ +VN +G++++ ++ G ++M + L+G P+ L + +
Sbjct: 247 DLVEEMDAIVNRDGELVKCEIYGEVQMNSQLTGFPDLTLSFANPSI-------------- 292
Query: 236 LEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPL----IWVEATVEKHSK 291
LED++FH CVR +E+ + +SF+PPDG F LM+YR VK L ++V+ + S
Sbjct: 293 LEDMRFHPCVRYRPWESHQVLSFVPPDGEFKLMSYR--CVVKKLKNTPVYVKPQITSDSG 350
Query: 292 S-RIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRS 350
+ RI ++V RS T ++ + +P + ++ ++ G+ K W I
Sbjct: 351 TCRISVLVGIRSD--PGKTIESITLSFQLPHCVSSADLSSNHGTVTILSNK-TCTWTIGR 407
Query: 351 FPGGKEYMLRAEFRL-PSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQ 409
P K L L P + P ++ F+I +SG++ IEK Q
Sbjct: 408 IPKDKTPCLSGTLALEPGLERLHVFP----TFKLGFKIMGIALSGLR------IEKLDLQ 457
Query: 410 ALP-----WVRYITMAGEYELRL 427
+P R T AGE+++RL
Sbjct: 458 TIPPRLYKGFRAQTRAGEFDVRL 480
>At4g35410 clathrin assembly protein AP19 homolog
Length = 162
Score = 46.2 bits (108), Expect = 3e-05
Identities = 24/104 (23%), Positives = 50/104 (48%)
Query: 57 GISYMFVQHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYEL 116
G ++ +++++Y + Q N +L +H V++ YF + E L NF Y +
Sbjct: 54 GYKVVYKRYASLYFCMCIDQEDNELEVLEIIHHYVEILDRYFGSVCELDLIFNFHKAYYI 113
Query: 117 LDEIMDFGYPQYTEAKILSEFIKTDAYRMEVTQRPPMAVTNAVS 160
LDE++ G Q + K ++ I +EV + +++N ++
Sbjct: 114 LDELLIAGELQESSKKTVARIISAQDQLVEVAKEEASSISNIIA 157
>At2g17380 clathrin assembly protein AP19, small subunit
Length = 161
Score = 44.3 bits (103), Expect = 1e-04
Identities = 23/104 (22%), Positives = 50/104 (47%)
Query: 57 GISYMFVQHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYEL 116
G ++ +++++Y + + N +L +H V++ YF + E L NF Y +
Sbjct: 54 GYKVVYKRYASLYFCMCIDEADNELEVLEIIHHYVEILDRYFGSVCELDLIFNFHKAYYI 113
Query: 117 LDEIMDFGYPQYTEAKILSEFIKTDAYRMEVTQRPPMAVTNAVS 160
LDE++ G Q + K ++ I +EV + +++N ++
Sbjct: 114 LDELLIAGELQESSKKTVARIISAQDQLVEVAKEEASSISNIIA 157
>At1g60970 coatomer zeta subunit like protein
Length = 177
Score = 34.3 bits (77), Expect = 0.14
Identities = 31/134 (23%), Positives = 60/134 (44%), Gaps = 8/134 (5%)
Query: 8 LFLLDIKGRVLIWRDYRGDVTALDAER-----FFTKLIEKQADAQSQDPVVHDNGISYMF 62
+ LLD +G+ + + Y D A+ FTK + A + + + +N + Y F
Sbjct: 10 ILLLDSEGKRVAVKYYSDDWPTNSAQEAFEKSVFTKTQKTNARTEVEVTALENNIVVYKF 69
Query: 63 VQHSNVYLMIATRQN-CNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIM 121
VQ + ++ +N AS+L L V + +++ DN +++ DEI+
Sbjct: 70 VQDLHFFVTGGEEENELILASVLEGLFDAVTLLLR--SNVDKREALDNLDLIFLSFDEII 127
Query: 122 DFGYPQYTEAKILS 135
D G T+A +++
Sbjct: 128 DGGIVLETDANVIA 141
>At1g47830 clathrin coat assembly protein AP17, putative
Length = 154
Score = 33.9 bits (76), Expect = 0.18
Identities = 19/76 (25%), Positives = 37/76 (48%)
Query: 61 MFVQHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEI 120
++ +++ ++ + N + L +H V++ H+F + E L NF VY +LDE
Sbjct: 70 IYRRYAGLFFSVCVDITDNELAYLESIHLFVEILDHFFSNVCELDLVFNFHKVYLILDEF 129
Query: 121 MDFGYPQYTEAKILSE 136
+ G Q T + + E
Sbjct: 130 ILAGELQETSKRAIIE 145
>At4g08521 putative coatomer protein
Length = 181
Score = 33.1 bits (74), Expect = 0.30
Identities = 29/135 (21%), Positives = 65/135 (47%), Gaps = 10/135 (7%)
Query: 8 LFLLDIKGRVLIWRDYRGD----VTALDAERF-FTKLIEKQADAQSQDPVVHDNGISYMF 62
+ LLD +G+ + + Y D + L E++ F+K + A +++ ++ N + Y F
Sbjct: 14 ILLLDSEGKRVAVKYYSDDWATNASKLAFEKYVFSKTSKTNARTEAEITLLESNIVVYKF 73
Query: 63 VQHSNVYLMIATRQNCNAASLLF--FLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEI 120
Q + ++ +N S + F V + ++ E++E +N +++ LDE+
Sbjct: 74 AQDLHFFVTGGENENELVLSSVLQGFFDAVALLLRNNVEKMEA---LENLDLIFLCLDEM 130
Query: 121 MDFGYPQYTEAKILS 135
+D G T+A +++
Sbjct: 131 VDQGMVLETDANVIA 145
>At2g20790 unknown protein
Length = 613
Score = 32.7 bits (73), Expect = 0.39
Identities = 34/131 (25%), Positives = 53/131 (39%), Gaps = 17/131 (12%)
Query: 151 PPMAVTNAVSWRSEGISYK-KNEVFLDVVESVNILVNSNGQIIRS-DVVGALKMRTYLSG 208
PP +W+ YK K + + E+V+ + +I + V G + R L G
Sbjct: 291 PPPQELKQPAWKP--YLYKGKQRLLFTIHETVSAAMYDRDEIPDNVSVAGQINCRAELEG 348
Query: 209 MPECKLGLNDRVLLEAQGRTTKGKAIDLEDIKFHQCVRLARFENDR-TISFIPPDGAFDL 267
+P+ L G +T +E I FH C ++ D+ I F PP G F L
Sbjct: 349 LPDVSFPL--------AGLSTA----HIEAISFHPCAQVPAHGIDKQNIVFQPPLGNFVL 396
Query: 268 MTYRLSTQVKP 278
M Y+ + P
Sbjct: 397 MRYQAGCGLGP 407
>At5g05010 coatomer delta subunit (delta-coat protein) (delta-COP)
Length = 527
Score = 32.0 bits (71), Expect = 0.67
Identities = 40/187 (21%), Positives = 66/187 (34%), Gaps = 31/187 (16%)
Query: 175 LDVVESVNILVNSNGQIIRSDVVGALKMR--TYLSGMPECKL--GLNDRVLLEAQGRTTK 230
L V E +N+ + +G + D+ G L ++ G + ++ G N +L
Sbjct: 287 LTVEEKLNVALRRDGGLSSFDMQGTLSLQILNQEDGFVQVQIATGENPEILF-------- 338
Query: 231 GKAIDLEDIKFHQCVRLARFENDRTISFIPPDGAFD---------LMTYRLSTQVKPLIW 281
K H + F N+ + PD F L+ +R+ + ++
Sbjct: 339 ---------KTHPNINRDMFNNENILGLKRPDQPFPTGQGGDGVGLLRWRMQRADESMVP 389
Query: 282 VEATVEKHSKSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEK 341
+ S S E V + TNV I +P+P P+VR G Y P
Sbjct: 390 LTINCWP-SVSGNETYVSLEYEASSMFDLTNVIISVPLPALREAPSVRQCDGEWRYDPRN 448
Query: 342 DALIWKI 348
L W I
Sbjct: 449 SVLEWSI 455
Score = 30.4 bits (67), Expect = 2.0
Identities = 27/166 (16%), Positives = 73/166 (43%), Gaps = 7/166 (4%)
Query: 15 GRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQHSNVYLMIAT 74
G+V++ R Y D++ + E K Q + + Y++ ++L++ T
Sbjct: 13 GKVIVSRHYV-DMSRIRIEGLLAAF-PKLVGMGKQHTYIETENVRYVYQPIEALFLLLVT 70
Query: 75 RQNCNAASLLFFLHRVVDVFKHYFEELEEESL-RDNFVVVYELLDEIMDFGYPQYTEAKI 133
+ N L L + + Y L+EE + R +F +++ DE++ G+ +
Sbjct: 71 TKQSNILEDLATLTLLSKLVPEYSMSLDEEGISRASFELIFAF-DEVISLGHKESVTVAQ 129
Query: 134 LSEFIKTDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFLDVVE 179
+ ++ + +++ ++ + + + + ++ + + K NE+ +E
Sbjct: 130 VKQYCEMESHEEKLHK---LVMQSKINDTKDVMKRKANEIDKSKIE 172
>At5g57460 unknown protein
Length = 646
Score = 31.6 bits (70), Expect = 0.88
Identities = 37/141 (26%), Positives = 58/141 (40%), Gaps = 8/141 (5%)
Query: 260 PPDGAFDLMTYRLSTQVKPLIWVEATVEKHSKSRIEIMVKARSQFKERSTATNVEIELPV 319
P + ++ Y L ++ PL V++ S + + +M++ S NV+ L +
Sbjct: 488 PSEEPIPILKYSLQPKLTPLPLRVRMVKRISGTLLSLMIQYVSNPDLPQPLKNVDFILKL 547
Query: 320 PVDAMNPNVRTSMGSAAYAPEKDALIWKIRSFP-GGKEYMLRAEFRLPSITAEEATPERK 378
PVD P + A L W+I P G LRA + S +EE E
Sbjct: 548 PVD---PTLLKVSPKAILNRTDRELKWQIPEIPLNGSPGRLRARMPIDSDNSEE---EPD 601
Query: 379 APIRVKFEIPYFT-VSGIQVR 398
VKF + T +SGI +R
Sbjct: 602 IICYVKFSVQGNTSLSGISLR 622
>At5g02250 ribonuclease II-like protein
Length = 803
Score = 31.2 bits (69), Expect = 1.1
Identities = 32/135 (23%), Positives = 56/135 (40%), Gaps = 13/135 (9%)
Query: 4 AASALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFV 63
A SA L D GR+ IW + D R+ T + +A+ + V +Y
Sbjct: 421 ALSATRLQD--GRIKIW------IHVADPARYVTPGSKVDREARRRGTSVFLPTATYPMF 472
Query: 64 QHSNVYLMIATRQ--NCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIM 121
++ RQ NCNA S+ L + ++ ++ +R +++ YE E++
Sbjct: 473 PEKLAMEGMSLRQGENCNAVSVSVVLRSDGCITEY---SVDNSIIRPTYMLTYESASELL 529
Query: 122 DFGYPQYTEAKILSE 136
+ E K+LSE
Sbjct: 530 HLNLEEEAELKLLSE 544
>At1g51780 hypothetical protein
Length = 424
Score = 29.6 bits (65), Expect = 3.3
Identities = 17/71 (23%), Positives = 34/71 (46%), Gaps = 4/71 (5%)
Query: 95 KHYFEELEEESLRDNFVVVYELLDEIMDFGYPQYTEAKILSEFIKTDAYRMEVTQRPPMA 154
K++ + E D V + + E + GY + +K+ +KT+ +M V+ + P+A
Sbjct: 33 KNFLSLAKREDFFDWMVGIRRRIHENPELGYEEVETSKL----VKTELDKMGVSYKNPVA 88
Query: 155 VTNAVSWRSEG 165
VT + + G
Sbjct: 89 VTGVIGYVGTG 99
>At2g22600 putative RNA-binding protein
Length = 649
Score = 29.3 bits (64), Expect = 4.4
Identities = 23/72 (31%), Positives = 38/72 (51%), Gaps = 5/72 (6%)
Query: 104 ESLRDNFVVVYELL---DEIMDFGYPQYTEAKILSEFIKT--DAYRMEVTQRPPMAVTNA 158
E L+ N V+ E+L DE+ DF PQ + L++ +K + +V+ PP +V
Sbjct: 509 EVLKSNSVMHTEVLKEVDELNDFTLPQSLLEEDLTQGMKQLQMSSNGDVSSLPPRSVLAK 568
Query: 159 VSWRSEGISYKK 170
RS+G+S +K
Sbjct: 569 QQRRSKGVSVRK 580
>At1g51900 Hypothetical protein
Length = 765
Score = 29.3 bits (64), Expect = 4.4
Identities = 21/90 (23%), Positives = 39/90 (43%), Gaps = 16/90 (17%)
Query: 118 DEIMDFGYPQYTEAKILSEFIKTDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFLDV 177
+EIM+ Y + + ++L + +K +R+E+++R Y K E+ + V
Sbjct: 464 EEIMEDDYIEALKCRMLDDILKKSGHRLEISRR----------------QYNKPEIEIQV 507
Query: 178 VESVNILVNSNGQIIRSDVVGALKMRTYLS 207
E V + IR D ++ TY S
Sbjct: 508 NEKEEKKVINTDMDIRYDDESPEEVETYSS 537
>At2g29360 putative tropinone reductase
Length = 271
Score = 28.5 bits (62), Expect = 7.4
Identities = 12/43 (27%), Positives = 23/43 (52%)
Query: 126 PQYTEAKILSEFIKTDAYRMEVTQRPPMAVTNAVSWRSEGISY 168
P + E +++E + + +R EV RPPM V+ S +++
Sbjct: 198 PWFIETPLVTESLSNEEFRKEVESRPPMGRVGEVNEVSSLVAF 240
>At1g24460 unknown protein
Length = 1791
Score = 28.5 bits (62), Expect = 7.4
Identities = 32/117 (27%), Positives = 52/117 (44%), Gaps = 18/117 (15%)
Query: 287 EKHSKSRIEIMVKARSQFKERSTATNVEIE------LPVPVDAMNPNVRTSMGSAAYAPE 340
E HS +I+ K RS +ER TNV E L V +D ++S+ S
Sbjct: 483 EGHS---FDIVEKVRSLAEERKELTNVSQEYNRLKDLIVSIDLPEEMSQSSLES------ 533
Query: 341 KDALIWKIRSFPGGKEYMLRAEFRLPSITAE-EATPERKAPIRVKFEIPYFTVSGIQ 396
L W SF GK+ + + R+ S++ A E K+ IR + + F++ ++
Sbjct: 534 --RLAWLRESFLQGKDEVNALQNRIESVSMSLSAEMEEKSNIRKELDDLSFSLKKME 588
>At5g39350 putative protein
Length = 677
Score = 28.1 bits (61), Expect = 9.7
Identities = 28/134 (20%), Positives = 57/134 (41%), Gaps = 5/134 (3%)
Query: 123 FGYPQYTEAKILSEFIKTDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFLDVVESVN 182
FG +Y + +L+ ++ M M + +SW + Y +N D + +
Sbjct: 148 FGRDKYVQNALLAMYMNFGKVEMARDVFDVMKNRDVISWNTMISGYYRNGYMNDALMMFD 207
Query: 183 ILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTTKGKAIDLEDIKFH 242
+VN + + + +V L + +L + ++G N L+E + K + + +
Sbjct: 208 WMVNESVDLDHATIVSMLPVCGHLK---DLEMGRNVHKLVEEKRLGDKIEVKNALVNMYL 264
Query: 243 QCVRL--ARFENDR 254
+C R+ ARF DR
Sbjct: 265 KCGRMDEARFVFDR 278
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,036,263
Number of Sequences: 26719
Number of extensions: 367848
Number of successful extensions: 814
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 788
Number of HSP's gapped (non-prelim): 23
length of query: 428
length of database: 11,318,596
effective HSP length: 102
effective length of query: 326
effective length of database: 8,593,258
effective search space: 2801402108
effective search space used: 2801402108
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0076a.11


