

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0075.5
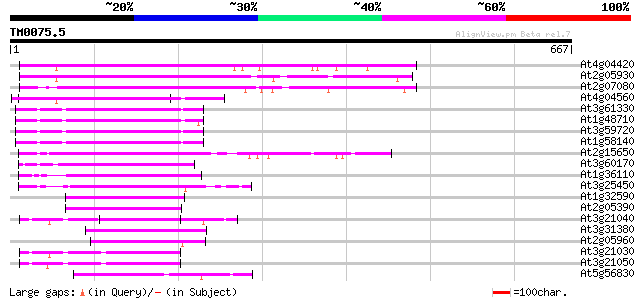
(667 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g04420 putative transposon protein 202 5e-52
At2g05930 copia-like retroelement pol polyprotein 183 3e-46
At2g07080 putative gag-protease polyprotein 168 1e-41
At4g04560 putative transposon protein 134 1e-31
At3g61330 copia-type polyprotein 124 2e-28
At1g48710 hypothetical protein 122 5e-28
At3g59720 copia-type reverse transcriptase-like protein 122 6e-28
At1g58140 hypothetical protein 122 6e-28
At2g15650 putative retroelement pol polyprotein 115 1e-25
At3g60170 putative protein 112 9e-25
At1g36110 hypothetical protein 95 1e-19
At3g25450 hypothetical protein 92 1e-18
At1g32590 hypothetical protein, 5' partial 90 5e-18
At2g05390 putative retroelement pol polyprotein 84 2e-16
At3g21040 hypothetical protein 80 4e-15
At3g31380 hypothetical protein 78 2e-14
At2g05960 putative retroelement pol polyprotein 77 4e-14
At3g21030 hypothetical protein 76 7e-14
At3g21050 hypothetical protein 70 3e-12
At5g56830 unknown protein 65 9e-11
>At4g04420 putative transposon protein
Length = 1008
Score = 202 bits (514), Expect = 5e-52
Identities = 154/547 (28%), Positives = 264/547 (48%), Gaps = 75/547 (13%)
Query: 12 ILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTE---LKPEDKWTKKEDDE 68
+L+ NY +WK +M ++ + AW A GWK PVI LK +D+W E+ +
Sbjct: 15 MLEKGNYGHWKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTKDQWNDAEEAK 74
Query: 69 ALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFET 128
A NS+AL++IFN V++N F+ I C AKEAW+ L A+EGTS V+ S++ +L +QFE
Sbjct: 75 AKANSRALSLIFNFVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQFEN 134
Query: 129 MKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDI 188
+ M E E+I EF +I +A+ LG+ ++KL +K+LR LP RF+ K TA+ + D
Sbjct: 135 LSMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSLDT 194
Query: 189 SNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVSLLNK 248
+I +E++G LQ +E+ + +K + ++ +++E Q KDT + +A+ S +
Sbjct: 195 DSIDFEEVVGMLQAYELEITSGKGGYSKGLALAASAKKNEIQELKDTMSMMAKDFSRAMR 254
Query: 249 AL--KSLGRMSNTNVLDNVS---DNVKNTEFQ-------------LKDKHENDTTKAIHV 290
+ K GR T+ + S D ++ E Q KD ++ H
Sbjct: 255 RVEKKGFGRNQGTDRYRDRSSKRDEIQCHECQGYGHIKAECPSLKRKDLKCSECNGLGHT 314
Query: 291 K-ALIG-------KCYSDAE--SSDGDEEELVETYKLLLAKWEESCMYGE-KMRKEVKDL 339
K +G C S++E S+DGD E+ ++ + + EE + + E +D
Sbjct: 315 KFDCVGSKSKPDKSCSSESESDSNDGDSEDYIKGFVSFVGIIEEKDESSDSEADGEDEDN 374
Query: 340 IAEKKQLQSNNSSLQEEVK-------TISKLR----EENEKLQITNAKLQEEVTLLNSK- 387
A++ + ++ EE + +SK + EE K+Q KL+ E+T N K
Sbjct: 375 SADEDSDIEKDVNINEEFRKLYDSWLMLSKEKVAWLEEKLKVQELTEKLKGELTAANQKN 434
Query: 388 --------------------LEGMKKSIRMMNKSTNVLEEILEVGKTVGDMEGIGF---- 423
L +K+I M+N T L+ IL G+ G+G+
Sbjct: 435 SELIQKCSVAEEKNRELSQELSDTRKNIHMLNSGTKDLDSILAAGRVGKSNFGLGYNGAG 494
Query: 424 -----SYKSANKSASSEKQTK-QPMSDPMLHHSVRHVYPQFRKSKKST-WRCHHCGKLGH 476
++ + +A ++ QT + D + V + ++ + T + C++CG+ GH
Sbjct: 495 SGTKTNFVRSEAAAPTKSQTGFRSNYDAVPARRVYQNHDHYQSRRTVTGYECYYCGRHGH 554
Query: 477 IRPYCYK 483
I+ YCY+
Sbjct: 555 IQRYCYR 561
>At2g05930 copia-like retroelement pol polyprotein
Length = 916
Score = 183 bits (464), Expect = 3e-46
Identities = 141/491 (28%), Positives = 234/491 (46%), Gaps = 41/491 (8%)
Query: 12 ILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTE---LKPEDKWTKKEDDE 68
IL+ NY +WK +M ++ + AW A GWK PVI LK ED+W E+ +
Sbjct: 27 ILEKGNYGHWKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTEDQWNDAEEAK 86
Query: 69 ALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFET 128
A NS+AL++IFN V++N F+ I C AKEAW+ L A+EGTS V+ S++ +L +QFE
Sbjct: 87 ATANSRALSLIFNSVNQNQFKQIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQFEN 146
Query: 129 MKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDI 188
+ M E E+I EF +I +A+ LG+ ++KL +K+LR LP RF+ K TA+ + D
Sbjct: 147 LTMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSLDT 206
Query: 189 SNIKVDELIGSLQTFEMSL-NGRSEKKAKSITFVSNTEEDEDQREKDTDANIA-EAVSLL 246
++I +E++G Q +E+ + +G+ S +D E +I + V
Sbjct: 207 NSIDFEEVVGMFQAYELEITSGKGGYGHIKAECPSLKRKDLKCSECKGLGHIKFDCVGSK 266
Query: 247 NKALKSLGRMSNTNVLDNVS-DNVKN-TEFQ--LKDKHENDTTKAIHVKALIGKCYSDAE 302
+K +S S ++ D S D +K F +++K E+ ++A G+ ++
Sbjct: 267 SKPDRSCSSESESDSNDGDSEDYIKGFVSFVGIIEEKDESSDSEA------DGEDEDNSA 320
Query: 303 SSDGDEEELV---ETYKLLLAKWEESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQEEVKT 359
D D E+ V E ++ L W + KE + EK ++Q L+ E+
Sbjct: 321 DEDSDIEKDVKINEEFRKLYDSW-------LMLSKEKVAWLEEKLKVQELTEKLKGELTA 373
Query: 360 ISKLREE-NEKLQITNAKLQEEVTLLNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGDM 418
++ E +K + K +E L+ +L +K I M+N T L+ IL G+
Sbjct: 374 ANQKNSELTQKCSVAEEKNRE----LSQELSDTRKKIHMLNSGTKDLDSILAAGRVGKSN 429
Query: 419 EGIGFS-YKSANKSASSEKQTKQPMSDPMLHHSVRHVYPQFR----------KSKKSTWR 467
G+G++ S K+ + P S P R + + +
Sbjct: 430 FGLGYNGVGSGTKTNFVRSEAGAPTKSQTGFRSNYDAVPARRMYQNHDHYHSRRTVTGYE 489
Query: 468 CHHCGKLGHIR 478
C++CG+ GH R
Sbjct: 490 CYYCGRHGHFR 500
>At2g07080 putative gag-protease polyprotein
Length = 627
Score = 168 bits (425), Expect = 1e-41
Identities = 144/558 (25%), Positives = 232/558 (40%), Gaps = 111/558 (19%)
Query: 12 ILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDDEALG 71
+LD Y YWK M ++ + G+K KP+ WT +E ++
Sbjct: 15 LLDTKRYGYWKVCMTQIIRGQED--------GFKIT--------KPKANWTAEEKLQSKF 58
Query: 72 NSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFETMKM 131
N++A+ IFNGVD++ F+LI C AK+AW+ L+ +HEGTS V+ ++L + TQFE +KM
Sbjct: 59 NARAMKAIFNGVDEDEFKLIQGCKSAKQAWDTLQKSHEGTSSVKRTRLDHIATQFEYLKM 118
Query: 132 NEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDISNI 191
DE I +F +I LAN +G+ ++KL +K+LR LP +F + A + I
Sbjct: 119 EPDEKIVKFSSKISALANEAEVMGKTYKDQKLVKKLLRCLPPKFAAHKAVMRVAGNTDKI 178
Query: 192 KVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVSLLNKALK 251
+L+G L+ EM + K +K+I F ++ ++ Q KD A +A K ++
Sbjct: 179 SFVDLVGMLKLEEMKADQDKVKPSKNIAFNADQGSEQFQEIKDGMALLARNFGKALKRVE 238
Query: 252 SLGRMSNTNVLDNVSDNV-KNTEFQLKD-------KHENDTTKAIHVKALIGKC------ 297
G S + +D++ K E Q + K E TK +K L KC
Sbjct: 239 IDGERSRGRFSRSENDDLRKKKEIQCYECGGFGHIKPECPITKRKEMKCL--KCKGVGHT 296
Query: 298 -----------------YSDAESSDGDEEEL----------------------------V 312
+SD+ES D EE L
Sbjct: 297 KFECPNKSKLKEKSLISFSDSESDDEGEELLNFVAFMASSDSSKFMSDTDSDCDEELNPK 356
Query: 313 ETYKLLLAKWEESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQ-EEVKTISKLREENEKLQ 371
+ Y++L W + + K+ L+ EK L++ +++ E+ + +S + +
Sbjct: 357 DKYRVLYDSWVQ-------LSKDKLKLVKEKLTLEAKLANVSTEDKQKLSGITVDGNSQD 409
Query: 372 ITNAKL----------QEEVTLLNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGDMEGI 421
KL ++ LL +L K IRM+NK L++IL +G+T G+
Sbjct: 410 YYQKKLDCLQKECHRERDRTKLLERELNDKYKQIRMLNKGLESLDKILAMGRTDSQQRGL 469
Query: 422 GFSYKSANKSASSEKQTKQPMSDPMLHHSVRHVYPQFRKSKKSTWR-------------- 467
G+ + + VR Y + +K KS
Sbjct: 470 GYQGYTGKIKKEEGRVINFVSGGSTSETVVRQSYIEPKKQVKSHVEIKRESVVRTRMVGV 529
Query: 468 --CHHCGKLGHIRPYCYK 483
C HCGK H+R CYK
Sbjct: 530 ICCDHCGKRFHMREQCYK 547
>At4g04560 putative transposon protein
Length = 590
Score = 134 bits (338), Expect = 1e-31
Identities = 75/258 (29%), Positives = 135/258 (52%), Gaps = 8/258 (3%)
Query: 3 ARRSIYMP-PI-LDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTE---LKP 57
A+R I +P P+ LD +Y YWK + ++S+D AW A+ GW P K
Sbjct: 336 AQRFIAIPKPLKLDAEHYGYWKVLIKRSIQSIDMDAWFAVEDGWMPPTTKDAKRDIVSKS 395
Query: 58 EDKWTKKEDDEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMS 117
+W E A NS+AL+VIF + +N F + C AKE WEIL+ + E T+ V+ +
Sbjct: 396 RTEWIADEKTAANHNSQALSVIFGSLLRNKFTQVQGCLSAKEVWEILQVSFECTNNVKRT 455
Query: 118 KLQLLTTQFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDM 177
+L +L ++FE + M +ES+ +F+ ++ + LG+ ++K+ +K LRSLP +F
Sbjct: 456 RLDMLASEFENLTMEAEESVDDFNGKLSSITQEAVVLGKTYKDKKMVKKFLRSLPDKFQS 515
Query: 178 KVTAIEEAQDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDA 237
+AI+ + + +K D+++G +Q ++ + E T+ E+D+ E+D
Sbjct: 516 HKSAIDVSLNSDQLKFDQVVGMMQAYD---TDKEEILNSYATYFGAIEDDDHTVEEDAQM 572
Query: 238 NIAEAVSLLNKALKSLGR 255
+++ L+ + G+
Sbjct: 573 GTIKSLILIQSDSEKEGK 590
Score = 58.5 bits (140), Expect = 1e-08
Identities = 43/183 (23%), Positives = 84/183 (45%), Gaps = 29/183 (15%)
Query: 11 PILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDDEA- 69
PI + NY +W+ +M ++ W+ + +G P + + PE K + A
Sbjct: 8 PIFNKENYGFWRIKMKTIFQTKK--LWEIVDEGVPKP--PAEGDHSPEAVQQKTRCEAAS 63
Query: 70 LGNSKALNVIFNGVDKNMF-RLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFET 128
L + AL ++ V ++F R+ + + WE +E
Sbjct: 64 LKDLTALQILQTAVSDSIFPRIAPASSALGKPWE-----------------------YEN 100
Query: 129 MKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDI 188
+KM E ++I F ++ ++ N GE S+ ++ +KIL SLPKRFD+ V +++ +D+
Sbjct: 101 LKMKESDNINTFMTKLIEMGNQLRVHGEEKSDYQIVQKILISLPKRFDIIVAMMKQTKDL 160
Query: 189 SNI 191
+++
Sbjct: 161 TSL 163
>At3g61330 copia-type polyprotein
Length = 1352
Score = 124 bits (310), Expect = 2e-28
Identities = 69/223 (30%), Positives = 122/223 (53%), Gaps = 7/223 (3%)
Query: 8 YMPPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDD 67
+ P+L +NYD W RM L + D W+ + KG+ P + +D D
Sbjct: 8 FQVPVLTKSNYDNWSLRMKAILGAHD--VWEIVEKGFIEPENEGSLSQTQKDGLR----D 61
Query: 68 EALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFE 127
+ KAL +I+ G+D++ F + T AKEAWE L+T+++G +V+ +LQ L +FE
Sbjct: 62 SRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFE 121
Query: 128 TMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQD 187
++M E E + ++ R+ + N+ GE + + ++ K+LRSL +F+ VT IEE +D
Sbjct: 122 ALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKD 181
Query: 188 ISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQ 230
+ + +++L+GSLQ +E + E A+ + + T+E+ Q
Sbjct: 182 LEAMTIEQLLGSLQAYE-EKKKKKEDIAEQVLNMQITKEENGQ 223
>At1g48710 hypothetical protein
Length = 1352
Score = 122 bits (307), Expect = 5e-28
Identities = 72/226 (31%), Positives = 123/226 (53%), Gaps = 13/226 (5%)
Query: 8 YMPPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDD 67
+ P+L +NYD W RM L + D W+ + KG+ P + +D D
Sbjct: 8 FQVPVLTKSNYDNWSLRMKAILGAHD--VWEIVEKGFIEPENEGSLSQTQKDGLR----D 61
Query: 68 EALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFE 127
+ KAL +I+ G+D++ F + T AKEAWE L+T+++G +V+ +LQ L +FE
Sbjct: 62 SRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFE 121
Query: 128 TMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQD 187
++M E E + ++ R+ + N+ GE + + ++ K+LRSL +F+ VT IEE +D
Sbjct: 122 ALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKD 181
Query: 188 ISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSN---TEEDEDQ 230
+ + +++L+GSLQ +E + +KK I V N T+E+ Q
Sbjct: 182 LEAMTIEQLLGSLQAYE----EKKKKKEDIIEQVLNMQITKEENGQ 223
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 122 bits (306), Expect = 6e-28
Identities = 68/223 (30%), Positives = 121/223 (53%), Gaps = 7/223 (3%)
Query: 8 YMPPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDD 67
+ P+L +NYD W RM L + D W+ + KG+ P + +D D
Sbjct: 8 FQVPVLTKSNYDNWSLRMKAILGAHD--VWEIVEKGFIEPENEGSLSQTQKDGLR----D 61
Query: 68 EALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFE 127
+ KAL +I+ G+D++ F + T AKEAWE L+T+++G +V+ +LQ L +FE
Sbjct: 62 SRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFE 121
Query: 128 TMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQD 187
++M E E + ++ R+ + N+ GE + + ++ K+LRSL +F+ VT IEE +D
Sbjct: 122 ALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKD 181
Query: 188 ISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQ 230
+ + +++L+GSLQ +E + E + + + T+E+ Q
Sbjct: 182 LEAMTIEQLLGSLQAYE-EKKKKKEDIVEQVLNMQITKEENGQ 223
>At1g58140 hypothetical protein
Length = 1320
Score = 122 bits (306), Expect = 6e-28
Identities = 68/223 (30%), Positives = 121/223 (53%), Gaps = 7/223 (3%)
Query: 8 YMPPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDD 67
+ P+L +NYD W RM L + D W+ + KG+ P + +D D
Sbjct: 8 FQVPVLTKSNYDNWSLRMKAILGAHD--VWEIVEKGFIEPENEGSLSQTQKDGLR----D 61
Query: 68 EALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFE 127
+ KAL +I+ G+D++ F + T AKEAWE L+T+++G +V+ +LQ L +FE
Sbjct: 62 SRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFE 121
Query: 128 TMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQD 187
++M E E + ++ R+ + N+ GE + + ++ K+LRSL +F+ VT IEE +D
Sbjct: 122 ALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKD 181
Query: 188 ISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQ 230
+ + +++L+GSLQ +E + E + + + T+E+ Q
Sbjct: 182 LEAMTIEQLLGSLQAYE-EKKKKKEDIVEQVLNMQITKEENGQ 223
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 115 bits (287), Expect = 1e-25
Identities = 112/480 (23%), Positives = 208/480 (43%), Gaps = 62/480 (12%)
Query: 11 PILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDDEAL 70
PI DG YD+W +M ++ W + +G PV E PE K +EA+
Sbjct: 10 PIFDGEKYDFWSIKMATIFRTRK--LWSVVEEGV--PVEPVQAEETPETARAKTLREEAV 65
Query: 71 GN-SKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFETM 129
N + AL ++ V +F I + +KEAW++LK ++G+ +VR+ KLQ L ++E +
Sbjct: 66 TNDTMALQILQTAVTDQIFSRIAAASSSKEAWDVLKDEYQGSPQVRLVKLQSLRREYENL 125
Query: 130 KMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDIS 189
KM ++++I F ++ L GE + +L +KIL SLP +FD V+ +E+ +D+
Sbjct: 126 KMYDNDNIKTFTDKLIVLEIQLTYHGEKKTNTQLIQKILISLPAKFDSIVSVLEQTRDLD 185
Query: 190 NIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVSLLNKA 249
+ + EL+G L+ E + R E + +V + + ++ +T+ + N+
Sbjct: 186 ALTMSELLGILKAQEARVTAREESTKEGAFYVRSKGRESGFKQDNTNNRV-------NQD 238
Query: 250 LKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHEND---TTKAIHVKAL----IGKCYSDAE 302
K G ++ K+TE + ++K +ND K ++K IG ++
Sbjct: 239 KKWCGFHKSS----------KHTEEECREKPKNDDHGKNKRSNIKCYKCGKIGHYANECR 288
Query: 303 SSDG-------DEEELVETYKLLLAKWEESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQE 355
S + +EE++ E + L A EES E + + + S++ +
Sbjct: 289 SKNKERAHVTLEEEDVNEDHMLFSASEEESTTLREDVWLVDSGCTNHMTKEERYFSNINK 348
Query: 356 EVKTISKLREENEKLQITNAKLQEEVTLLNSK--------LEGMKKS------------- 394
+K +R N + +T K V + K + G++K+
Sbjct: 349 SIKV--PIRVRNGDIVMTAGKGDITVMTRHGKRIIKNVFLVPGLEKNLLSVPQIISSGYW 406
Query: 395 IRMMNKSTNVLEEILEVGKTVGDMEGIGFSYKSANKSASSEKQTKQPMSDPMLHHSVRHV 454
+R +K + + GK + ++E S+K S E T ++ H + HV
Sbjct: 407 VRFQDKRCIIQD---ANGKEIMNIEMTDKSFKIKLSSVEEEAMTANVQTEETWHKRLGHV 463
Score = 30.4 bits (67), Expect = 3.3
Identities = 17/65 (26%), Positives = 28/65 (42%), Gaps = 9/65 (13%)
Query: 426 KSANKSASSEKQTKQPMSDPMLHHSVRHVYPQFR---------KSKKSTWRCHHCGKLGH 476
+S K ++ + Q H S +H + R K+K+S +C+ CGK+GH
Sbjct: 223 ESGFKQDNTNNRVNQDKKWCGFHKSSKHTEEECREKPKNDDHGKNKRSNIKCYKCGKIGH 282
Query: 477 IRPYC 481
C
Sbjct: 283 YANEC 287
>At3g60170 putative protein
Length = 1339
Score = 112 bits (279), Expect = 9e-25
Identities = 71/211 (33%), Positives = 112/211 (52%), Gaps = 9/211 (4%)
Query: 11 PILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDDEA- 69
P DG YD+W M FL+S + W+ + +G V+ +T P + + +EA
Sbjct: 13 PRFDGY-YDFWSMTMENFLRSRE--LWRLVEEGIPAIVVGTT----PVSEAQRSAVEEAK 65
Query: 70 LGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFETM 129
L + K N +F +D+ + I + +K WE +K ++G++KV+ ++LQ L +FE +
Sbjct: 66 LKDLKVKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEFELL 125
Query: 130 KMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDIS 189
M E E I F R + N GE M + + KILRSL +F+ V +IEE+ D+S
Sbjct: 126 AMKEGEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEESNDLS 185
Query: 190 NIKVDELIGSLQTFEMSLNGR-SEKKAKSIT 219
+ +DEL GSL E LNG E++A +T
Sbjct: 186 TLSIDELHGSLLVHEQRLNGHVQEEQALKVT 216
>At1g36110 hypothetical protein
Length = 745
Score = 95.1 bits (235), Expect = 1e-19
Identities = 57/220 (25%), Positives = 110/220 (49%), Gaps = 26/220 (11%)
Query: 11 PILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDDEAL 70
P+L TNY W RM + +S++ + W+ I G D+
Sbjct: 20 PMLTSTNYTVWAIRMQI-ARSVNKM-WETIEPGI----------------------DDGY 55
Query: 71 GNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFETMK 130
N+ A +IF + +++ + T AK W+ +KT + G +V+ ++LQ L +FE MK
Sbjct: 56 KNTMARGLIFQSIPESLTLQVGTLATAKLVWDSIKTRYVGADRVKEARLQTLMAEFEKMK 115
Query: 131 MNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLP-KRFDMKVTAIEEAQDIS 189
M E E I F R+ +LA + ALG + KL +K L +LP +++ + ++E+ D++
Sbjct: 116 MKESEKIDVFAGRLAELATRSDALGSNIETSKLVKKFLNALPLRKYIHIIASLEQVLDLN 175
Query: 190 NIKVDELIGSLQTFEMSL-NGRSEKKAKSITFVSNTEEDE 228
N ++++G ++ +E + +G ++ + +NT+ +
Sbjct: 176 NTSFEDIVGRIKVYEERVWDGEEQEDDQGKLMYANTDTQD 215
>At3g25450 hypothetical protein
Length = 1343
Score = 91.7 bits (226), Expect = 1e-18
Identities = 68/281 (24%), Positives = 132/281 (46%), Gaps = 44/281 (15%)
Query: 11 PILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDDEAL 70
P+L+ NY W RM L+ W I G D+E
Sbjct: 23 PMLNSVNYTVWTMRMEAVLRVHK--LWGTIEPG--------------------SADEEK- 59
Query: 71 GNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFETMK 130
N A ++F + +++ + + WE +K+ + G +V+ ++LQ L +F+ +K
Sbjct: 60 -NDMARALLFQSIPESLILQVGKQKTSSAVWEAIKSRNLGAERVKEARLQTLMAEFDKLK 118
Query: 131 MNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLP-KRFDMKVTAIEEAQDIS 189
M + E+I ++ RI ++ ALGE + E K+ +K L+SLP K++ V A+E+ D+
Sbjct: 119 MKDSETIDDYVGRISEITTKAAALGEDIEESKIVKKFLKSLPRKKYIHIVAALEQVLDLK 178
Query: 190 NIKVDELIGSLQTFEMSL---NGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVSLL 246
+++ G ++T+E + + E + K +T V D+ + E++ ++
Sbjct: 179 TTTFEDIAGRIKTYEDRVWDDDDSHEDQGKLMTEVEEEVVDDLEEEEE---------EVI 229
Query: 247 NKALKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKA 287
NK +K + ++V+D + ++ E K+K E+DT +A
Sbjct: 230 NKEIK-----AKSHVIDRLLKLIRLQE--QKEKEEDDTHEA 263
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 89.7 bits (221), Expect = 5e-18
Identities = 46/142 (32%), Positives = 80/142 (55%)
Query: 67 DEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQF 126
++ + + K N +F +DK + + I +K+ WE +K ++G +V+ ++LQ L F
Sbjct: 20 EKTVKDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQGNDRVQSAQLQRLRRSF 79
Query: 127 ETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQ 186
E ++M E+I + R+ ++ N LGE M + K+ KILR+L ++F V AIEE+
Sbjct: 80 EVLEMKIGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTLVEKFTYVVCAIEESN 139
Query: 187 DISNIKVDELIGSLQTFEMSLN 208
+I + VD L SL E +L+
Sbjct: 140 NIKELTVDGLQSSLMVHEQNLS 161
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 84.3 bits (207), Expect = 2e-16
Identities = 41/139 (29%), Positives = 82/139 (58%), Gaps = 1/139 (0%)
Query: 67 DEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQF 126
D+ N A ++F V ++ + +K WE +KT + G +V+ +KLQ L +F
Sbjct: 19 DDMEKNDMARALLFQSVPESTILQVGKHKTSKAMWEAIKTRNLGAERVKEAKLQTLMAEF 78
Query: 127 ETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLP-KRFDMKVTAIEEA 185
+ + M ++E+I EF RI +++ + +LGE + E K+ +K L+SLP K++ + A+E+
Sbjct: 79 DRLNMKDNETIDEFVGRISEISTKSESLGEEIEESKIVKKFLKSLPRKKYIHIIAALEQI 138
Query: 186 QDISNIKVDELIGSLQTFE 204
D++ ++++G ++T+E
Sbjct: 139 LDLNTTGFEDIVGRMKTYE 157
>At3g21040 hypothetical protein
Length = 534
Score = 80.1 bits (196), Expect = 4e-15
Identities = 57/270 (21%), Positives = 121/270 (44%), Gaps = 21/270 (7%)
Query: 12 ILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWK-----HPVIASTTELKPEDKWTKKED 66
+ D +Y++W + + +++ W + G P +A+T +++ +W +
Sbjct: 9 VSDDFDYEHWSP--IAETRLVENGVWDVVQNGVSPNPTTDPEVAATIKVEDLTQWRNR-- 64
Query: 67 DEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQF 126
+ ++KAL ++ + + ++FR + +KE W++LK +G KL+ L Q
Sbjct: 65 --MIEDNKALKIMQSALPDSVFRKTISVASSKELWDLLK---KGNDNEEAKKLRRLEKQL 119
Query: 127 ETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQ 186
E + M E E + + R+ + F G P+S+EK+ K+L SLP+ +D +T ++E
Sbjct: 120 ENLTMYEGERMKSYLKRVEKIIEEFFVWGNPISDEKVIAKLLTSLPRPYDDSITVLKEFM 179
Query: 187 DISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDE------DQREKDTDANIA 240
+ ++ +L+ + + F + ++ K I + D D+ DA +
Sbjct: 180 TLPDLTHRDLLKAFEMFGSNPKTMPQELMKFINILRKAHSDRLPCRGYDKNNPIKDAKLL 239
Query: 241 EAVSLLNKALKSLGRMSNTNVLDNVSDNVK 270
K + G S + L+ V D ++
Sbjct: 240 RLEKQFEKLMMYDGE-SMDSYLERVLDTIE 268
Score = 50.8 bits (120), Expect = 2e-06
Identities = 27/96 (28%), Positives = 52/96 (54%)
Query: 108 HEGTSKVRMSKLQLLTTQFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKI 167
++ + ++ +KL L QFE + M + ES+ + R+ D A G P+S++ + K+
Sbjct: 227 YDKNNPIKDAKLLRLEKQFEKLMMYDGESMDSYLERVLDTIEQFRASGNPLSDDDVIAKL 286
Query: 168 LRSLPKRFDMKVTAIEEAQDISNIKVDELIGSLQTF 203
L SL +D + +EE ++ ++ +LIG L+ F
Sbjct: 287 LTSLSWPYDDAIPVLEELMNLPDMTPHDLIGVLEMF 322
>At3g31380 hypothetical protein
Length = 262
Score = 77.8 bits (190), Expect = 2e-14
Identities = 41/145 (28%), Positives = 75/145 (51%), Gaps = 1/145 (0%)
Query: 91 INTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFETMKMNEDESIYEFHMRIRDLANS 150
+ AKE W+ +KT H G +V+ +++Q L FE MKM E E I +F R+ +L+
Sbjct: 18 VGNLQTAKEVWDSIKTRHVGAERVKEARVQTLMADFEKMKMKEAEKIDDFAGRLSELSTK 77
Query: 151 TFALGEPMSEEKLARKILRSLP-KRFDMKVTAIEEAQDISNIKVDELIGSLQTFEMSLNG 209
+ LG + KL +K L SLP K++ + ++E+ D++N ++++G ++ E +
Sbjct: 78 SAPLGVNIEVPKLVKKFLNSLPRKKYIHIIASLEQVLDLNNTTFEDIVGCMKVNEERVYD 137
Query: 210 RSEKKAKSITFVSNTEEDEDQREKD 234
E+ + + T D +D
Sbjct: 138 PEEETNEDQNKLMYTSSDSKSGNRD 162
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 76.6 bits (187), Expect = 4e-14
Identities = 42/144 (29%), Positives = 83/144 (57%), Gaps = 8/144 (5%)
Query: 97 AKEAWEILKTAHEGTSKVRMSKLQLLTTQFETMKMNEDESIYEFHMRIRDLANSTFALGE 156
+K W+ +++ + G +V+ +KL+ L +F+ +KM ++E+I E R+ +++ + +LGE
Sbjct: 43 SKAVWDKIQSRNLGAERVKEAKLKTLMAEFDKLKMKDNETIDECAGRLSEISTKSTSLGE 102
Query: 157 PMSEEKLARKILRSLP-KRFDMKVTAIEEAQDISNIKVDELIGSLQTFE-------MSLN 208
+ E K+ +K L+SLP K++ V A+E+ D+ N +++G ++T+E L
Sbjct: 103 DIEETKVVKKFLKSLPTKKYIHIVAALEQVLDLKNTTFKDIVGRIKTYEDKIWVLITCLK 162
Query: 209 GRSEKKAKSITFVSNTEEDEDQRE 232
+E++ KS+ V EE RE
Sbjct: 163 KEAEEEEKSVVGVEAEEELVISRE 186
>At3g21030 hypothetical protein
Length = 411
Score = 75.9 bits (185), Expect = 7e-14
Identities = 48/197 (24%), Positives = 98/197 (49%), Gaps = 15/197 (7%)
Query: 12 ILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWK-----HPVIASTTELKPEDKWTKKED 66
+ D +Y+ W MV + +++ W + G +P +A+T + +W
Sbjct: 9 VSDDFDYEDWSP--MVKTRLVENGVWDVVQNGVSPNPTTNPELAATIKAVDLAQWRNL-- 64
Query: 67 DEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQF 126
+ ++KAL ++ + + ++FR + AKE W++LK ++ + +KL+ L QF
Sbjct: 65 --VVKDNKALKIMQSSLPDSVFRKTISIASAKELWDLLKKGND----TKEAKLRRLEKQF 118
Query: 127 ETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQ 186
E + M E E + + R+ + F LG P+S++K+ K+L SLP+ +D V ++E
Sbjct: 119 ENLMMYEGERMKLYLKRLEKIIEELFVLGNPISDDKVIAKLLTSLPRSYDDSVPVLKEFM 178
Query: 187 DISNIKVDELIGSLQTF 203
+ + +L+ + + F
Sbjct: 179 TLPELTHRDLLKAFEMF 195
>At3g21050 hypothetical protein
Length = 420
Score = 70.5 bits (171), Expect = 3e-12
Identities = 44/197 (22%), Positives = 99/197 (49%), Gaps = 15/197 (7%)
Query: 12 ILDGTNYDYWKARMMVFLKSMDSIAWKAIVKG-----WKHPVIASTTELKPEDKWTKKED 66
+ D +Y++W + + +++ W + G K P +A+T + +W +
Sbjct: 9 VSDDFDYEHWSP--IAKKRLVENGVWDVVQNGVSPNPTKIPQLAATIQAVDLAQWRNR-- 64
Query: 67 DEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQF 126
+ ++KAL ++ + + ++FR + AKE W++LK ++ + +KL+ L QF
Sbjct: 65 --VIKDNKALKIMQSSLPDSVFRKTISIASAKELWDLLKKGND----TKEAKLRRLEKQF 118
Query: 127 ETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQ 186
E + M E E + + R+ ++ LG P+S++K+ K+L SL +D + ++E
Sbjct: 119 EKLMMYEGEPMDLYLKRVEEITERFEVLGNPISDDKVITKLLTSLSWPYDDSIPVLKEFM 178
Query: 187 DISNIKVDELIGSLQTF 203
+ ++ + +L+ + + F
Sbjct: 179 TLPDLTLRDLLKAFELF 195
>At5g56830 unknown protein
Length = 242
Score = 65.5 bits (158), Expect = 9e-11
Identities = 58/226 (25%), Positives = 99/226 (43%), Gaps = 21/226 (9%)
Query: 76 LNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFETMKMNEDE 135
L++I +G+ +F I T +KEAWEIL G + KL L ++ M++ E
Sbjct: 14 LSLIQSGLSTEIFSWIAYATSSKEAWEILMVRSVGGPIFQARKLLDLKKEYNNTTMSDAE 73
Query: 136 SIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDISNIKVDE 195
+++EF ++ +L + G + E+ L +KIL SLP RFD + + E + V+
Sbjct: 74 TVHEFTGKLLELVFRMKSCGWKVEEKILVKKILSSLPPRFDKVLVEVAE-----KLSVNV 128
Query: 196 LIGSLQTFEMSLNGRSEKKAKSITFVSNTEE-------------DEDQREKDTDANIAEA 242
+I L E ++ E +S+ N EE D Q + +E
Sbjct: 129 IIDCLLVHEYNMRPDQESFQESVIQSVNVEESFEAEAEILYSLNDITQSQVAMMERQSEV 188
Query: 243 VSLLNKALKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAI 288
+SL+ K K L +M ++ K+ + +E + TK I
Sbjct: 189 LSLVQKNQKMLMQMQKQMMM---LMQQKDHVLAHSESYEEEMTKVI 231
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.127 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,826,318
Number of Sequences: 26719
Number of extensions: 651215
Number of successful extensions: 3456
Number of sequences better than 10.0: 359
Number of HSP's better than 10.0 without gapping: 104
Number of HSP's successfully gapped in prelim test: 255
Number of HSP's that attempted gapping in prelim test: 2906
Number of HSP's gapped (non-prelim): 651
length of query: 667
length of database: 11,318,596
effective HSP length: 106
effective length of query: 561
effective length of database: 8,486,382
effective search space: 4760860302
effective search space used: 4760860302
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0075.5


