

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0075.14
(356 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
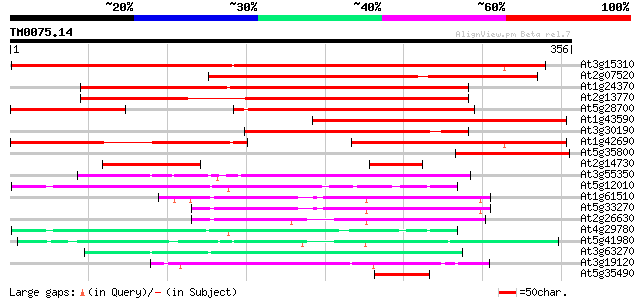
Score E
Sequences producing significant alignments: (bits) Value
At3g15310 unknown protein 407 e-114
At2g07520 pseudogene 351 4e-97
At1g24370 hypothetical protein 318 4e-87
At2g13770 hypothetical protein 244 7e-65
At5g28700 putative protein 201 7e-52
At1g43590 hypothetical protein 172 3e-43
At3g30190 hypothetical protein 161 6e-40
At1g42690 unknown protein 151 5e-37
At5g35800 putative protein 117 7e-27
At2g14730 hypothetical protein 70 2e-12
At3g55350 unknown protein 57 2e-08
At5g12010 putative protein 55 8e-08
At1g61510 hypothetical protein 52 5e-07
At5g33270 putative protein 50 2e-06
At2g26630 En/Spm-like transposon protein 49 3e-06
At4g29780 unknown protein 46 3e-05
At5g41980 unknown protein 45 5e-05
At3g63270 unknown protein 45 6e-05
At3g19120 unknown protein 45 8e-05
At5g35490 putative protein 44 1e-04
>At3g15310 unknown protein
Length = 415
Score = 407 bits (1047), Expect = e-114
Identities = 200/342 (58%), Positives = 252/342 (73%), Gaps = 4/342 (1%)
Query: 2 FRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADAV 61
FRRR+RM+K +FLRIV LSS +F R DA + G SP+ KCT A+R+LAYG A+DAV
Sbjct: 75 FRRRFRMKKPLFLRIVDRLSSELMFFQHRRDATGRFGHSPIQKCTAAIRLLAYGYASDAV 134
Query: 62 DEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSIDC 121
DEY+++G TTA+ CL F GII + EYLRAPT +L+R+L + + RGFPGMIGS+DC
Sbjct: 135 DEYLRMGETTAMSCLENFTKGIISFFGDEYLRAPTATNLRRLLNIGKIRGFPGMIGSLDC 194
Query: 122 MHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPV 181
MHWEWKNCP AW+GQ+TRG G T++LEA+AS DLWIWH FFG PGTLNDIN+LDRSP+
Sbjct: 195 MHWEWKNCPTAWKGQYTRGS-GKPTIVLEAIASQDLWIWHVFFGPPGTLNDINILDRSPI 253
Query: 182 FDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERC 241
FDD+ QG+AP V + VN R Y++AYYL DGIYP + TF++SIRLPQ+ LFA QE
Sbjct: 254 FDDILQGRAPNVKYKVNGREYHLAYYLTDGIYPKWATFIQSIRLPQNRKATLFATHQEAD 313
Query: 242 RKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDFE 301
RKD+ERAFGVLQARF II+ PA +WD +G IM++CIILHNMIVEDERD + ++ E
Sbjct: 314 RKDVERAFGVLQARFHIIKNPALVWDKEKIGNIMKACIILHNMIVEDERDGYNIQFDVSE 373
Query: 302 QSGEGGSSTPQ---PYSTEVLPAFANHVRARSELRDSNVHHE 340
G+ TPQ YST + N + R++LRD N+H +
Sbjct: 374 FLQVEGNQTPQVDLSYSTGMPLNIENMMGMRNQLRDQNMHQQ 415
>At2g07520 pseudogene
Length = 222
Score = 351 bits (900), Expect = 4e-97
Identities = 169/209 (80%), Positives = 181/209 (85%), Gaps = 6/209 (2%)
Query: 127 KNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDDVE 186
KNCP AWEGQFTRGDKGT+T+ EAVASHDLWIWH FFGC TLNDIN LDRSPVFDD+E
Sbjct: 16 KNCPIAWEGQFTRGDKGTSTISFEAVASHDLWIWHTFFGCASTLNDINFLDRSPVFDDIE 75
Query: 187 QGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERCRKDIE 246
QG P V+F V QR YNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLF Q+QE CRKDIE
Sbjct: 76 QGNTPRVNFFVYQRQYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFVQLQEGCRKDIE 135
Query: 247 RAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDFEQSGEG 306
RAFGVL ARFKII W+IADL IIMRSCIILHNMIVE+ERDT+AQ WTD++QS E
Sbjct: 136 RAFGVLHARFKII------WNIADLAIIMRSCIILHNMIVENERDTYAQHWTDYDQSKES 189
Query: 307 GSSTPQPYSTEVLPAFANHVRARSELRDS 335
GSSTPQP+STEVL AFANHV ARS+LRDS
Sbjct: 190 GSSTPQPFSTEVLHAFANHVHARSKLRDS 218
>At1g24370 hypothetical protein
Length = 413
Score = 318 bits (814), Expect = 4e-87
Identities = 146/246 (59%), Positives = 187/246 (75%), Gaps = 1/246 (0%)
Query: 46 TTAMRMLAYGVAADAVDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQ 105
T A RMLAYGV AD+ DEYIKIG +TALE L+RFC I+ ++ YLR+P D+ R+L
Sbjct: 2 TAAFRMLAYGVPADSTDEYIKIGESTALESLKRFCRAIVEVFACRYLRSPDANDVARLLH 61
Query: 106 VSEQRGFPGMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFG 165
+ E RGFP M+GS+DCMHW+WKNCP AW GQ+ G + T+ILEAVA +DLWIWHA+FG
Sbjct: 62 IGESRGFPRMLGSLDCMHWKWKNCPTAWGGQYA-GRSRSPTIILEAVADYDLWIWHAYFG 120
Query: 166 CPGTLNDINVLDRSPVFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRL 225
PG+ NDINVL+ S +F ++ +G AP S+++N++PYNM+YYLADGIYP + T V++I
Sbjct: 121 LPGSNNDINVLEASHLFANLAEGTAPPASYVINEKPYNMSYYLADGIYPKWSTLVQTIHD 180
Query: 226 PQSEPDKLFAQVQERCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMI 285
P+ KLFA QE CRKD+ERAFGVLQ+RF I+ P+RLW+ L IM SCII+HNMI
Sbjct: 181 PRGPKKKLFAMKQEACRKDVERAFGVLQSRFAIVAGPSRLWNKTVLHDIMTSCIIMHNMI 240
Query: 286 VEDERD 291
+EDERD
Sbjct: 241 IEDERD 246
>At2g13770 hypothetical protein
Length = 244
Score = 244 bits (622), Expect = 7e-65
Identities = 124/246 (50%), Positives = 156/246 (63%), Gaps = 36/246 (14%)
Query: 46 TTAMRMLAYGVAADAVDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQ 105
T A RMLAYGV AD+ DEYIKIG +TALE L+RFC I+ ++ YLR+P D+ R+L
Sbjct: 2 TAAFRMLAYGVPADSTDEYIKIGESTALESLKRFCRAIVEVFACCYLRSPDANDVARLLH 61
Query: 106 VSEQRGFPGMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFG 165
+ E RGFP EAVA +DLWIWHA+FG
Sbjct: 62 IGESRGFP------------------------------------EAVADYDLWIWHAYFG 85
Query: 166 CPGTLNDINVLDRSPVFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRL 225
PG+ N INVL+ S +F ++ +G AP S+++N +PYNM YYLADGIYP + T V++I
Sbjct: 86 LPGSNNGINVLEASHLFANLAEGTAPPASYVINGKPYNMGYYLADGIYPKWSTLVQTIHD 145
Query: 226 PQSEPDKLFAQVQERCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMI 285
P+ KLFA QE CRKD+ERAFGVLQ RF I+ P+RLW+ L IM SCII+HNMI
Sbjct: 146 PRGPKKKLFAMKQEACRKDVERAFGVLQLRFAIVAGPSRLWNKTVLHDIMTSCIIMHNMI 205
Query: 286 VEDERD 291
+EDERD
Sbjct: 206 IEDERD 211
>At5g28700 putative protein
Length = 292
Score = 201 bits (510), Expect = 7e-52
Identities = 95/153 (62%), Positives = 116/153 (75%), Gaps = 2/153 (1%)
Query: 143 GTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDDVEQGKAPTVSFIVNQRPY 202
G +T+++ VAS DLWIWHA FG PGTLNDINVLDRSPVFDD+ QG+A V ++VN + Y
Sbjct: 105 GESTLLV--VASQDLWIWHAIFGPPGTLNDINVLDRSPVFDDILQGRASKVKYVVNGKDY 162
Query: 203 NMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERCRKDIERAFGVLQARFKIIREP 262
N+AYYL DGIYP + TF++SI PQ + LF Q+ CRKD+ERAFGV QARF I++ P
Sbjct: 163 NLAYYLTDGIYPKWATFIQSISNPQGDKASLFVTTQKACRKDVERAFGVFQARFSIVKHP 222
Query: 263 ARLWDIADLGIIMRSCIILHNMIVEDERDTFAQ 295
A D +G IMR+CIILHNMIVEDERD + Q
Sbjct: 223 ALFHDKVKIGNIMRACIILHNMIVEDERDGYTQ 255
Score = 78.6 bits (192), Expect = 5e-15
Identities = 40/73 (54%), Positives = 51/73 (69%)
Query: 1 MFRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADA 60
+FR R+RM K +F+RIV S+ YF QR DA + S L K T A+RMLAYG+AADA
Sbjct: 38 IFRHRFRMNKPLFMRIVERFSNEVPYFKQRRDATGRLDFSALQKSTAAIRMLAYGIAADA 97
Query: 61 VDEYIKIGGTTAL 73
VDEY++IG +T L
Sbjct: 98 VDEYLRIGESTLL 110
>At1g43590 hypothetical protein
Length = 168
Score = 172 bits (435), Expect = 3e-43
Identities = 88/163 (53%), Positives = 109/163 (65%), Gaps = 2/163 (1%)
Query: 193 VSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERCRKDIERAFGVL 252
++F+VN YN+AYYL D IYP + TF++SI LPQ E LFA QE CRKD+ERAFGVL
Sbjct: 1 MAFVVNGNEYNLAYYLTDEIYPKWATFIQSISLPQDEKASLFATNQEACRKDVERAFGVL 60
Query: 253 QARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQ-RWTDFEQSGEGGSSTP 311
QARF I++ PA +WD +G IMR+CIILHNMIVEDERD + Q +DF SS
Sbjct: 61 QARFAIVKHPALIWDKIKIGNIMRACIILHNMIVEDERDGYTQYDVSDFAHPESASSSQV 120
Query: 312 Q-PYSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIWTKF 353
YST++ N + R+ LRD H L+ADLV+HIW KF
Sbjct: 121 DFTYSTDMPSNLGNMMATRTRLRDRTKHEHLKADLVEHIWQKF 163
>At3g30190 hypothetical protein
Length = 263
Score = 161 bits (407), Expect = 6e-40
Identities = 76/142 (53%), Positives = 100/142 (69%), Gaps = 6/142 (4%)
Query: 150 EAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDDVEQGKAPTVSFIVNQRPYNMAYYLA 209
+AVA +D+WIWHA+FG PG+ NDINVL+ +F ++ + AP S++ N +PYNM YYLA
Sbjct: 104 QAVADYDMWIWHAYFGLPGSNNDINVLEAYHLFANLAEDTAPPASYVSNGKPYNMGYYLA 163
Query: 210 DGIYPSYPTFVKSIRLPQSEPDKLFAQVQERCRKDIERAFGVLQARFKIIREPARLWDIA 269
DGIY + T V++I P+ KLFA E CRKD+E AF VL +RF I+ EP+RLW+
Sbjct: 164 DGIYSKWSTLVQTIHDPRGPKKKLFAMKLETCRKDVEPAFEVLHSRFAIVAEPSRLWN-- 221
Query: 270 DLGIIMRSCIILHNMIVEDERD 291
M SCII+HNMI+EDERD
Sbjct: 222 ----KMTSCIIMHNMIIEDERD 239
>At1g42690 unknown protein
Length = 333
Score = 151 bits (382), Expect = 5e-37
Identities = 78/151 (51%), Positives = 95/151 (62%), Gaps = 31/151 (20%)
Query: 1 MFRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADA 60
+FRRR+RM K +F+RIV S+ YF QR DA ++ G S L K T A+RMLAYG+AADA
Sbjct: 70 IFRRRFRMNKSLFMRIVERFSNEVPYFKQRRDATRRLGFSALQKSTAAIRMLAYGIAADA 129
Query: 61 VDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSID 120
YLR PT++DL R+L + EQRGFPGMIGSID
Sbjct: 130 ------------------------------YLRRPTRDDLIRLLHIGEQRGFPGMIGSID 159
Query: 121 CMHWEWKNCPKAWEGQFTRGDKGTTTVILEA 151
CMHWEWKNCP AW+GQ+TRG G T++LEA
Sbjct: 160 CMHWEWKNCPTAWKGQYTRG-SGKPTIVLEA 189
Score = 118 bits (296), Expect = 4e-27
Identities = 65/138 (47%), Positives = 84/138 (60%), Gaps = 2/138 (1%)
Query: 218 TFVKSIRLPQSEPDKLFAQVQERCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRS 277
TF++SI + Q LFA QE CRKD+ERAFGVLQARF II+ PA D +G IMR+
Sbjct: 191 TFIQSISILQGNKASLFATTQEACRKDVERAFGVLQARFAIIKHPALFHDKVKIGNIMRA 250
Query: 278 CIILHNMIVEDERDTFAQRWTDFEQSGEGGSSTPQ--PYSTEVLPAFANHVRARSELRDS 335
CIILHNMIVED+RD + Q E SS+ Y+T + N + + +RD
Sbjct: 251 CIILHNMIVEDKRDGYTQFDVSEFVHPESASSSQVDFTYATVMPSNLGNMMATGARVRDR 310
Query: 336 NVHHELQADLVKHIWTKF 353
H EL+ADLV+H+W +
Sbjct: 311 IKHEELKADLVEHVWQHY 328
>At5g35800 putative protein
Length = 73
Score = 117 bits (294), Expect = 7e-27
Identities = 54/72 (75%), Positives = 62/72 (86%)
Query: 284 MIVEDERDTFAQRWTDFEQSGEGGSSTPQPYSTEVLPAFANHVRARSELRDSNVHHELQA 343
MIV DER+T+AQ W+D+ QS GSSTPQP+S EVLP FA+HVRARSELRDSN+HHELQA
Sbjct: 1 MIVNDERETYAQHWSDYNQSEASGSSTPQPFSIEVLPEFASHVRARSELRDSNLHHELQA 60
Query: 344 DLVKHIWTKFAN 355
LVK+IWTKF N
Sbjct: 61 GLVKYIWTKFRN 72
>At2g14730 hypothetical protein
Length = 117
Score = 70.1 bits (170), Expect = 2e-12
Identities = 32/62 (51%), Positives = 44/62 (70%)
Query: 60 AVDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSI 119
AVD+Y++IG T + C+ II L+ EYLR PT++DL+R+L++ E RGF GM GSI
Sbjct: 19 AVDKYLRIGENTLMSCMIHSVEAIIYLFGKEYLRRPTRQDLKRLLRIGELRGFLGMTGSI 78
Query: 120 DC 121
DC
Sbjct: 79 DC 80
Score = 50.1 bits (118), Expect = 2e-06
Identities = 23/34 (67%), Positives = 26/34 (75%)
Query: 229 EPDKLFAQVQERCRKDIERAFGVLQARFKIIREP 262
+ D LFA QE CRKD+ERAFGVLQARF I+ P
Sbjct: 81 DKDSLFATNQEVCRKDVERAFGVLQARFAIVTNP 114
>At3g55350 unknown protein
Length = 406
Score = 56.6 bits (135), Expect = 2e-08
Identities = 66/257 (25%), Positives = 108/257 (41%), Gaps = 16/257 (6%)
Query: 44 KCTTAMRMLAYGVAADAVDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRI 103
+ A+R L G + + E + +T + RF + H +L P++ D +
Sbjct: 114 RVAVALRRLGSGESLSVIGETFGMNQSTVSQITWRFVESMEERAIH-HLSWPSKLDEIKS 172
Query: 104 LQVSEQRGFPGMIGSIDCMHWEWKNCP------KAWEGQFTRGDKGTTTVILEAVASHDL 157
+ + G P G+ID H N P K W G+K + + L+AV D+
Sbjct: 173 -KFEKISGLPNCCGAIDITHIVM-NLPAVEPSNKVW----LDGEKNFS-MTLQAVVDPDM 225
Query: 158 WIWHAFFGCPGTLNDINVLDRSPVFDDVEQGKAPTVSFI-VNQRPYNMAYYLADGIYPSY 216
G PG+LND VL S + VE+GK + +++R Y + D +P
Sbjct: 226 RFLDVIAGWPGSLNDDVVLKNSGFYKLVEKGKRLNGEKLPLSERTELREYIVGDSGFPLL 285
Query: 217 PTFVKSIR-LPQSEPDKLFAQVQERCRKDIERAFGVLQARFKIIREPARLWDIADLGIIM 275
P + + P S P F + K + A L+ R++II + D L I+
Sbjct: 286 PWLLTPYQGKPTSLPQTEFNKRHSEATKAAQMALSKLKDRWRIINGVMWMPDRNRLPRII 345
Query: 276 RSCIILHNMIVEDERDT 292
C +LHN+I++ E T
Sbjct: 346 FVCCLLHNIIIDMEDQT 362
>At5g12010 putative protein
Length = 502
Score = 54.7 bits (130), Expect = 8e-08
Identities = 62/290 (21%), Positives = 123/290 (42%), Gaps = 23/290 (7%)
Query: 2 FRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADAV 61
F++ +RM K F I +L+S+ + D A + I + + LA G V
Sbjct: 175 FKKAFRMSKSTFELICDELNSA----VAKEDTALRNAIPVRQRVAVCIWRLATGEPLRLV 230
Query: 62 DEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQ-RGFPGMIGSID 120
+ +G +T + + C I + +YL+ P E L+ I + E G P ++GS+
Sbjct: 231 SKKFGLGISTCHKLVLEVCKAIKDVLMPKYLQWPDDESLRNIRERFESVSGIPNVVGSMY 290
Query: 121 CMHWEWKNCPKAWEGQF------TRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDIN 174
H PK + R K + ++ ++AV + G PG++ D
Sbjct: 291 TTHIPII-APKISVASYFNKRHTERNQKTSYSITIQAVVNPKGVFTDLCIGWPGSMPDDK 349
Query: 175 VLDRSPVFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLF 234
VL++S ++ G ++ + L D + Y +++ Q ++
Sbjct: 350 VLEKSLLYQRANNGGLLKGMWVAG----GPGHPLLDWVLVPYTQ--QNLTWTQHAFNEKM 403
Query: 235 AQVQERCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNM 284
++VQ ++ AFG L+ R+ +++ + + DL ++ +C +LHN+
Sbjct: 404 SEVQGVAKE----AFGRLKGRWACLQKRTEV-KLQDLPTVLGACCVLHNI 448
>At1g61510 hypothetical protein
Length = 608
Score = 52.0 bits (123), Expect = 5e-07
Identities = 58/232 (25%), Positives = 100/232 (43%), Gaps = 35/232 (15%)
Query: 95 PTQEDLQRI--LQVSEQRGFP---GMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVIL 149
PT++D+ I V+++R P IG++D H + P + + + RG K T+ +
Sbjct: 356 PTRDDVTGISPFLVNDKRYMPYFIDCIGALDGTHVSVR--PPSGDVERYRGRKSEATMNI 413
Query: 150 EAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDDVEQGKAPTVSFIVNQRPYNMAYYLA 209
AV + + A+ G PG +D VL + SF P YYL
Sbjct: 414 LAVCNFSMKFNIAYVGVPGRAHDTKVLTYCATHE---------ASF---PHPPAGKYYLV 461
Query: 210 DGIYPSYPTFVKSIRL------------PQSEPDKLFAQVQERCRKDIERAFGVLQARFK 257
D YP+ ++ R P + +LF + R IER FGV +A+++
Sbjct: 462 DSGYPTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHSSLRSVIERTFGVWKAKWR 521
Query: 258 IIREPARLWDIADLGIIMRSCIILHNMIVEDER-DTFAQRW---TDFEQSGE 305
I+ +++ I+ S + LHN I + ++ D+ + W +EQ G+
Sbjct: 522 ILDRKHPKYEVKKWIKIVTSTMALHNYIRDSQQEDSDFRHWEIVESYEQHGD 573
>At5g33270 putative protein
Length = 343
Score = 50.1 bits (118), Expect = 2e-06
Identities = 50/206 (24%), Positives = 88/206 (42%), Gaps = 30/206 (14%)
Query: 116 IGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINV 175
IG++D H + P + + + RG K T+ + A+ + + +A+ G PG +D V
Sbjct: 117 IGALDGTHVSVR--PPSGDVERYRGRKSEATMNILALCNFSMKFTYAYVGVPGRAHDTKV 174
Query: 176 LDRSPVFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRL---------- 225
L + SF P YYL D YP+ ++ R
Sbjct: 175 LTYCATHE---------ASF---PHPPAGKYYLVDSGYPTRSGYLGPHRRTRYHLELFNR 222
Query: 226 --PQSEPDKLFAQVQERCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHN 283
P + +LF + R IER FGV +A+++I+ +++ I+ S + LHN
Sbjct: 223 GGPPTNSRELFNRRHSSLRSVIERTFGVWKAKWRILDRKHLKYEVKKWIKIVTSTMALHN 282
Query: 284 MIVEDER-DTFAQRW---TDFEQSGE 305
I + ++ D+ + W +EQ G+
Sbjct: 283 YIRDSQQEDSDFRHWEIVESYEQHGD 308
>At2g26630 En/Spm-like transposon protein
Length = 292
Score = 49.3 bits (116), Expect = 3e-06
Identities = 49/204 (24%), Positives = 83/204 (40%), Gaps = 35/204 (17%)
Query: 116 IGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINV 175
+G++D H + P + + +G K T+ + A+ + D+ +A+ G PG +D V
Sbjct: 45 VGALDGTHVPVR--PPSQTAKKYKGRKLEPTMNVLAICNFDMKFIYAYVGVPGRAHDTKV 102
Query: 176 LD----RSPVFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRL------ 225
L+ P F GK YYL D YP+ ++ R
Sbjct: 103 LNYCATNEPYFSHPPNGK----------------YYLVDSGYPTRTGYLGPHRRMRYHLG 146
Query: 226 ------PQSEPDKLFAQVQERCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCI 279
P +LF + R IER FGV +A+++I+ + +A I+ + +
Sbjct: 147 QFGRGGPPVTARELFNRKHSGLRSVIERTFGVWKAKWRIVDRKHPKYGLAKWIKIVTATM 206
Query: 280 ILHNMIVEDER-DTFAQRWTDFEQ 302
LHN I + R D +W E+
Sbjct: 207 ALHNFIRDSHREDHDFLQWQSIEE 230
>At4g29780 unknown protein
Length = 540
Score = 46.2 bits (108), Expect = 3e-05
Identities = 59/290 (20%), Positives = 114/290 (38%), Gaps = 23/290 (7%)
Query: 2 FRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADAV 61
FRR +RM K F I +L ++ + + ++ I + + LA G V
Sbjct: 213 FRREFRMSKSTFNLICEELDTT----VTKKNTMLRDAIPAPKRVGVCVWRLATGAPLRHV 268
Query: 62 DEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQ-RGFPGMIGSID 120
E +G +T + + C I + +YL P+ ++ E P ++GSI
Sbjct: 269 SERFGLGISTCHKLVIEVCRAIYDVLMPKYLLWPSDSEINSTKAKFESVHKIPNVVGSIY 328
Query: 121 CMHWEWKNCPKAWEGQF------TRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDIN 174
H PK + R K + ++ ++ V + D G PG+L D
Sbjct: 329 TTHIP-IIAPKVHVAAYFNKRHTERNQKTSYSITVQGVVNADGIFTDVCIGNPGSLTDDQ 387
Query: 175 VLDRSPVFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLF 234
+L++S + S+IV + + YL P +++ Q ++
Sbjct: 388 ILEKSSLSRQRAARGMLRDSWIVGNSGFPLTDYLL------VPYTRQNLTWTQHAFNESI 441
Query: 235 AQVQERCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNM 284
++Q ER L+ R+ +++ + + DL ++ +C +LHN+
Sbjct: 442 GEIQGIATAAFER----LKGRWACLQKRTEV-KLQDLPYVLGACCVLHNI 486
>At5g41980 unknown protein
Length = 374
Score = 45.4 bits (106), Expect = 5e-05
Identities = 73/352 (20%), Positives = 131/352 (36%), Gaps = 40/352 (11%)
Query: 6 YRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADAVDEYI 65
+RM K VF ++ L + + + K E A+ + ++ + + AV E
Sbjct: 46 FRMDKPVFYKLCDLLQTRG--LLRHTNRIKIE-----AQLAIFLFIIGHNLRTRAVQELF 98
Query: 66 KIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSIDCMHWE 125
G T N +I + + + + L+ ++ F +G +D H
Sbjct: 99 CYSGETISRHFNNVLNAVIAISKDFFQPNSNSDTLE-----NDDPYFKDCVGVVDSFHIP 153
Query: 126 WKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDD- 184
+G F G+ G T + A +S DL + G G+ +D VL+ + +
Sbjct: 154 VMVGVDE-QGPFRNGN-GLLTQNVLAASSFDLRFNYVLAGWEGSASDQQVLNAALTRRNK 211
Query: 185 --VEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIR----LPQSEPDKLFAQVQ 238
V QGK YY+ D YP+ P F+ + E ++F +
Sbjct: 212 LQVPQGK----------------YYIVDNKYPNLPGFIAPYHGVSTNSREEAKEMFNERH 255
Query: 239 ERCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWT 298
+ + I R FG L+ RF I+ + +++ +C LHN + ++ D R
Sbjct: 256 KLLHRAIHRTFGALKERFPILLSAPPYPLQTQVKLVIAAC-ALHNYVRLEKPDDLVFRMF 314
Query: 299 DFEQSGEGGSSTPQPYSTEVLPAFA-NHVRARSELRDS-NVHHELQADLVKH 348
+ E E G E + H E+ DS + E+ ++L H
Sbjct: 315 EEETLAEAGEDREVALEEEQVEIVGQEHGFRPEEVEDSLRLRDEIASELWNH 366
>At3g63270 unknown protein
Length = 396
Score = 45.1 bits (105), Expect = 6e-05
Identities = 55/243 (22%), Positives = 97/243 (39%), Gaps = 5/243 (2%)
Query: 48 AMRMLAYGVAADAVDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRIL-QV 106
A+R LA G + +V +G +T + RF + +H +LR P + ++ I +
Sbjct: 112 ALRRLASGDSQVSVGAAFGVGQSTVSQVTWRFIEALEERAKH-HLRWPDSDRIEEIKSKF 170
Query: 107 SEQRGFPGMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGC 166
E G P G+ID H P + ++ L+ V H++ + G
Sbjct: 171 EEMYGLPNCCGAIDTTHIIM-TLPAVQASDDWCDQEKNYSMFLQGVFDHEMRFLNMVTGW 229
Query: 167 PGTLNDINVLDRSPVFDDVEQGKAPTVSFIVNQRPYNMAYYLADGI-YPSYPTFVKSIRL 225
PG + +L S F E + + + + Y+ GI YP P +
Sbjct: 230 PGGMTVSKLLKFSGFFKLCENAQILDGNPKTLSQGAQIREYVVGGISYPLLPWLITPHDS 289
Query: 226 PQ-SEPDKLFAQVQERCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNM 284
S+ F + E+ R AF L+ ++I+ + D L I+ C +LHN+
Sbjct: 290 DHPSDSMVAFNERHEKVRSVAATAFQQLKGSWRILSKVMWRPDRRKLPSIILVCCLLHNI 349
Query: 285 IVE 287
I++
Sbjct: 350 IID 352
>At3g19120 unknown protein
Length = 446
Score = 44.7 bits (104), Expect = 8e-05
Identities = 49/222 (22%), Positives = 95/222 (42%), Gaps = 13/222 (5%)
Query: 90 EYLRAPTQEDLQRILQVS----EQRGFPGMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTT 145
E+++ P + +R+++ + E P + G+ID + + K G
Sbjct: 197 EFIKIPVGK--RRLIETTQGFEELTSLPNICGAIDSTPVKLRRRTKLNPRNIYGCKYGYD 254
Query: 146 TVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDDVEQGKAPTVSFIVNQRPYNMA 205
V+L+ VA H W PG +D + S ++ + G I + +
Sbjct: 255 AVLLQVVADHKKIFWDVCVKAPGGEDDSSHFRDSLLYKRLTSGDIVWEKVINIRGHHVRP 314
Query: 206 YYLADGIYPSYPTFVKSIRLPQSE---PDKLFAQVQERCRKDIERAFGVLQARFKIIREP 262
Y + D YP +F+ + P P+ LF + + R + A G+L+AR+KI++
Sbjct: 315 YIVGDWCYPLL-SFLMTPFSPNGSGTPPENLFDGMLMKGRSVVVEAIGLLKARWKILQSL 373
Query: 263 ARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDFEQSG 304
+ A I+ +C +LHN + + R+ + W D +++G
Sbjct: 374 NVGVNHAPQTIV--ACCVLHN-LCQIAREPEPEIWKDPDEAG 412
>At5g35490 putative protein
Length = 106
Score = 44.3 bits (103), Expect = 1e-04
Identities = 19/35 (54%), Positives = 24/35 (68%)
Query: 232 KLFAQVQERCRKDIERAFGVLQARFKIIREPARLW 266
K+FA QE CRKD+ER F VLQ++F II + W
Sbjct: 70 KMFAAKQEACRKDVERVFRVLQSKFAIIARSSNCW 104
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.139 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,081,488
Number of Sequences: 26719
Number of extensions: 338257
Number of successful extensions: 1035
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 998
Number of HSP's gapped (non-prelim): 35
length of query: 356
length of database: 11,318,596
effective HSP length: 100
effective length of query: 256
effective length of database: 8,646,696
effective search space: 2213554176
effective search space used: 2213554176
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0075.14


