

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0073.8
(1156 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
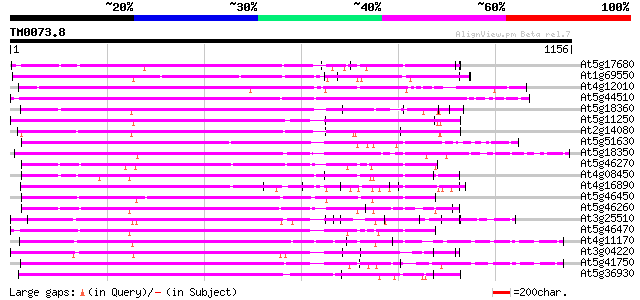
Score E
Sequences producing significant alignments: (bits) Value
At5g17680 disease resistance protein RPP1-WsB - like protein 565 e-161
At1g69550 putative disease resistance protein 481 e-136
At4g12010 like disease resistance protein (TMV N-like) 474 e-134
At5g44510 disease resistance protein-like 473 e-133
At5g18360 disease resistance protein -like 469 e-132
At5g11250 RPP1 disease resistance protein - like 459 e-129
At2g14080 putative disease resistance protein 458 e-129
At5g51630 putative protein 457 e-128
At5g18350 disease resistance protein -like 456 e-128
At5g46270 disease resistance protein-like 454 e-127
At4g08450 putative protein 449 e-126
At4g16890 disease resistance RPP5 like protein 447 e-125
At5g46450 disease resistance protein-like 446 e-125
At5g46260 disease resistance protein-like 437 e-122
At3g25510 unknown protein 435 e-122
At5g46470 disease resistance protein-like 431 e-120
At4g11170 RPP1-WsA-like disease resistance protein 427 e-119
At3g04220 putative disease resistance protein 423 e-118
At5g41750 disease resistance protein-like 422 e-118
At5g36930 disease resistance like protein 415 e-116
>At5g17680 disease resistance protein RPP1-WsB - like protein
Length = 1294
Score = 565 bits (1455), Expect = e-161
Identities = 355/955 (37%), Positives = 547/955 (57%), Gaps = 90/955 (9%)
Query: 4 SSSSSSAAAAAMAPPPNPLKYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLET 61
SSSSSS++ K DVF+SF GED KTF SHL R + F DD L+
Sbjct: 6 SSSSSSSSTV--------WKTDVFVSFRGEDVRKTFVSHLFCEFDRMGIKAFRDDLDLQR 57
Query: 62 GD----EICKAIEASTIYVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPS 117
G E+ AI+ S ++V S NYA+S CLDEL+KIMEC + ++PIFY VDPS
Sbjct: 58 GKSISPELIDAIKGSRFAIVVVSRNYAASSWCLDELLKIMECNKD---TIVPIFYEVDPS 114
Query: 118 TVRHQTNSYGDAFDEHQNRLVPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQD 177
VR Q S+G+ + H ++ G +WK+AL + A +SG S +S LI +I +D
Sbjct: 115 DVRRQRGSFGEDVESHSDKEKVG----KWKEALKKLAAISGEDS-RNWDDSKLIKKIVKD 169
Query: 178 IFKKLNPPF---SQGMLGIDKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHK 234
I KL S+G++G+ H+ +QS++ + + VR++GI GMGG+GK+T+A+ LY++
Sbjct: 170 ISDKLVSTSWDDSKGLIGMSSHMDFLQSMISIVDKDVRMLGIWGMGGVGKTTIAKYLYNQ 229
Query: 235 LGTQFSSRCLIVNAQQKIDRYGIYSLRKKYLSKLLGEDIQS-----NGLNYAIERVKRAK 289
L QF C + N ++ +RYG+ L+ ++L ++ E + + N ER +
Sbjct: 230 LSGQFQVHCFMENVKEVCNRYGVRRLQVEFLCRMFQERDKEAWSSVSCCNIIKERFRHKM 289
Query: 290 VLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSL 349
V ++LDD+ S + EL+ G FG GSRIIVT+R RH+L + + +Y+V+ + +++L
Sbjct: 290 VFIVLDDVDRSEQLNELVKETGWFGPGSRIIVTTRDRHLLLSHGINLVYKVKCLPKKEAL 349
Query: 350 ELFRLNAFKEDYPL-EGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKE 408
+LF AF+E+ L G+ L V+ + YA G+PLAL+VLGS LY + + WES L +LK
Sbjct: 350 QLFCNYAFREEIILPHGFEELSVQAVNYASGLPLALRVLGSFLYRRSQIEWESTLARLKT 409
Query: 409 LPNVDIFDVLKLSYDGLDDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKD 468
P+ DI +VL++SYDGLD++ K IFL I+CFY VD V +LLD CG++A+IG+ +L +
Sbjct: 410 YPHSDIMEVLRVSYDGLDEQEKAIFLYISCFYNMKQVDYVRKLLDLCGYAAEIGITILTE 469
Query: 469 RGLISILEDRIAVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQC 528
+ LI + +HDL+ +MGRE+VRQQ V++P + LW+ ++I H++ +N GT ++
Sbjct: 470 KSLIVESNGCVKIHDLLEQMGRELVRQQAVNNPAQRLLLWDPEDICHLLSENSGTQLVEG 529
Query: 529 IFLDMCQIEKVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHW 588
I L++ +I +V F+ + N+++L F+ S G++ V +P+ L LP L+ L W
Sbjct: 530 ISLNLSEISEVFASDRAFEGLSNLKLLNFYDLSF--DGETRVHLPNGLSYLPRKLRYLRW 587
Query: 589 DEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQE 648
D +P ++ P F PE LV+L M S LE+LW Q L+
Sbjct: 588 DGYPLKTMPSRFFPEFLVELCMSNSNLEKLWDGIQPLR---------------------- 625
Query: 649 LPNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLTR-------------------- 688
NL+++DLS LV +PDLSK+ N++E+ LS C+SL
Sbjct: 626 --NLKKMDLSRCKYLVEVPDLSKATNLEELNLSYCQSLVEVTPSIKNLKGLSCFYLTNCI 683
Query: 689 ----LPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLV 744
+PI + LK LE + +SGCS++++ PEI N L L T I+ELPSS++ L
Sbjct: 684 QLKDIPIGII-LKSLETVGMSGCSSLKHFPEIS---WNTRRLYLSSTKIEELPSSISRLS 739
Query: 745 GLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDL-KLTKLDLNDCSR 803
L +L + C +L ++PS +G L L L+L C LE P ++ +L L L+++ C
Sbjct: 740 CLVKLDMSDCQRLRTLPSYLGHLVSLKSLNLDGCRRLENLPDTLQNLTSLETLEVSGCLN 799
Query: 804 LKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKL 863
+ FP + + S + ++ET+I+++P+ + NL L++L + E L SLP SI L+
Sbjct: 800 VNEFPRV---STSIEVLRISETSIEEIPARICNLSQLRSLDISENKRLASLPVSISELRS 856
Query: 864 LSELDCSGCRKLTGIPNDI-GCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVS 917
L +L SGC L P +I +S LR L T + LPE+I +L +LE L S
Sbjct: 857 LEKLKLSGCSVLESFPLEICQTMSCLRWFDLDRTSIKELPENIGNLVALEVLQAS 911
Score = 92.8 bits (229), Expect = 1e-18
Identities = 84/295 (28%), Positives = 132/295 (44%), Gaps = 36/295 (12%)
Query: 642 VFSLHQELPNLERLDLSNSWKL-------VRIPD-LSKSPNIKEIILSGCKSLTRLPINL 693
VF+ + L L L N + L V +P+ LS P + L +P
Sbjct: 540 VFASDRAFEGLSNLKLLNFYDLSFDGETRVHLPNGLSYLPRKLRYLRWDGYPLKTMPSRF 599
Query: 694 FKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKF 753
F +FL L +S SN+E + + + + NL + L + L+ LEEL+L +
Sbjct: 600 FP-EFLVELCMSN-SNLEKLWDGIQPLRNLKKMDLSRCKYLVEVPDLSKATNLEELNLSY 657
Query: 754 CSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEP 813
C L + SI L L LT C L+ P I L + ++ CS LK FP I
Sbjct: 658 CQSLVEVTPSIKNLKGLSCFYLTNCIQLKDIPIGIILKSLETVGMSGCSSLKHFPEI--- 714
Query: 814 AESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCR 873
+ + + L+ T I++LPSS+ L L L + +C L +LP+ + +L L L+ GCR
Sbjct: 715 SWNTRRLYLSSTKIEELPSSISRLSCLVKLDMSDCQRLRTLPSYLGHLVSLKSLNLDGCR 774
Query: 874 KLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHL 928
+L NLP+++ +L+SLE+L+VS C + P +
Sbjct: 775 RLE-----------------------NLPDTLQNLTSLETLEVSGCLNVNEFPRV 806
Score = 62.8 bits (151), Expect = 1e-09
Identities = 68/234 (29%), Positives = 103/234 (43%), Gaps = 18/234 (7%)
Query: 702 LNLSGCSNVENIPEIKETMENLAVLVLKQTAIQ-----ELPSSLNHLVGLEELSLKFCSK 756
LNLS S V E + NL +L + LP+ L++L
Sbjct: 532 LNLSEISEVFASDRAFEGLSNLKLLNFYDLSFDGETRVHLPNGLSYLPRKLRYLRWDGYP 591
Query: 757 LESIPSSIGT--LTKLCKLDLTYCESLETFPGSIFDLK-LTKLDLNDCSRLKTFPAILEP 813
L+++PS L +LC + +LE I L+ L K+DL+ C L P + +
Sbjct: 592 LKTMPSRFFPEFLVELCMSN----SNLEKLWDGIQPLRNLKKMDLSRCKYLVEVPDLSKA 647
Query: 814 AE-SFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGC 872
++S ++ ++ PS ++NL GL L C L+ +P I LK L + SGC
Sbjct: 648 TNLEELNLSYCQSLVEVTPS-IKNLKGLSCFYLTNCIQLKDIPIGII-LKSLETVGMSGC 705
Query: 873 RKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
L P + R L L T + LP SI+ LS L LD+S C++L +P
Sbjct: 706 SSLKHFPE---ISWNTRRLYLSSTKIEELPSSISRLSCLVKLDMSDCQRLRTLP 756
Score = 40.4 bits (93), Expect = 0.006
Identities = 32/113 (28%), Positives = 51/113 (44%), Gaps = 3/113 (2%)
Query: 699 LERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLE 758
L L+LS N+ IP + NL L L + +P+S+ L L L+L C +L+
Sbjct: 957 LRALSLSNM-NMTEIPNSIGNLWNLLELDLSGNNFEFIPASIKRLTRLNRLNLNNCQRLQ 1015
Query: 759 SIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAIL 811
++P + L + + C SL + G L KL ++C +L IL
Sbjct: 1016 ALPDELP--RGLLYIYIHSCTSLVSISGCFNQYCLRKLVASNCYKLDQAAQIL 1066
>At1g69550 putative disease resistance protein
Length = 1398
Score = 481 bits (1239), Expect = e-136
Identities = 344/1004 (34%), Positives = 541/1004 (53%), Gaps = 99/1004 (9%)
Query: 6 SSSSAAAAAMAPPPNPLK----YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDD--- 56
SSS++ ++ PP+ L + VF SF G+D + F SH+ RRK + FID+
Sbjct: 56 SSSTSHPSSSTSPPSSLSCTGTHHVFPSFRGDDVRRNFLSHIQKEFRRKGITPFIDNEIR 115
Query: 57 RSLETGDEICKAIEASTIYVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDP 116
R G E+ KAI S I +++ S NYASS+ CL+ELV+IM+CK + V IFY VDP
Sbjct: 116 RGESIGPELIKAIRESKIAIVLLSRNYASSKWCLEELVEIMKCKKEFGLTVFAIFYEVDP 175
Query: 117 STVRHQTNSYGDAFDEHQNRLVPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQ 176
S V+ T +G F + +M RW++A E A ++GY S +E+ +I+ IA
Sbjct: 176 SHVKKLTGEFGAVFQKTCKGRTKENIM-RWRQAFEEVATIAGYDSRNWENEAAMIEEIAI 234
Query: 177 DIFKKL--NPPFS--QGMLGIDKHIAQIQSLLQLDS-EAVRIIGICGMGGIGKSTLAEAL 231
+I K+L + PFS +G++G+ HI +++ LL LDS + R +GI G GIGKST+A L
Sbjct: 235 EISKRLINSSPFSGFEGLIGMKAHIEKMKQLLCLDSTDERRTVGISGPSGIGKSTIARVL 294
Query: 232 YHKLGTQFSSRCLIVNAQQKI------DRYGIYSLRKKYLSKLLG-EDIQSNGLNYAIER 284
++++ F + D L +++L++L+ EDI+ + L A
Sbjct: 295 HNQISDGFQMSVFMKFKPSYTRPICSDDHDVKLQLEQQFLAQLINQEDIKIHQLGTAQNF 354
Query: 285 VKRAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMS 344
V KVL++LD + + +L + G GSRII+T++ + +LK + IY V
Sbjct: 355 VMGKKVLIVLDGVDQLVQLLAMPKAVC-LGPGSRIIITTQDQQLLKAFQIKHIYNVDFPP 413
Query: 345 YQDSLELFRLNAFKEDYPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELK 404
++L++F ++AF D P +G+ L +V + A +PL L+V+GS +E W+ EL
Sbjct: 414 DHEALQIFCIHAFGHDSPDDGFEKLATKVTRLAGNLPLGLRVMGSHFRGMSKEDWKGELP 473
Query: 405 KLKELPNVDIFDVLKLSYDGLDDEPKDIFLDITCFYAGDFVDKVVELLDSCGFS-ADIGM 463
+L+ + +I +LK SYD LDDE KD+FL I CF+ + +D E FS G+
Sbjct: 474 RLRIRLDGEIGSILKFSYDVLDDEDKDLFLHIACFFNDEGIDHTFEDTLRHKFSNVQRGL 533
Query: 464 DVLKDRGLISILEDRI-AVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKG 522
VL R LIS ED +H+L++++GREIVR Q V +PGK L + KEI V+ + G
Sbjct: 534 QVLVQRSLIS--EDLTQPMHNLLVQLGREIVRNQSVYEPGKRQFLVDGKEICEVLTSHTG 591
Query: 523 TDAIQCIFLDM-CQIEKVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPN 581
++++ I ++ ++++ + +F+ M N++ F ++S + +P L LP
Sbjct: 592 SESVIGINFEVYWSMDELNISDRVFEGMSNLQFFRFDENS-----YGRLHLPQGLNYLPP 646
Query: 582 GLKVLHWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLI 641
L++LHWD +P S P F + LVK+ ++ S LE+LW Q L +L + + HL
Sbjct: 647 KLRILHWDYYPMTSLPSKFNLKFLVKIILKHSELEKLWEGIQPLVNL-KVMDLRYSSHL- 704
Query: 642 VFSLHQELPNLE------RLDLSNSWKLVRIP-DLSKSPNIKEIILSGCKSLTRLPINLF 694
+ELPNL + LS+ L+ +P + + NIK + + GC SL +LP ++
Sbjct: 705 -----KELPNLSTAINLLEMVLSDCSSLIELPSSIGNATNIKSLDIQGCSSLLKLPSSIG 759
Query: 695 KLKFLERLNLSGCSNVENIP---------------------EIKETMEN----------- 722
L L RL+L GCS++ +P E+ ++ N
Sbjct: 760 NLITLPRLDLMGCSSLVELPSSIGNLINLPRLDLMGCSSLVELPSSIGNLINLEAFYFHG 819
Query: 723 ----------------LAVLVLKQ-TAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIG 765
L +L LK+ +++ E+PSS+ +L+ L+ L+L CS L +PSSIG
Sbjct: 820 CSSLLELPSSIGNLISLKILYLKRISSLVEIPSSIGNLINLKLLNLSGCSSLVELPSSIG 879
Query: 766 TLTKLCKLDLTYCESLETFPGSIFDL-KLTKLDLNDCSRLKTFPAILEPAESFAHISLAE 824
L L KLDL+ C SL P SI +L L +L L++CS L P+ + + ++L+E
Sbjct: 880 NLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSSLVELPSSIGNLINLKTLNLSE 939
Query: 825 -TAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIG 883
+++ +LPSS+ NL+ LQ L L EC L LP+SI NL L +LD SGC L +P IG
Sbjct: 940 CSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLINLKKLDLSGCSSLVELPLSIG 999
Query: 884 CLSSLRTLSLQD-TGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
L +L+TL+L + + +V LP SI +L +L+ L +S C L +P
Sbjct: 1000 NLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELP 1043
Score = 178 bits (452), Expect = 1e-44
Identities = 120/308 (38%), Positives = 179/308 (57%), Gaps = 16/308 (5%)
Query: 649 LPNLERLDLSNSWKLVRIPD-LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGC 707
L NL+ L+LS LV +P + N++E+ LS C SL LP ++ L L++L+LSGC
Sbjct: 929 LINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLINLKKLDLSGC 988
Query: 708 SNVENIPEIKETMENLAVLVLKQ-TAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGT 766
S++ +P + NL L L + +++ ELPSS+ +L+ L+EL L CS L +PSSIG
Sbjct: 989 SSLVELPLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGN 1048
Query: 767 LTKLCKLDLTYCESLETFPGSIFDL-KLTKLDLNDCSRLKTFPAILEPAESFAHISLAE- 824
L L KLDL+ C SL P SI +L L L+L+ CS L P+ S +++L +
Sbjct: 1049 LINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPS------SIGNLNLKKL 1102
Query: 825 -----TAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIP 879
+++ +LPSS+ NL+ L+ L L C L LP SI NL L EL S C L +P
Sbjct: 1103 DLSGCSSLVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSSLVELP 1162
Query: 880 NDIGCLSSLRTLSLQD-TGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAF 938
+ IG L +L+ L L + + +V LP SI +L +L+ LD++ C KL +P LP + L+A
Sbjct: 1163 SSIGNLINLQELYLSECSSLVELPSSIGNLINLKKLDLNKCTKLVSLPQLPDSLSVLVAE 1222
Query: 939 HCPSIRSV 946
C S+ ++
Sbjct: 1223 SCESLETL 1230
Score = 63.9 bits (154), Expect = 5e-10
Identities = 74/281 (26%), Positives = 114/281 (40%), Gaps = 39/281 (13%)
Query: 675 IKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQ 734
I E++ S S + + IN ++ LN+S + E M NL + +
Sbjct: 582 ICEVLTSHTGSESVIGINFEVYWSMDELNISD--------RVFEGMSNLQFFRFDENSYG 633
Query: 735 EL--PSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLK 792
L P LN+L + + S+PS L L K+ L + E + + G +
Sbjct: 634 RLHLPQGLNYLPPKLRILHWDYYPMTSLPSKFN-LKFLVKIILKHSELEKLWEGIQPLVN 692
Query: 793 LTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLE 852
L +DL S LK P +L + L + L +C L
Sbjct: 693 LKVMDLRYSSHLKELP------------------------NLSTAINLLEMVLSDCSSLI 728
Query: 853 SLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQD-TGVVNLPESIAHLSSL 911
LP+SI N + LD GC L +P+ IG L +L L L + +V LP SI +L +L
Sbjct: 729 ELPSSIGNATNIKSLDIQGCSSLLKLPSSIGNLITLPRLDLMGCSSLVELPSSIGNLINL 788
Query: 912 ESLDVSYCRKLECIPHLPPFMKQLLAFH---CPSIRSVMSN 949
LD+ C L +P + L AF+ C S+ + S+
Sbjct: 789 PRLDLMGCSSLVELPSSIGNLINLEAFYFHGCSSLLELPSS 829
>At4g12010 like disease resistance protein (TMV N-like)
Length = 1219
Score = 474 bits (1221), Expect = e-134
Identities = 362/1106 (32%), Positives = 575/1106 (51%), Gaps = 105/1106 (9%)
Query: 19 PNPLKYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEICKA---IEAST 73
P+ ++DVFLSF G DT FT HL ALR + + FIDDR L GD + IE S
Sbjct: 6 PSSAEFDVFLSFRGFDTRNNFTGHLQKALRLRGIDSFIDDR-LRRGDNLTALFDRIEKSK 64
Query: 74 IYVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEH 133
I +IVFS NYA+S CL ELVKI+EC+ + + V+PIFY+VD S V Q NS+ F +
Sbjct: 65 IAIIVFSTNYANSAWCLRELVKILECRNSNQQLVVPIFYKVDKSDVEKQRNSFAVPF-KL 123
Query: 134 QNRLVPGVVMQR---WKKALLEAADLSGYH-SIAARSESMLIDRIAQDIFKKLN---PPF 186
PGV + WK AL A+++ GY + SE+ L+D IA D FKKLN P
Sbjct: 124 PELTFPGVTPEEISSWKAALASASNILGYVVKEISTSEAKLVDEIAVDTFKKLNDLAPSG 183
Query: 187 SQGMLGIDKHIAQIQSLLQL-DSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLI 245
++G++GI+ + ++ LL D + V IIGI GM GIGK+TLA+ LY ++ QF C +
Sbjct: 184 NEGLVGIESRLKNLEKLLSWEDLDTVHIIGIVGMVGIGKTTLADCLYGRMRGQFDGSCFL 243
Query: 246 VNAQQKIDRYGIYSLRKKYLSKLLGE-DIQSNGLNYAIERVKRA----KVLLILDDLKIS 300
N ++ R G+ SL +K S +L + D++ A ER +R ++L++LDD+
Sbjct: 244 TNIRENSGRSGLESLLQKLFSTVLNDRDLEIGAPGNAHERFERRLKSKRLLIVLDDVNDE 303
Query: 301 IPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKED 360
I L+G + GSRII+T+R +++ + + Y + ++ +++L+LF LNAF
Sbjct: 304 KQIRYLMGHCKWYQGGSRIIITTRDSKLIETIKGRK-YVLPKLNDREALKLFSLNAFSNS 362
Query: 361 YPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKL 420
+PL+ + L VL YAKG PLAL+VLGS L +++ WE++L +LK + DI++VL+
Sbjct: 363 FPLKEFEGLTNMVLDYAKGHPLALKVLGSDLCERDDLYWEAKLDRLKSRSHGDIYEVLET 422
Query: 421 SYDGLDDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISILEDRIA 480
SY+ L E K++FLDI CF+ + VD V LL+S G + L D+ LI++ ++RI
Sbjct: 423 SYEELTTEQKNVFLDIACFFRSENVDYVTSLLNSHGVDVSGVVKDLVDKCLITLSDNRIE 482
Query: 481 VHDLVLEMGREIVRQ---------QCVSDPGK----HSHLWNHKEIYHVVRKNKGTDAIQ 527
+HD++ M +EI + + +S G H LW+ ++I ++ + GTD I+
Sbjct: 483 MHDMLQTMAKEISLKVETIGIRDCRWLSRHGNQCQWHIRLWDSEDICDLLTEGLGTDKIR 542
Query: 528 CIFLDMCQIEKVQLHPEIFKSMPNIRML--YFHKHSSFKQGQSNVIVPDLLESLPNGLKV 585
IFLD ++ ++L + F+ M N++ L Y S + + + + L LPN L
Sbjct: 543 GIFLDTSKLRAMRLSAKAFQGMYNLKYLKIYDSHCSRGCEAEFKLHLRRGLSFLPNELTY 602
Query: 586 LHWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSL 645
LHW +P +S PLDF P+ LV L + S+LE++W D++ + L ++ H I +L
Sbjct: 603 LHWHGYPLQSIPLDFDPKNLVDLKLPHSQLEEIWDDEKDVGM----LKWVDLSHSI--NL 656
Query: 646 HQEL-----PNLERLDLSNSWKLVRIPD-LSKSPNIKEIILSGCKSLTRLPINLFKLKFL 699
Q L NLERL+L L ++P ++ + + L C SL LP + K + L
Sbjct: 657 RQCLGLANAHNLERLNLEGCTSLKKLPSTINCLEKLIYLNLRDCTSLRSLPKGI-KTQSL 715
Query: 700 ERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLES 759
+ L LSGCS+++ P I EN+ VL+L T I+ LP S+ L L+LK C KL+
Sbjct: 716 QTLILSGCSSLKKFPLIS---ENVEVLLLDGTVIKSLPESIQTFRRLALLNLKNCKKLKH 772
Query: 760 IPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAE---- 815
+ S + L L +L L+ C LE FP D++ ++ L D + + P ++ +
Sbjct: 773 LSSDLYKLKCLQELILSGCSQLEVFPEIKEDMESLEILLMDDTSITEMPKMMHLSNIKTF 832
Query: 816 ----SFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSG 871
+ +H+S+ ++ +P +L L L L C L LP++I L L L SG
Sbjct: 833 SLCGTSSHVSV---SMFFMPPTL-GCSRLTDLYLSRC-SLYKLPDNIGGLSSLQSLCLSG 887
Query: 872 CRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPF 931
+ +P L++L+ L+ +C+ L+ +P LP
Sbjct: 888 -NNIENLPESFNQLNNLKWFDLK-----------------------FCKMLKSLPVLPQN 923
Query: 932 MKQLLAFHCPSIRSVMSNSRFLSDSKEGTFELYFTNNEKQDPGAQRDVVRNA--EHRITD 989
++ L A C S+ ++ + L+ + F+N K + AQ +V +A + ++
Sbjct: 924 LQYLDAHECESLETLANPLTPLTVGERIHSMFIFSNCYKLNQDAQASLVGHARIKSQLMA 983
Query: 990 VACRFAFY-----------CFPGSEVPNWFHHHCKRNSVTVDKDSLNLSNDNRIIGFAMC 1038
A +Y C+P +E+P+WF H S+ + D +G A+
Sbjct: 984 NASAKRYYRGFVPEPLVGICYPATEIPSWFCHQRLGRSLEIPLPPHWC--DINFVGLALS 1041
Query: 1039 FVLQLEDMNYTGYRFSNFRCRLTFES 1064
V+ +D + RFS +C FE+
Sbjct: 1042 VVVSFKDYEDSAKRFS-VKCCGNFEN 1066
>At5g44510 disease resistance protein-like
Length = 1187
Score = 473 bits (1218), Expect = e-133
Identities = 362/1100 (32%), Positives = 577/1100 (51%), Gaps = 62/1100 (5%)
Query: 1 MSESSSSSSAAAAAMAPPPNPLK----YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FI 54
+S S SSSS PP+ L + VFLSF GED K SH+ +R + FI
Sbjct: 21 ISSSLSSSS--------PPSSLSQNWLHPVFLSFRGEDVRKGLLSHIQKEFQRNGITPFI 72
Query: 55 DD---RSLETGDEICKAIEASTIYVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIF 111
D+ R G E+ +AI S I +I+ S NY SS+ CLDELV+IM+C+ + V+ +F
Sbjct: 73 DNEMKRGGSIGPELLQAIRGSKIAIILLSRNYGSSKWCLDELVEIMKCREELGQTVMTVF 132
Query: 112 YRVDPSTVRHQTNSYGDAFDEHQNRLVPGVVMQRWKKALLEAADLSGYHSIAARSESMLI 171
Y VDPS VR Q +G F + P ++QRWK+AL AA++ G S +E+ +I
Sbjct: 133 YDVDPSDVRKQKGDFGKVFKKTCVGR-PEEMVQRWKQALTSAANILGEDSRNWENEADMI 191
Query: 172 DRIAQDIFKKLNPPFSQGM---LGIDKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLA 228
+I++D+ L+ S+ +GI+ H +I SLLQLD E VR+IGI G GIGK+T++
Sbjct: 192 IKISKDVSDVLSFTPSKDFDEFVGIEAHTTEITSLLQLDLEEVRMIGIWGPAGIGKTTIS 251
Query: 229 EALYHKLGTQFSSRCLIVNAQQKIDR--YGIYS----LRKKYLSKLLGE-DIQSNGLNYA 281
LY+KL QF +I N + + R + YS L+K+ LS+++ + D+ L A
Sbjct: 252 RVLYNKLFHQFQLGAIIDNIKVRYPRPCHDEYSAKLQLQKELLSQMINQKDMVVPHLGVA 311
Query: 282 IERVKRAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQ 341
ER+K KVLL+LDD+ + + + FG GSRIIV ++ +LK IY+V
Sbjct: 312 QERLKDKKVLLVLDDVDGLVQLDAMAKDVQWFGLGSRIIVVTQDLKLLKAHGIKYIYKVD 371
Query: 342 GMSYQDSLELFRLNAFKEDYPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWES 401
+ ++LE+F + AF E P G+ + V A +PL L+V+GS L ++ W
Sbjct: 372 FPTSDEALEIFCMYAFGEKSPKVGFEQIARTVTTLAGKLPLGLRVMGSYLRRMSKQEWAK 431
Query: 402 ELKKLKELPNVDIFDVLKLSYDGLDDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADI 461
+ +L+ + DI VLK SY+ L ++ KD+FL ITCF+ + ++ + L
Sbjct: 432 SIPRLRTSLDDDIESVLKFSYNSLAEQEKDLFLHITCFFRRERIETLEVFLAKKSVDMRQ 491
Query: 462 GMDVLKDRGLISILEDRIAVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNK 521
G+ +L D+ L+S+ I +H+L++++G +IVR+Q + PGK L + ++I V+ +
Sbjct: 492 GLQILADKSLLSLNLGNIEMHNLLVQLGLDIVRKQSIHKPGKRQFLVDTEDICEVLTDDT 551
Query: 522 GTDAIQCIFLDMCQIEK--VQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESL 579
GT + I L++ + + + + F+ M N++ L FH H + + +P L +
Sbjct: 552 GTRTLIGIDLELSGVIEGVINISERAFERMCNLQFLRFH-HPYGDRCHDILYLPQGLSHI 610
Query: 580 PNGLKVLHWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGH 639
L++LHW+ +P P F PE LVK++MR+S LE+LW ++ +++L + F +
Sbjct: 611 SRKLRLLHWERYPLTCLPPKFNPEFLVKINMRDSMLEKLWDGNEPIRNL-KWMDLSFCVN 669
Query: 640 LIVFSLHQELPNLERLDLSNSWKLVRIP-DLSKSPNIKEIILSGCKSLTRLPINLFKLKF 698
L NL+ L L N LV +P + + N+ E+ L C SL +LP ++ L
Sbjct: 670 LKELPDFSTATNLQELRLINCLSLVELPSSIGNATNLLELDLIDCSSLVKLPSSIGNLTN 729
Query: 699 LERLNLSGCSNVENIPEIKETMENLAVLVLKQ-TAIQELPSSLNHLVGLEELSLKFCSKL 757
L++L L+ CS++ +P + +L L L +++ E+PSS+ ++V L+++ CS L
Sbjct: 730 LKKLFLNRCSSLVKLPSSFGNVTSLKELNLSGCSSLLEIPSSIGNIVNLKKVYADGCSSL 789
Query: 758 ESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDL-KLTKLDLNDCSRLKTFPAILEPAES 816
+PSSIG T L +L L C SL P S+ +L +L L+L+ C L P+I
Sbjct: 790 VQLPSSIGNNTNLKELHLLNCSSLMECPSSMLNLTRLEDLNLSGCLSLVKLPSIGNVINL 849
Query: 817 FAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLT 876
+ +++ +LP +++N L TL L C +L LP+SI N+ L L +GC L
Sbjct: 850 QSLYLSDCSSLMELPFTIENATNLDTLYLDGCSNLLELPSSIWNITNLQSLYLNGCSSLK 909
Query: 877 GIPNDIGCLSSLRTLSLQD-TGVVNLPESIAHLSSLESLDVSYCR---KLECIPH-LPPF 931
+P+ + +L++LSL + +V LP SI +S+L LDVS C +L + H + P
Sbjct: 910 ELPSLVENAINLQSLSLMKCSSLVELPSSIWRISNLSYLDVSNCSSLLELNLVSHPVVPD 969
Query: 932 MKQLLAFHCPSIRSVMSNSRFLSDSKEGTFELYFTNNEKQDPGAQRDVVRNAEHRITDVA 991
L A C S+ V F + K L F N K + A RD++ I A
Sbjct: 970 SLILDAGDCESL--VQRLDCFFQNPK---IVLNFANCFKLNQEA-RDLI------IQTSA 1017
Query: 992 CRFAFYCFPGSEVPNWFHHHCKRNSVTVDKDSLNLSNDNRIIGFAMCFVLQLEDMNYTGY 1051
CR A PG +VP +F + +S+TV LN + + F C +L +E N
Sbjct: 1018 CRNAI--LPGEKVPAYFTYRATGDSLTV---KLNQKYLLQSLRFKACLLL-VEGQN---- 1067
Query: 1052 RFSNFRCRLTFESDGQTHIL 1071
++ N+ L + HI+
Sbjct: 1068 KWPNWGMNLVTSREPDGHIV 1087
>At5g18360 disease resistance protein -like
Length = 900
Score = 469 bits (1206), Expect = e-132
Identities = 312/877 (35%), Positives = 482/877 (54%), Gaps = 76/877 (8%)
Query: 23 KYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDD---RSLETGDEICKAIEASTIYVI 77
++ VF SFSG+D +TF SHL RRK + FID+ RS E+ +AI S I V+
Sbjct: 15 RHHVFPSFSGKDVRRTFLSHLLKEFRRKGIRTFIDNDIKRSQMISSELVRAIRESRIAVV 74
Query: 78 VFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRL 137
V S YASS CL+ELV+I + + ++P+FY VDPS VR +T +G AF+E R
Sbjct: 75 VLSRTYASSSWCLNELVEIKKVS----QMIMPVFYEVDPSDVRKRTGEFGKAFEEACERQ 130
Query: 138 VPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKLNPPFSQ---GMLGID 194
V Q+W++AL+ A+++G S +E+ LID+IA I +LN S+ ++GID
Sbjct: 131 PDEEVKQKWREALVYIANIAGESSQNWDNEADLIDKIAMSISYELNSTLSRDSYNLVGID 190
Query: 195 KHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQ----- 249
H+ ++ SLL L+S V+++GI G GIGK+T+A AL+++L F + N +
Sbjct: 191 NHMRELDSLLCLESTEVKMVGIWGPAGIGKTTIARALFNRLSENFQHTIFMENVKGSSRT 250
Query: 250 QKIDRYGIY-SLRKKYLSKLLG-EDIQSNGLNYAIERVKRAKVLLILDDLKISIPILELI 307
++D YG L++++LS+++ + ++ + L ER++ KVL++LDD+ + L+
Sbjct: 251 SELDAYGFQLRLQEQFLSEVIDHKHMKIHDLGLVKERLQDLKVLVVLDDVDKLEQLDALV 310
Query: 308 GGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYA 367
FG GSRIIVT+ ++ +L+ IYE+ S DSL++F AF E +G
Sbjct: 311 KQSQWFGSGSRIIVTTENKQLLRAHGITCIYELGFPSRSDSLQIFCQYAFGESSAPDGCI 370
Query: 368 NLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLDD 427
L E+ K A +PLAL+VLGS L ++ +S L +L+ N DI +VL++ YDG+ D
Sbjct: 371 ELATEITKLAGYLPLALKVLGSSLRGMSKDEQKSALPRLRTSLNEDIRNVLRVGYDGIHD 430
Query: 428 EPKDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISI--LEDRIAVHDLV 485
+ K IFL I C + G+ VD V ++L S G G+ VL R LI I I +H+L+
Sbjct: 431 KDKVIFLHIACLFNGENVDYVKQILASSGLDVTFGLQVLTSRSLIHISRCNRTITMHNLL 490
Query: 486 LEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIEKVQLHPEI 545
++GREIV +Q +++PGK L + EIY V+ N GT A+ I LD+ +I ++ L+
Sbjct: 491 EQLGREIVCEQSIAEPGKRQFLMDASEIYDVLADNTGTGAVLGISLDISKINELFLNERA 550
Query: 546 FKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCPEKL 605
F M N+ L F+K SS K Q + +P L+ LP L++LHWD FP S PL FCP+ L
Sbjct: 551 FGGMHNLLFLRFYKSSSSKD-QPELHLPRGLDYLPRKLRLLHWDAFPMTSMPLSFCPQFL 609
Query: 606 VKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVR 665
V +++RES+LE+LW Q L+S L+++DLS S L
Sbjct: 610 VVINIRESQLEKLWEGTQPLRS------------------------LKQMDLSKSENLKE 645
Query: 666 IPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAV 725
IPDLSK+ NI+E+ LS C SL LP ++ L L L++ CS +E IP +E+L++
Sbjct: 646 IPDLSKAVNIEELCLSYCGSLVMLPSSIKNLNKLVVLDMKYCSKLEIIP-CNMDLESLSI 704
Query: 726 LVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFP 785
L L + E ++ +G LS + +E IP+++ + L LD++ C++L+TFP
Sbjct: 705 LNLDGCSRLESFPEISSKIGFLSLSE---TAIEEIPTTVASWPCLAALDMSGCKNLKTFP 761
Query: 786 GSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCL 845
P +E + L+ T I+++P + L L L +
Sbjct: 762 --------------------CLPKTIE------WLDLSRTEIEEVPLWIDKLSKLNKLLM 795
Query: 846 KECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDI 882
C L S+ + I L+ + LD GC+ + P +I
Sbjct: 796 NSCMKLRSISSGISTLEHIKTLDFLGCKNIVSFPVEI 832
Score = 94.4 bits (233), Expect = 3e-19
Identities = 75/242 (30%), Positives = 124/242 (50%), Gaps = 28/242 (11%)
Query: 686 LTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVL-KQTAIQELPSSLNHLV 744
+T +P++ F +FL +N+ S +E + E + + +L + L K ++E+P L+ V
Sbjct: 597 MTSMPLS-FCPQFLVVINIRE-SQLEKLWEGTQPLRSLKQMDLSKSENLKEIPD-LSKAV 653
Query: 745 GLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRL 804
+EEL L +C L +PSSI L KL LD+ YC LE P ++ L+ L+L+ CSRL
Sbjct: 654 NIEELCLSYCGSLVMLPSSIKNLNKLVVLDMKYCSKLEIIPCNMDLESLSILNLDGCSRL 713
Query: 805 KTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDL------------- 851
++FP I + +SL+ETAI+++P+++ + L L + C +L
Sbjct: 714 ESFPEI---SSKIGFLSLSETAIEEIPTTVASWPCLAALDMSGCKNLKTFPCLPKTIEWL 770
Query: 852 -------ESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLS-LQDTGVVNLPE 903
E +P I L L++L + C KL I + I L ++TL L +V+ P
Sbjct: 771 DLSRTEIEEVPLWIDKLSKLNKLLMNSCMKLRSISSGISTLEHIKTLDFLGCKNIVSFPV 830
Query: 904 SI 905
I
Sbjct: 831 EI 832
Score = 61.6 bits (148), Expect = 2e-09
Identities = 45/144 (31%), Positives = 65/144 (44%), Gaps = 20/144 (13%)
Query: 812 EPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSG 871
+P S + L+++ K L V ++ LCL C L LP+SI NL L LD
Sbjct: 627 QPLRSLKQMDLSKSENLKEIPDLSKAVNIEELCLSYCGSLVMLPSSIKNLNKLVVLDMKY 686
Query: 872 CRKLTGIP-----------NDIGC---------LSSLRTLSLQDTGVVNLPESIAHLSSL 911
C KL IP N GC S + LSL +T + +P ++A L
Sbjct: 687 CSKLEIIPCNMDLESLSILNLDGCSRLESFPEISSKIGFLSLSETAIEEIPTTVASWPCL 746
Query: 912 ESLDVSYCRKLECIPHLPPFMKQL 935
+LD+S C+ L+ P LP ++ L
Sbjct: 747 AALDMSGCKNLKTFPCLPKTIEWL 770
>At5g11250 RPP1 disease resistance protein - like
Length = 1189
Score = 459 bits (1181), Expect = e-129
Identities = 321/973 (32%), Positives = 519/973 (52%), Gaps = 78/973 (8%)
Query: 1 MSESSSSSSAAAAAMAPPPNPL--KYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDD 56
+S SSS SS + +++ PP + + VF SFSGED + F SH+ +R + F+D+
Sbjct: 37 LSPSSSPSSLSPSSVPPPSSSCIWTHQVFPSFSGEDVRRDFLSHIQMEFQRMGITPFVDN 96
Query: 57 ---RSLETGDEICKAIEASTIYVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYR 113
R G E+ +AI S I +I+ S NYASS+ CLDELV+IM+C+ Y + V+ IFY+
Sbjct: 97 EIKRGESIGPELLRAIRGSKIAIILLSRNYASSKWCLDELVEIMKCREEYGQTVMAIFYK 156
Query: 114 VDPSTVRHQTNSYGDAFDEHQNRLVPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDR 173
VDPS V++ T +G F + P + RW++A + A ++GYHSI +E+ +I +
Sbjct: 157 VDPSDVKNLTGDFGKVFRKTCAGK-PKKDIGRWRQAWEKVATVAGYHSINWDNEAAMIKK 215
Query: 174 IAQDIFKKL-NPPFSQ---GMLGIDKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAE 229
IA DI L N S+ G++G+ H+ +++ LL LD++ VRIIGI G GIGK+T+A
Sbjct: 216 IATDISNILINSTPSRDFDGLVGMRAHLEKMKPLLCLDTDEVRIIGIWGPPGIGKTTIAR 275
Query: 230 ALYHKLGTQFSSRCLIVNAQQKIDR-------YGIYSLRKKYLSKLLGE-DIQSNGLNYA 281
+Y++L F + N + R L++ ++S++ + DI+ L A
Sbjct: 276 VVYNQLSHSFQLSVFMENIKANYTRPTGSDDYSAKLQLQQMFMSQITKQKDIEIPHLGVA 335
Query: 282 IERVKRAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQ 341
+R+K KVL++LD + S+ + + FG GSRII+T++ + + + + IY+V
Sbjct: 336 QDRLKDKKVLVVLDGVNQSVQLDAMAKEAWWFGPGSRIIITTQDQKLFRAHGINHIYKVD 395
Query: 342 GMSYQDSLELFRLNAFKEDYPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWES 401
+++L++F + AF ++ P +G+ NL +V+ A +PL L+++GS RE W+
Sbjct: 396 FPPTEEALQIFCMYAFGQNSPKDGFQNLAWKVINLAGNLPLGLRIMGSYFRGMSREEWKK 455
Query: 402 ELKKLKELPNVDIFDVLKLSYDGLDDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADI 461
L +L+ + DI +LK SYD LDDE K++FL I CF+ G + + E L
Sbjct: 456 SLPRLESSLDADIQSILKFSYDALDDEDKNLFLHIACFFNGKEIKILEEHLAKKFVEVRQ 515
Query: 462 GMDVLKDRGLISILE-DRIAVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKN 520
++VL ++ LIS I +H L+ ++G EIVR Q + +PG+ L++ +EI V+ +
Sbjct: 516 RLNVLAEKSLISFSNWGTIEMHKLLAKLGGEIVRNQSIHEPGQRQFLFDGEEICDVLNGD 575
Query: 521 -KGTDAIQCIFLDMCQIEKVQLHPEIFKSMPNIRMLYFH-KHSSFKQGQSNVIVPDLLES 578
G+ ++ I E+ ++ +F+ M N++ L F H + + + L
Sbjct: 576 AAGSKSVIGIDFHYIIEEEFDMNERVFEGMSNLQFLRFDCDHDTLQLSRG-------LSY 628
Query: 579 LPNGLKVLHWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFG 638
L L++L W FP P E L++L++ S+L+ LW G+
Sbjct: 629 LSRKLQLLDWIYFPMTCLPSTVNVEFLIELNLTHSKLDMLW----------EGV------ 672
Query: 639 HLIVFSLHQELPNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKF 698
+ L NL ++DLS S L +PDLS + N++++ILS C SL +LP +
Sbjct: 673 --------KPLHNLRQMDLSYSVNLKELPDLSTAINLRKLILSNCSSLIKLPSCIGNAIN 724
Query: 699 LERLNLSGCSNVENIPEIKETMENLAVLVLKQTA-IQELPSSLNHLVGLEELSLKFCSKL 757
LE L+L+GCS++ +P + + NL L+L+ + + ELPSS+ + + L EL L +CS L
Sbjct: 725 LEDLDLNGCSSLVELPSFGDAI-NLQKLLLRYCSNLVELPSSIGNAINLRELDLYYCSSL 783
Query: 758 ESIPSSIGTLTKLCKLDLTYCESLETFPGSIFD-LKLTKLDLNDCSRLKTFPAILEPAES 816
+PSSIG L LDL C +L P SI + + L KLDL C++L P+ + A +
Sbjct: 784 IRLPSSIGNAINLLILDLNGCSNLLELPSSIGNAINLQKLDLRRCAKLLELPSSIGNAIN 843
Query: 817 FAHISLAE-TAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKL 875
++ L + +++ +LPSS+ N L + L C +L LP SI NL+ L EL GC KL
Sbjct: 844 LQNLLLDDCSSLLELPSSIGNATNLVYMNLSNCSNLVELPLSIGNLQKLQELILKGCSKL 903
Query: 876 TGIP-------------NDIGCL-------SSLRTLSLQDTGVVNLPESIAHLSSLESLD 915
+P ND L +++R L L T + +P SI L+ L
Sbjct: 904 EDLPININLESLDILVLNDCSMLKRFPEISTNVRALYLCGTAIEEVPLSIRSWPRLDELL 963
Query: 916 VSYCRKLECIPHL 928
+SY L PH+
Sbjct: 964 MSYFDNLVEFPHV 976
Score = 67.0 bits (162), Expect = 6e-11
Identities = 60/226 (26%), Positives = 93/226 (40%), Gaps = 55/226 (24%)
Query: 651 NLERLDLSNSWKLVRIP-DLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSN 709
NL ++LSN LV +P + ++E+IL GC L LPIN+ L+ L+ L L+ CS
Sbjct: 867 NLVYMNLSNCSNLVELPLSIGNLQKLQELILKGCSKLEDLPINI-NLESLDILVLNDCSM 925
Query: 710 VENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTK 769
++ PEI N+ L L TAI+E+P S+ L+EL + + L P + +T
Sbjct: 926 LKRFPEIST---NVRALYLCGTAIEEVPLSIRSWPRLDELLMSYFDNLVEFPHVLDIITN 982
Query: 770 LCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKK 829
L L+ I++
Sbjct: 983 L--------------------------------------------------DLSGKEIQE 992
Query: 830 LPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKL 875
+P ++ + LQTL LK + SLP +LK + DC +L
Sbjct: 993 VPPLIKRISRLQTLILKGYRKVVSLPQIPDSLKWIDAEDCESLERL 1038
>At2g14080 putative disease resistance protein
Length = 1215
Score = 458 bits (1178), Expect = e-129
Identities = 319/962 (33%), Positives = 504/962 (52%), Gaps = 76/962 (7%)
Query: 16 APPPNPL-------KYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FID---DRSLETGD 63
+PP +P K+DVF SF G D K+F SH+ +RK + FID +RS G
Sbjct: 41 SPPTSPQSSLSCNQKHDVFPSFHGADVRKSFLSHILKEFKRKGIDTFIDNNIERSKSIGP 100
Query: 64 EICKAIEASTIYVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQT 123
E+ +AI+ S I V++ S++YASS CL+ELV+IM+C+ D+ V+ IFY VDP+ V+ QT
Sbjct: 101 ELIEAIKGSKIAVVLLSKDYASSSWCLNELVEIMKCRKMLDQTVMTIFYEVDPTDVKKQT 160
Query: 124 NSYGDAFDEHQNRLVPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKLN 183
+G F + V ++W +AL E A ++G HSI +E+ +I++I+ DI KLN
Sbjct: 161 GDFGKVFKKTCMGKT-NAVSRKWIEALSEVATIAGEHSINWDTEAAMIEKISTDISNKLN 219
Query: 184 PPFS----QGMLGIDKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQF 239
G++G+ H+ +++ LL LDS VR+IGI G GIGK+T+ LY++L + F
Sbjct: 220 NSTPLRDFDGLVGMGAHMEKLELLLCLDSCEVRMIGIWGPPGIGKTTIVRFLYNQLSSSF 279
Query: 240 SSRCLIVNAQ-------QKIDRYGIYSLRKKYLSKLLG-EDIQSNGLNYAIERVKRAKVL 291
+ N + D L++++LSK+L +DI+ L ER+ KVL
Sbjct: 280 ELSIFMENIKTMHTILASSDDYSAKLILQRQFLSKILDHKDIEIPHLRVLQERLYNKKVL 339
Query: 292 LILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLEL 351
++LDD+ S+ + L FG SRI++T++ R +LK + IY+V + D+L++
Sbjct: 340 VVLDDVDQSVQLDALAKETRWFGPRSRILITTQDRKLLKAHRINNIYKVDLPNSDDALQI 399
Query: 352 FRLNAFKEDYPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPN 411
F + AF + P +G+ L +V PL L+V+GS + ++ W E+ +L+ +
Sbjct: 400 FCMYAFGQKTPYDGFYKLARKVTWLVGNFPLGLRVVGSYFREMSKQEWRKEIPRLRARLD 459
Query: 412 VDIFDVLKLSYDGLDDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGL 471
I VLK SYD L DE KD+FL I CF+ + ++K+ + L VL ++ L
Sbjct: 460 GKIESVLKFSYDALCDEDKDLFLHIACFFNHESIEKLEDFLGKTFLDIAQRFHVLAEKSL 519
Query: 472 ISILEDRIAVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKN-KGTDAIQCIF 530
ISI + + +HD + ++G+EIVR+Q V +PG+ L + ++I V+ + G ++ I+
Sbjct: 520 ISINSNFVEMHDSLAQLGKEIVRKQSVREPGQRQFLVDARDISEVLADDTAGGRSVIGIY 579
Query: 531 LDMCQIEKV-QLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWD 589
LD+ + + V + + F+ M N++ L + + V +P L + L++L W
Sbjct: 580 LDLHRNDDVFNISEKAFEGMSNLQFLRVKNFGNL--FPAIVCLPHCLTYISRKLRLLDWM 637
Query: 590 EFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQEL 649
FP FP F PE LV+L+M S+LE+LW + Q L+
Sbjct: 638 YFPMTCFPSKFNPEFLVELNMWGSKLEKLWEEIQPLR----------------------- 674
Query: 650 PNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSN 709
NL+R+DL +S L +PDLS + N++ + L+GC SL LP ++ L +L LSGCS+
Sbjct: 675 -NLKRMDLFSSKNLKELPDLSSATNLEVLNLNGCSSLVELPFSIGNATKLLKLELSGCSS 733
Query: 710 VENIPEIKETMENLAVLVLKQTA-IQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLT 768
+ +P NL + + ELPSS+ + L+EL L CS L+ +PSSIG T
Sbjct: 734 LLELPSSIGNAINLQTIDFSHCENLVELPSSIGNATNLKELDLSCCSSLKELPSSIGNCT 793
Query: 769 KLCKLDLTYCESLETFPGSIFD-LKLTKLDLNDCSRLKTFPAILEPAESFAHISLAE-TA 826
L KL L C SL+ P SI + L +L L CS L P+ + A + + LA +
Sbjct: 794 NLKKLHLICCSSLKELPSSIGNCTNLKELHLTCCSSLIKLPSSIGNAINLEKLILAGCES 853
Query: 827 IKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIG--- 883
+ +LPS + L+ L L L LP+ I NL LSEL GC+KL +P +I
Sbjct: 854 LVELPSFIGKATNLKILNLGYLSCLVELPSFIGNLHKLSELRLRGCKKLQVLPTNINLEF 913
Query: 884 --------CL---------SSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
C+ ++++ L L+ T + +P S+ LE L + Y L
Sbjct: 914 LNELDLTDCILLKTFPVISTNIKRLHLRGTQIEEVPSSLRSWPRLEDLQMLYSENLSEFS 973
Query: 927 HL 928
H+
Sbjct: 974 HV 975
Score = 57.0 bits (136), Expect = 6e-08
Identities = 54/196 (27%), Positives = 77/196 (38%), Gaps = 46/196 (23%)
Query: 651 NLERLDLSNSWKLVRIPD-LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGC-- 707
NL+ L+L LV +P + + E+ L GCK L LP N+ L+FL L+L+ C
Sbjct: 866 NLKILNLGYLSCLVELPSFIGNLHKLSELRLRGCKKLQVLPTNI-NLEFLNELDLTDCIL 924
Query: 708 --------SNV-------------------------------ENIPEIKETMENLAVLVL 728
+N+ EN+ E +E + VL L
Sbjct: 925 LKTFPVISTNIKRLHLRGTQIEEVPSSLRSWPRLEDLQMLYSENLSEFSHVLERITVLEL 984
Query: 729 KQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSI 788
I+E+ LN + L L L C KL S+P L LD C SLE S
Sbjct: 985 SDINIREMTPWLNRITRLRRLKLSGCGKLVSLPQ---LSDSLIILDAENCGSLERLGCSF 1041
Query: 789 FDLKLTKLDLNDCSRL 804
+ + LD +C +L
Sbjct: 1042 NNPNIKCLDFTNCLKL 1057
>At5g51630 putative protein
Length = 1239
Score = 457 bits (1176), Expect = e-128
Identities = 364/1131 (32%), Positives = 551/1131 (48%), Gaps = 174/1131 (15%)
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRK--NVFID---DRSLETGDEICKAIEASTIYVIV 78
YDVF SFSGED K+F SHL L RK N FID +RS ++ AI S I ++V
Sbjct: 11 YDVFPSFSGEDVRKSFLSHLLKKLHRKSINTFIDNNIERSHAIAPDLLSAINNSMISIVV 70
Query: 79 FSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRLV 138
FS+ YASS CL+ELV+I +C + VIPIFY VDPS VR QT +G+ F
Sbjct: 71 FSKKYASSTWCLNELVEIHKCYKELTQIVIPIFYEVDPSDVRKQTREFGEFFKVTCVGKT 130
Query: 139 PGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKLNPPFSQG----MLGID 194
V Q+W +AL E A ++G+ S +E+ +I+ IA+D+ KL S ++GI+
Sbjct: 131 EDV-KQQWIEALEEVASIAGHDSKNWPNEANMIEHIAKDVLNKLIATSSSNCFGDLVGIE 189
Query: 195 KHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQKIDR 254
H+ ++S+L L+SE R++GI G GIGK+T+A LY KL +QF + D
Sbjct: 190 AHLKAVKSILCLESEEARMVGILGPSGIGKTTIARILYSKLSSQFDYHVFGSFKRTNQDN 249
Query: 255 YGI-YSLRKKYLSKLLGE-DIQSNGLNYAIERVKRAKVLLILDDLKISIPILELIGGHGN 312
YG+ S +++LS++L + D++ + L +R+K KVL++LDD+ + L+G G
Sbjct: 250 YGMKLSWEEQFLSEILDQKDLKISQLGVVKQRLKHKKVLIVLDDVDNLELLKTLVGQTGW 309
Query: 313 FGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYANLIVE 372
FG GSRIIVT++ R +LK+ + D IYEV S + +L + +AF + P +G+ L E
Sbjct: 310 FGPGSRIIVTTQDRILLKSHKIDHIYEVGYPSRKLALRILCRSAFDRNSPPDGFMQLANE 369
Query: 373 VLKYAKGVPLALQVLGSLLYDKEREVWESELKKLK-ELPNVDIFDVLKLSYDGLDDEPKD 431
V + +PLAL ++GS L +++E W + L+ L + +I L++SYD L ++
Sbjct: 370 VTELVGNLPLALNIMGSSLKGRDKEEWIEMMPSLRNSLVDGEILKTLRVSYDRLHGNYQE 429
Query: 432 IFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISI--LEDRIAVHDLVLEMG 489
IFL I C V+ ++ +L G +A IG+ +L ++ LI I L+ + +H L+ ++G
Sbjct: 430 IFLYIACLLNCCGVEYIISML---GDNAIIGLKILAEKSLIHISPLDKTVEMHSLLQKLG 486
Query: 490 REIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIE-KVQLHPEIFKS 548
R+IVR + +PGK L + ++I V N GT+ + I L+ +I + + + F+
Sbjct: 487 RKIVRDESFGNPGKRRFLLDAEDICDVFTDNTGTETVLGISLNTLEINGTLSVDDKSFQG 546
Query: 549 MPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCPEKLVKL 608
M N++ L ++ G+ + +P L SLP L++LHW +FP R P +F E LV L
Sbjct: 547 MHNLQFLKVFENWRRGSGEGILSLPQGLNSLPRKLRLLHWYKFPLRCMPSNFKAEYLVNL 606
Query: 609 HMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIPD 668
M S+LE+LW Q+L +L+++DLS S L IPD
Sbjct: 607 EMAYSQLERLWEGT------------------------QQLGSLKKMDLSKSENLKEIPD 642
Query: 669 LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIP-------------- 714
LS + N++E+ L CKSL LP ++ L L L +S CSNVE +P
Sbjct: 643 LSYAVNLEEMDLCSCKSLVTLPSSVRNLDKLRVLRMSSCSNVEVLPTDLNLESLDLLNLE 702
Query: 715 ------EIKETMENLAVLVLKQTAIQE------------------------LPSSL--NH 742
+ N+++L L TAI E LPS+ H
Sbjct: 703 DCSQLRSFPQISRNISILNLSGTAIDEESSLWIENMSRLTHLRWDFCPLKSLPSNFRQEH 762
Query: 743 LVGLE--------------------ELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLE 782
LV L + L KL+ P ++ +T L LDL C+SL
Sbjct: 763 LVSLHMTHSKLEKLWEGAQPFGNLVNIDLSLSEKLKEFP-NLSKVTNLDTLDLYGCKSLV 821
Query: 783 TFPGSIFDL-KLTK-----------------------LDLNDCSRLKTFPAILEPAESFA 818
T P SI L KLT+ LDL+ CS+L TFP I + +
Sbjct: 822 TVPSSIQSLSKLTELNMRRCTGLEALPTDVNLESLHTLDLSGCSKLTTFPKI---SRNIE 878
Query: 819 HISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGI 878
+ L +TAI+++PS + + L TL +K C L ++ SIC LK + + S C +LT
Sbjct: 879 RLLLDDTAIEEVPSWIDDFFELTTLSMKGCKRLRNISTSICELKCIEVANFSDCERLTEF 938
Query: 879 PNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAF 938
+ LRT+ ++ L E + L ++ L CRKL I + F
Sbjct: 939 DDASMVRRILRTID----DLIALYEEASFLHAIFVL----CRKLVSI--------CAMVF 982
Query: 939 HCPSIRSVMSNSRFLSDSKEGTFELYFTNNEKQDPGAQRDVVRNAEHRITDVACRFAFYC 998
P S NS +L F N D R+AE I +
Sbjct: 983 KYPQALSYFFNS--------PEADLIFANCSSLD--------RDAETLILE--SNHGCAV 1024
Query: 999 FPGSEVPNWFHHHCKRNSVTVDKDSLNLSNDNRIIGFAMCFVLQL-EDMNY 1048
PG +VPN F + +SV++ S + +GF C VL+ D+N+
Sbjct: 1025 LPGGKVPNCFMNQACGSSVSIPLHESYYSEE--FLGFKACIVLETPPDLNF 1073
>At5g18350 disease resistance protein -like
Length = 1193
Score = 456 bits (1174), Expect = e-128
Identities = 388/1248 (31%), Positives = 587/1248 (46%), Gaps = 185/1248 (14%)
Query: 10 AAAAAMAPPPNPLKYDVFLSFSGEDTGKTFTSHLHAALRRKNVF--IDD---RSLETGDE 64
A+++ + P + VFLSF GED K F SH+ RK +F +D R G
Sbjct: 6 ASSSLSSTPTRTWTHHVFLSFRGEDVRKGFLSHIQKEFERKGIFPFVDTKMKRGSSIGPV 65
Query: 65 ICKAIEASTIYVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTN 124
+ AI S I +++ S+NYASS CL+ELV IM+C+ + + V+ +FY VDPS VR QT
Sbjct: 66 LSDAIIVSKIAIVLLSKNYASSTWCLNELVNIMKCREEFGQTVMTVFYEVDPSDVRKQTG 125
Query: 125 SYGDAFDEHQNRLVPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKLNP 184
+G AF+ V Q W++AL++ +++ G ES LID+IA+D+ +LN
Sbjct: 126 DFGIAFETTCVGKTEEV-KQSWRQALIDVSNIVGEVYRIWSKESDLIDKIAEDVLDELNY 184
Query: 185 PFSQ---GMLGIDKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSS 241
S+ G +GI +H+ +++SLL L+S VR+IGI G GIGK+T+A AL ++ F
Sbjct: 185 TMSRDFDGYVGIGRHMRKMKSLLCLESGDVRMIGIVGPPGIGKTTIARALRDQISENFQL 244
Query: 242 RCLIVNAQQKIDR--YGIYSLRK----------------KYLSKLLGE-DIQSNGLNYAI 282
I + + R YG L+ +LS++L + DI + LN A
Sbjct: 245 TAFIDDIRLTYPRRCYGESGLKPPTAFMNDDRRKIVLQTNFLSEILNQKDIVIHNLNAAP 304
Query: 283 ERVKRAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQG 342
+K KVL+ILDD+ + + G FG GSRII+T++ R +LK D IYEV
Sbjct: 305 NWLKDRKVLVILDDVDHLEQLDAMAKETGWFGYGSRIIITTQDRKLLKAHNIDYIYEVGL 364
Query: 343 MSYQDSLELFRLNAFKEDYPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESE 402
D+L++F L+AF +++P + + L EV + A +PL L+VLGS L E W++
Sbjct: 365 PRKDDALQIFCLSAFGQNFPHDDFQYLACEVTQLAGELPLGLKVLGSYLKGMSLEEWKNA 424
Query: 403 LKKLKELPNVDIFDVLKLSYDGLDDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADIG 462
L +LK + DI L+ SYD L + + +FL I C + G V V + L D G
Sbjct: 425 LPRLKTCLDGDIEKTLRYSYDALSRKDQALFLHIACLFRGYEVGHVKQWLGKSDLDVDHG 484
Query: 463 MDVLKDRGLISILEDRIAVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKN-K 521
+DVL+ + LISI + +H L+ ++G EIVR Q +P + L + +I V N
Sbjct: 485 LDVLRQKSLISIDMGFLNMHSLLQQLGVEIVRNQSSQEPRERQFLVDVNDISDVFTYNTA 544
Query: 522 GTDAIQCIFLDMCQI-EKVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLP 580
GT +I I L++ +I EK+ + +F M N++ L+ ++ K + +P L LP
Sbjct: 545 GTKSILGIRLNVPEIEEKIVIDELVFDGMTNLQFLFVNEGFGDK-----LSLPRGLNCLP 599
Query: 581 NGLKVLHWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHL 640
L+VLHW+ P R +P F LV+L MR + E+LW +++L
Sbjct: 600 GKLRVLHWNYCPLRLWPSKFSANFLVELVMRGNNFEKLW--EKIL--------------- 642
Query: 641 IVFSLHQELPNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLE 700
L +L+R+DLS+S L IPDLS + N++E+ LS C L L ++ K L+
Sbjct: 643 -------PLKSLKRMDLSHSKDLKEIPDLSNATNLEELDLSSCSGLLELTDSIGKATNLK 695
Query: 701 RLNLSGCSNVENIPEIKETMENLAVLVLKQ-TAIQELPSSLNHLVGLEELSLKFCSKLES 759
RL L+ CS ++ +P NL VL L + +ELP S+ L L+ L L C KL +
Sbjct: 696 RLKLACCSLLKKLPSSIGDATNLQVLDLFHCESFEELPKSIGKLTNLKVLELMRCYKLVT 755
Query: 760 IPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAH 819
+P+SI T KL L ++ CE L+ FP T ++L DC++LK FP I + +
Sbjct: 756 LPNSIKT-PKLPVLSMSECEDLQAFP--------TYINLEDCTQLKMFPEI---STNVKE 803
Query: 820 ISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNS--------------------IC 859
+ L TAI+ +PSS+ + L L + EC +L+ PN I
Sbjct: 804 LDLRNTAIENVPSSICSWSCLYRLDMSECRNLKEFPNVPVSIVELDLSKTEIEEVPSWIE 863
Query: 860 NLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGV--------------------- 898
NL LL L GC++L I +I L +L L L GV
Sbjct: 864 NLLLLRTLTMVGCKRLNIISPNISKLKNLEDLELFTDGVSGDAASFYAFVEFSDRHDWTL 923
Query: 899 ----------------------------VNLPESIAHLSSLESLDVSYCRKLECIPHLPP 930
+P+ I L L LDVS CR L +P LP
Sbjct: 924 ESDFQVHYILPICLPKMAISLRFWSYDFETIPDCINCLPGLSELDVSGCRNLVSLPQLPG 983
Query: 931 FMKQLLAFHCPSIRSVMSNSRFLSDSKEGTFELYFTNNEKQDPGAQRDVVRNAEHRITDV 990
+ L A +C S+ + N F + F N++ A I
Sbjct: 984 SLLSLDANNCESLERI--NGSFQNPEICLNFANCINLNQE------------ARKLIQTS 1029
Query: 991 ACRFAFYCFPGSEVPNWFHHHCKRNSVTVDKDSLNLSNDNRIIGFAMCFVLQLEDMNYTG 1050
AC +A PG+EVP F S+T++ + L + R + C +L ++N
Sbjct: 1030 ACEYAI--LPGAEVPAHFTDQDTSGSLTINITTKTLPSRLR---YKACILLSKGNINLED 1084
Query: 1051 Y---RFSNFRCRLTFESDGQTHILPIDDQLNNSLHQIDLRVGHLVRDHTFIWNYYLMDSA 1107
F + C +T G+ +IL + + DH +I++Y S
Sbjct: 1085 EDEDSFMSVSCHVT----GKQNILILPSPVLRG-----------YTDHLYIFDY----SF 1125
Query: 1108 SNHNGIIHAHNFTF-EISNGLYLKSSSL-VKECGIYPLYRKEKDDDDG 1153
S H A TF E+ + + S VK CG++ L+ K G
Sbjct: 1126 SLHEDFPEAKEATFSELMFDFIVHTKSWNVKSCGVH-LFEKSSSMPSG 1172
>At5g46270 disease resistance protein-like
Length = 1145
Score = 454 bits (1167), Expect = e-127
Identities = 317/936 (33%), Positives = 482/936 (50%), Gaps = 117/936 (12%)
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FID---DRSLETGDEICKAIEASTIYVIV 78
YDVFLSF G D TF SH L RK + F D +RS ++ +AI+ S I V++
Sbjct: 12 YDVFLSFRGGDVRVTFRSHFLKELDRKLITAFRDNEIERSHSLWPDLEQAIKDSRIAVVI 71
Query: 79 FSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRLV 138
FS+NYASS CL+EL++I+ C D+ VIP+FY VDPS VRHQ +G F++ R
Sbjct: 72 FSKNYASSSWCLNELLEIVNCN---DKIVIPVFYGVDPSQVRHQIGDFGKIFEKTCKRQT 128
Query: 139 PGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKL---NPPFSQGMLGIDK 195
V Q WKKAL + A++ G+ S E+ +I+ IA D+ KL P + +GI+
Sbjct: 129 EQVKNQ-WKKALTDVANMLGFDSATWDDEAKMIEEIANDVLAKLLLTTPKDFENFVGIED 187
Query: 196 HIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHKLG-----TQFSSRCLIVNAQQ 250
HIA + LL+L++E VR++GI G GIGK+T+A AL+++L ++F R + +++
Sbjct: 188 HIANMSVLLKLEAEEVRMVGIWGSSGIGKTTIARALFNQLSRHFPVSKFIDRAFVYKSRE 247
Query: 251 KIDRYGI------YSLRKKYLSKLLG-EDIQSNGLNYAIERVKRAKVLLILDDLKISIPI 303
R L++K LS++L DI+ + L ER++ KVL+I+DDL + +
Sbjct: 248 IFSRANPDDHNMKLHLQEKLLSEILRMPDIKIDHLGVLGERLQHQKVLIIVDDLDDQVIL 307
Query: 304 LELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPL 363
L+G FG GSRII + ++H L+ E D IYEV + Q +L + +AF++ P
Sbjct: 308 DSLVGQTQWFGSGSRIIAVTNNKHFLRAHEIDHIYEVSLPTQQHALAMLCQSAFRKKSPP 367
Query: 364 EGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYD 423
EG+ L+V+V ++ +PL L VLGS L +++E W L +L+ + I +L++SYD
Sbjct: 368 EGFEMLVVQVARHVDSLPLGLNVLGSYLRGRDKEYWMEMLPRLENGLHDKIEKILRISYD 427
Query: 424 GL-DDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISILEDRIAVH 482
GL +E K IF I C + V + LL G +IG+ L D+ +I + + +H
Sbjct: 428 GLGSEEDKAIFRHIACLFNHMEVTTITSLLTDLGI--NIGLKNLVDKSIIHVRRGCVEMH 485
Query: 483 DLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIEKVQLH 542
++ EMGR+IVR Q + PGK L + +I V+ + GT + I L+ +I+++ +H
Sbjct: 486 RMLQEMGRKIVRTQSIDKPGKREFLVDPNDISDVLSEGIGTQKVLGISLNTGEIDELYVH 545
Query: 543 PEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCP 602
FK M N+R L + K G+ + +P+ L+ LP LK+L W FP R P +F P
Sbjct: 546 ESAFKGMSNLRFLEIDSKNFGKAGR--LYLPESLDYLPPRLKLLCWPNFPMRCMPSNFRP 603
Query: 603 EKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWK 662
E LV L M S+L +LW + V C L+ +D+ S
Sbjct: 604 ENLVTLKMPNSKLHKLW--EGVASLTC----------------------LKEMDMVGSSN 639
Query: 663 LVRIPDLSKSPNIKEIILSGCKSLTRLPINL-----------------------FKLKFL 699
L IPDLS N++ + L CKSL LP ++ F LK L
Sbjct: 640 LKEIPDLSMPTNLEILKLGFCKSLVELPSSIRNLNKLLKLDMEFCHSLEILPTGFNLKSL 699
Query: 700 ERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLV--------------- 744
+ LN CS + PE N++VL+L T I+E P +L +LV
Sbjct: 700 DHLNFRYCSELRTFPEFS---TNISVLMLFGTNIEEFP-NLENLVELSLSKEESDGKQWD 755
Query: 745 --------------GLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFD 790
L+ L L+ L +PSS L +L +L +TYC +LET P I
Sbjct: 756 GVKPLTPFLEMLSPTLKSLKLENIPSLVELPSSFQNLNQLKELSITYCRNLETLPTGINL 815
Query: 791 LKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPD 850
L L CS+L++FP I + + + ++L ET I+++P ++N L L ++ C
Sbjct: 816 KSLNYLCFKGCSQLRSFPEI---STNISVLNLEETGIEEVPWQIENFFNLTKLTMRSCSK 872
Query: 851 LESLPNSICNLKLLSELDCSGCRKLT-----GIPND 881
L+ L +I +K L ++D S C LT G P+D
Sbjct: 873 LKCLSLNIPKMKTLWDVDFSDCAALTVVNLSGYPSD 908
>At4g08450 putative protein
Length = 1234
Score = 449 bits (1154), Expect = e-126
Identities = 315/921 (34%), Positives = 489/921 (52%), Gaps = 87/921 (9%)
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNVF-IDDRSLETGD----EICKAIEASTIYVIV 78
YDVF SFSGED TF +H L RK + D +E G+ E+ +AI+ S I V+V
Sbjct: 11 YDVFTSFSGEDIRVTFLTHFLKELDRKMIIAFKDNEIERGNSIGTELIQAIKDSRIAVVV 70
Query: 79 FSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEH-QNRL 137
FS+ Y+SS CL+ELV+I+ CK VIP+FY +DPS VR Q +G++F E +NR
Sbjct: 71 FSKKYSSSSWCLNELVEIVNCK----EIVIPVFYDLDPSDVRKQEGEFGESFKETCKNRT 126
Query: 138 VPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKL-----NPPFSQGMLG 192
+QRW +AL A+++GYH+ +E+ LI+ I D+ KL + F + G
Sbjct: 127 --DYEIQRWGQALTNVANIAGYHTRKPNNEAKLIEEITNDVLDKLMKLTPSKDFDE-FFG 183
Query: 193 IDKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLI------- 245
I+ HI ++ LL L+SE VR++GI G GIGK+T+A AL++++ F R I
Sbjct: 184 IEDHIKELSLLLCLESEEVRMVGIWGPTGIGKTTIARALFNRIYRHFQGRVFIDRAFISK 243
Query: 246 ---VNAQQKIDRYGI-YSLRKKYLSKLLGE-DIQSNGLNYAIERVKRAKVLLILDDLKIS 300
+ ++ D Y + L++K LSKLL + +++ N L+ ER+++ KVL+ +DDL
Sbjct: 244 SMAIYSRANSDDYNLKLHLQEKLLSKLLDKKNLEINHLDAVKERLRQMKVLIFIDDLDDQ 303
Query: 301 IPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKED 360
+ + L FG GSRIIV ++ +H+L+ D IYEV S ++++F +AF++D
Sbjct: 304 VVLEALACQTQWFGHGSRIIVITKDKHLLRAYGIDHIYEVLLPSKDLAIKMFCRSAFRKD 363
Query: 361 YPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKL 420
P G+ L +V+K A +PL L +LGS L + +E W + L+ + I L++
Sbjct: 364 SPPNGFIELAYDVVKRAGSLPLGLNILGSYLRGRSKEDWIDMMPGLRNKLDGKIQKTLRV 423
Query: 421 SYDGL-DDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISI--LED 477
SYDGL ++ + IF I C + + + +LL+ G + G+ L D+ LI I +
Sbjct: 424 SYDGLASEDDQAIFRHIACIFNFEACSDIKKLLEDSGLNVTNGLINLVDKSLIRIEPKQK 483
Query: 478 RIAVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIE 537
+ +H L+ E REI+R Q DPGK L + K+I V+ GT + I LDM +IE
Sbjct: 484 TVEMHCLLQETAREIIRAQSFDDPGKREFLVDGKDIADVLDNCSGTRKVLGISLDMDEIE 543
Query: 538 KVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFP 597
++ L + FK M N+R L + +++ + + +++P LPN L++L W FP R P
Sbjct: 544 ELHLQVDAFKKMLNLRFLKLYTNTNISEKEDKLLLPKEFNYLPNTLRLLSWQRFPMRCMP 603
Query: 598 LDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDL 657
DF P+ LVKL M S+LE+LW LQ L ++ +L F NLE L L
Sbjct: 604 SDFFPKYLVKLLMPGSKLEKLWDGVMPLQCL-KNMNLFGSENLKEFPNLSLATNLETLSL 662
Query: 658 SNSWKLVRIPDLSKSPN-IKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEI 716
LV +P + N + + +SGC +L + P ++ LK L L L+GCS ++ P I
Sbjct: 663 GFCLSLVEVPSTIGNLNKLTYLNMSGCHNLEKFPADV-NLKSLSDLVLNGCSRLKIFPAI 721
Query: 717 KETMENLAVLVLKQTAIQELPSSLN------------------------------HL--- 743
N++ L L A++E PS+L+ HL
Sbjct: 722 S---SNISELCLNSLAVEEFPSNLHLENLVYLLIWGMTSVKLWDGVKVLTSLKTMHLRDS 778
Query: 744 VGLEE------------LSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDL 791
L+E L+L+ C + +PSSI L L +LD++ C +LETFP I
Sbjct: 779 KNLKEIPDLSMASNLLILNLEQCISIVELPSSIRNLHNLIELDMSGCTNLETFPTGINLQ 838
Query: 792 KLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDL 851
L +++L CSRLK FP I + + + + L++TAI+++P ++N L+ L + +C L
Sbjct: 839 SLKRINLARCSRLKIFPDI---STNISELDLSQTAIEEVPLWIENFSKLKYLIMGKCNML 895
Query: 852 ESLPNSICNLKLLSELDCSGC 872
E + +I LK L +D S C
Sbjct: 896 EYVFLNISKLKHLKSVDFSDC 916
Score = 99.8 bits (247), Expect = 8e-21
Identities = 86/281 (30%), Positives = 133/281 (46%), Gaps = 36/281 (12%)
Query: 649 LPNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCS 708
LPN RL + + +P + ++++ G K L +L + L+ L+ +NL G
Sbjct: 585 LPNTLRLLSWQRFPMRCMPSDFFPKYLVKLLMPGSK-LEKLWDGVMPLQCLKNMNLFGSE 643
Query: 709 NVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLT 768
N++ P +L+ LE LSL FC L +PS+IG L
Sbjct: 644 NLKEFP------------------------NLSLATNLETLSLGFCLSLVEVPSTIGNLN 679
Query: 769 KLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIK 828
KL L+++ C +LE FP + L+ L LN CSRLK FPAI + + + + L A++
Sbjct: 680 KLTYLNMSGCHNLEKFPADVNLKSLSDLVLNGCSRLKIFPAI---SSNISELCLNSLAVE 736
Query: 829 KLPSSL--QNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLS 886
+ PS+L +NLV L + L + + L L + + L IP D+ S
Sbjct: 737 EFPSNLHLENLVYLLIWGMTSV----KLWDGVKVLTSLKTMHLRDSKNLKEIP-DLSMAS 791
Query: 887 SLRTLSLQDT-GVVNLPESIAHLSSLESLDVSYCRKLECIP 926
+L L+L+ +V LP SI +L +L LD+S C LE P
Sbjct: 792 NLLILNLEQCISIVELPSSIRNLHNLIELDMSGCTNLETFP 832
>At4g16890 disease resistance RPP5 like protein
Length = 1301
Score = 447 bits (1150), Expect = e-125
Identities = 339/1003 (33%), Positives = 508/1003 (49%), Gaps = 118/1003 (11%)
Query: 23 KYDVFLSFSGEDTGKTFTSHLHAALRRKNV-FIDD---RSLETGDEICKAIEASTIYVIV 78
+YDVF SF GED +F SHL LR K + FIDD RS G E+ AI+ S I +++
Sbjct: 11 RYDVFPSFRGEDVRDSFLSHLLKELRGKAITFIDDEIERSRSIGPELLSAIKESRIAIVI 70
Query: 79 FSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRLV 138
FS+NYASS CL+ELV+I +C N ++ VIPIF+ VD S V+ QT +G F+E +
Sbjct: 71 FSKNYASSTWCLNELVEIHKCYTNLNQMVIPIFFHVDASEVKKQTGEFGKVFEE-TCKAK 129
Query: 139 PGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKLNPPFSQ--GMLGIDKH 196
Q WK+AL A ++GY SE+ +I+ +A+D+ +K P ++GI+ H
Sbjct: 130 SEDEKQSWKQALAAVAVMAGYDLRKWPSEAAMIEELAEDVLRKTMTPSDDFGDLVGIENH 189
Query: 197 IAQIQSLLQLDSEAVRI-IGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQKIDRY 255
I I+S+L L+S+ RI +GI G GIGKST+ ALY KL QF R I
Sbjct: 190 IEAIKSVLCLESKEARIMVGIWGQSGIGKSTIGRALYSKLSIQFHHRAFITYKSTSGSDV 249
Query: 256 GIYSLR--KKYLSKLLGE-DIQSNGLNYAIERVKRAKVLLILDDLKISIPILELIGGHGN 312
LR K+ LS++LG+ DI+ +R+K+ KVL++LDD+ + L+G
Sbjct: 250 SGMKLRWEKELLSEILGQKDIKIEHFGVVEQRLKQQKVLILLDDVDSLEFLKTLVGKAEW 309
Query: 313 FGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYANLIVE 372
FG GSRIIV ++ R +LK E D IYEV+ S +L + +AF +D P + + L E
Sbjct: 310 FGSGSRIIVITQDRQLLKAHEIDLIYEVEFPSEHLALTMLCRSAFGKDSPPDDFKELAFE 369
Query: 373 VLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLDDEPKDI 432
V K A +PL L VLGS L + +E W + +L+ N DI L++SYD L + +D+
Sbjct: 370 VAKLAGNLPLGLSVLGSSLKGRTKEWWMEMMPRLRNGLNGDIMKTLRVSYDRLHQKDQDM 429
Query: 433 FLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISILED-RIAVHDLVLEMGRE 491
FL I C + G V V +LL ++G +L ++ LI I D I +H+L+ ++GRE
Sbjct: 430 FLYIACLFNGFEVSYVKDLLKD-----NVGFTMLTEKSLIRITPDGYIEMHNLLEKLGRE 484
Query: 492 IVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTD---AIQCIFLDMCQIEKVQLHPEIFKS 548
I R + +PGK L N ++I+ VV + GT+ I+ F + + + E FK
Sbjct: 485 IDRAKSKGNPGKRRFLTNFEDIHEVVTEKTGTETLLGIRLPFEEYFSTRPLLIDKESFKG 544
Query: 549 MPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCPEKLVKL 608
M N++ L + +P L LP L++L WD+ P +S P F E LV L
Sbjct: 545 MRNLQYLEIGYYGD---------LPQSLVYLPLKLRLLDWDDCPLKSLPSTFKAEYLVNL 595
Query: 609 HMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELP------NLERLDLSNSWK 662
M+ S+LE+LW L SL ++ + +L +E+P NLE LDL
Sbjct: 596 IMKYSKLEKLWEGTLPLGSL-KEMNLRYSNNL------KEIPDLSLAINLEELDLVGCKS 648
Query: 663 LVRIP-DLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIK---- 717
LV +P + + + + +S CK L P +L L+ LE LNL+GC N+ N P IK
Sbjct: 649 LVTLPSSIQNATKLIYLDMSDCKKLESFPTDL-NLESLEYLNLTGCPNLRNFPAIKMGCS 707
Query: 718 ------------------------------------------ETMENLAVLVLKQTAIQE 735
E + L V K + E
Sbjct: 708 DVDFPEGRNEIVVEDCFWNKNLPAGLDYLDCLTRCMPCEFRPEQLAFLNVRGYKHEKLWE 767
Query: 736 LPSSLNHLVGL---------EELSLKFCSKLES-----------IPSSIGTLTKLCKLDL 775
SL L G+ E L +KLES +PS+IG L +L +L++
Sbjct: 768 GIQSLGSLEGMDLSESENLTEIPDLSKATKLESLILNNCKSLVTLPSTIGNLHRLVRLEM 827
Query: 776 TYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQ 835
C LE P + L LDL+ CS L++FP I + + + L TAI+++PS++
Sbjct: 828 KECTGLEVLPTDVNLSSLETLDLSGCSSLRSFPLI---STNIVWLYLENTAIEEIPSTIG 884
Query: 836 NLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQD 895
NL L L +K+C LE LP + NL L LD SGC L P S++ L L++
Sbjct: 885 NLHRLVRLEMKKCTGLEVLPTDV-NLSSLETLDLSGCSSLRSFPL---ISESIKWLYLEN 940
Query: 896 TGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAF 938
T + +P+ ++ ++L++L ++ C+ L +P +++L++F
Sbjct: 941 TAIEEIPD-LSKATNLKNLKLNNCKSLVTLPTTIGNLQKLVSF 982
Score = 148 bits (374), Expect = 2e-35
Identities = 136/464 (29%), Positives = 209/464 (44%), Gaps = 98/464 (21%)
Query: 524 DAIQCIFLDMCQIEKVQLHPEIFK----------SMPNIRMLYFHKHS----SFKQGQSN 569
+A + I+LDM +K++ P PN+R K F +G++
Sbjct: 658 NATKLIYLDMSDCKKLESFPTDLNLESLEYLNLTGCPNLRNFPAIKMGCSDVDFPEGRNE 717
Query: 570 VIVPDLL--ESLPNGLKVLHWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQS 627
++V D ++LP GL L + R P +F PE+L L++R + E+LW
Sbjct: 718 IVVEDCFWNKNLPAGLDYL---DCLTRCMPCEFRPEQLAFLNVRGYKHEKLW-------- 766
Query: 628 LCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLT 687
G+ Q L +LE +DLS S L IPDLSK+ ++ +IL+ CKSL
Sbjct: 767 --EGI--------------QSLGSLEGMDLSESENLTEIPDLSKATKLESLILNNCKSLV 810
Query: 688 RLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQ-TAIQELPSSLNHLVGL 746
LP + L L RL + C+ +E +P + +L L L ++++ P ++V L
Sbjct: 811 TLPSTIGNLHRLVRLEMKECTGLEVLPT-DVNLSSLETLDLSGCSSLRSFPLISTNIVWL 869
Query: 747 EELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKT 806
+ + +E IPS+IG L +L +L++ C LE P + L LDL+ CS L++
Sbjct: 870 YLEN----TAIEEIPSTIGNLHRLVRLEMKKCTGLEVLPTDVNLSSLETLDLSGCSSLRS 925
Query: 807 FPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSE 866
FP I +ES + L TAI+++P L L+ L L C L +LP +I NL+ L
Sbjct: 926 FPLI---SESIKWLYLENTAIEEIPD-LSKATNLKNLKLNNCKSLVTLPTTIGNLQKLVS 981
Query: 867 LDCSGCRKLTGIPNDI-----------GCLSSLRT----------LSLQDTGVVNLPESI 905
+ C L +P D+ GC SSLRT L L++T + +P +I
Sbjct: 982 FEMKECTGLEVLPIDVNLSSLMILDLSGC-SSLRTFPLISTNIVWLYLENTAIEEIPSTI 1040
Query: 906 AHL-----------------------SSLESLDVSYCRKLECIP 926
+L SSL LD+S C L P
Sbjct: 1041 GNLHRLVKLEMKECTGLEVLPTDVNLSSLMILDLSGCSSLRTFP 1084
Score = 67.4 bits (163), Expect = 5e-11
Identities = 77/264 (29%), Positives = 115/264 (43%), Gaps = 77/264 (29%)
Query: 604 KLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKL 663
+LV+L M++ ++ D L SL L L F L E +++ L L N+ +
Sbjct: 888 RLVRLEMKKCTGLEVLPTDVNLSSL-ETLDLSGCSSLRSFPLISE--SIKWLYLENT-AI 943
Query: 664 VRIPDLSKSPNIKEIILSGCKSLTRLPI---NLFKLKFLER------------------- 701
IPDLSK+ N+K + L+ CKSL LP NL KL E
Sbjct: 944 EEIPDLSKATNLKNLKLNNCKSLVTLPTTIGNLQKLVSFEMKECTGLEVLPIDVNLSSLM 1003
Query: 702 -LNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESI 760
L+LSGCS++ P I N+ L L+ TAI+E+PS++ +L L +L +K C+ LE +
Sbjct: 1004 ILDLSGCSSLRTFPLIST---NIVWLYLENTAIEEIPSTIGNLHRLVKLEMKECTGLEVL 1060
Query: 761 PSSIGTLTKLCKLDLTYCESL--------------------------------------- 781
P+ + L+ L LDL+ C SL
Sbjct: 1061 PTDVN-LSSLMILDLSGCSSLRTFPLISTRIECLYLQNTAIEEVPCCIEDFTRLTVLMMY 1119
Query: 782 -----ETFPGSIFDLKLTKLDLND 800
+T +IF +LT+L+L D
Sbjct: 1120 CCQRLKTISPNIF--RLTRLELAD 1141
Score = 51.2 bits (121), Expect = 3e-06
Identities = 34/99 (34%), Positives = 51/99 (51%), Gaps = 4/99 (4%)
Query: 683 CKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNH 742
C L LP ++ L L L+LSGCS++ P I +E L L+ TAI+E+P +
Sbjct: 1054 CTGLEVLPTDV-NLSSLMILDLSGCSSLRTFPLISTRIE---CLYLQNTAIEEVPCCIED 1109
Query: 743 LVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESL 781
L L + C +L++I +I LT+L D T C +
Sbjct: 1110 FTRLTVLMMYCCQRLKTISPNIFRLTRLELADFTDCRGV 1148
>At5g46450 disease resistance protein-like
Length = 1123
Score = 446 bits (1148), Expect = e-125
Identities = 314/911 (34%), Positives = 495/911 (53%), Gaps = 80/911 (8%)
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FID---DRSLETGDEICKAIEASTIYVIV 78
YDVF SFSGED KTF SH L RK++ F D +RS E+ +AI+ S I VIV
Sbjct: 13 YDVFPSFSGEDVRKTFLSHFLRELERKSIITFKDNEMERSQSIAPELVEAIKDSRIAVIV 72
Query: 79 FSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEH-QNRL 137
FS+NYASS CL+EL++IM C + VIP+FY +DPS +R Q+ +G+AF + QN+
Sbjct: 73 FSKNYASSSWCLNELLEIMRCNKYLGQQVIPVFYYLDPSHLRKQSGEFGEAFKKTCQNQT 132
Query: 138 VPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKLNPPFS---QGMLGID 194
V +WK+AL + +++ GYHS SE+ +I+ I+ I KL+ S + +GI
Sbjct: 133 EE--VKNQWKQALTDVSNILGYHSKNCNSEATMIEEISSHILGKLSLTPSNDFEEFVGIK 190
Query: 195 KHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNA--QQKI 252
HI +++ LL L+S+ VR++GI G GIGK+T+A AL+ L +QF S I A + +
Sbjct: 191 DHIEKVRLLLHLESDEVRMVGIWGTSGIGKTTIARALFSNLSSQFQSSVYIDRAFISKSM 250
Query: 253 DRYGIYS---------LRKKYLSKLLGEDIQSNGLNYAIERVKRAKVLLILDDLKISIPI 303
+ YG + LR+ +L ++LG+ G ER+K KVL+I+DDL +
Sbjct: 251 EGYGRANPDDYNMKLRLRENFLFEILGKKNMKIGAME--ERLKHQKVLIIIDDLDDQDVL 308
Query: 304 LELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPL 363
L+G FG GSRIIV ++++H L+ D +YE S + +LE+F AF+++ P
Sbjct: 309 DALVGRTQWFGSGSRIIVVTKNKHFLRAHGIDHVYEACLPSEELALEMFCRYAFRKNSPP 368
Query: 364 EGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYD 423
+G+ L EV A +PL L+VLGS L ++ E W + +L+ + I L++SYD
Sbjct: 369 DGFMELSSEVALRAGNLPLGLKVLGSYLRGRDIEDWMDMMPRLQNDLDGKIEKTLRVSYD 428
Query: 424 GLDDEPKD-IFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISILEDRIAVH 482
GL+++ + IF I C + G+ V+ + LL +IG+ L D+ LI + ED I +H
Sbjct: 429 GLNNKKDEAIFRHIACLFNGEKVNDIKLLLAESDLDVNIGLKNLVDKSLIFVREDTIEMH 488
Query: 483 DLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIEKVQLH 542
L+ +MG+EIVR Q ++PG+ L + K IY V+ N GT + I LD+ + + + +H
Sbjct: 489 RLLQDMGKEIVRAQS-NEPGEREFLVDSKHIYDVLEDNTGTKKVLGIALDINETDGLYIH 547
Query: 543 PEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCP 602
FK M N+ L F+ + ++ + + + LP L++L W+++P R P +F P
Sbjct: 548 ESAFKGMRNLLFLNFY---TKQKKDVTWHLSEGFDHLPPKLRLLSWEKYPLRCMPSNFRP 604
Query: 603 EKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELP------NLERLD 656
E LVKL M ES+LE+LW H L+ + L +E+P NL++LD
Sbjct: 605 ENLVKLQMCESKLEKLWDG-------VHSLTGLRNMDLRGSENLKEIPDLSLATNLKKLD 657
Query: 657 LSNSWKLVRIPDLSKSPN-IKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPE 715
+SN LV + ++ N ++E+ + C++L LPI + L+ L LNL+GCS + + P+
Sbjct: 658 VSNCTSLVELSSTIQNLNQLEELQMERCENLENLPIGI-NLESLYCLNLNGCSKLRSFPD 716
Query: 716 IKETMENLAVLVLKQTAIQELPSSLN----HLVG-------------------------- 745
I T ++ L L +TAI+E P+ L+ + +G
Sbjct: 717 ISTT---ISELYLSETAIEEFPTELHLENLYYLGLYDMKSEKLWKRVQPLTPLMTMLSPS 773
Query: 746 LEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLK 805
L +L L L +PSS L L L++ C +LET P + L +LD + CSRL+
Sbjct: 774 LTKLFLSDIPSLVELPSSFQNLHNLEHLNIARCTNLETLPTGVNLELLEQLDFSGCSRLR 833
Query: 806 TFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLS 865
+FP I + + + L T I+++P +++ L L + C +L+ + +I L+ L
Sbjct: 834 SFPDI---STNIFSLVLDGTGIEEVPWWIEDFYRLSFLSMIGCNNLQGVSLNISKLEKLE 890
Query: 866 ELDCSGCRKLT 876
+D S C L+
Sbjct: 891 TVDFSDCEALS 901
>At5g46260 disease resistance protein-like
Length = 1205
Score = 437 bits (1124), Expect = e-122
Identities = 311/971 (32%), Positives = 491/971 (50%), Gaps = 117/971 (12%)
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FID---DRSLETGDEICKAIEASTIYVIV 78
YDVFLSF G D TF SH RK + F D +RS ++ +AI+ S I V+V
Sbjct: 12 YDVFLSFRGGDVRVTFRSHFLKEFDRKLITAFRDNEIERSHSLWPDLEQAIKESRIAVVV 71
Query: 79 FSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRLV 138
FS+NYASS CL+EL++I+ C D+ +IP+FY VDPS VR+Q +G F++ R
Sbjct: 72 FSKNYASSSWCLNELLEIVNCN---DKIIIPVFYGVDPSQVRYQIGEFGKIFEKTCKRQT 128
Query: 139 PGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKLNPPFS---QGMLGIDK 195
V Q WKKAL A++ G+ S E+ +I+ IA D+ +KL S + +G++
Sbjct: 129 EEVKNQ-WKKALTHVANMLGFDSSKWDDEAKMIEEIANDVLRKLLLTTSKDFEDFVGLED 187
Query: 196 HIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLI---------- 245
HIA + +LL L+S+ V+++GI G GIGK+T+A AL++ L F R I
Sbjct: 188 HIANMSALLDLESKEVKMVGIWGSSGIGKTTIARALFNNLFRHFQVRKFIDRSFAYKSRE 247
Query: 246 VNAQQKIDRYGI-YSLRKKYLSKLLG-EDIQSNGLNYAIERVKRAKVLLILDDLKISIPI 303
+++ D + + L++ +LS++L +I+ + L ER++ KVL+I+DD+ + +
Sbjct: 248 IHSSANPDDHNMKLHLQESFLSEILRMPNIKIDHLGVLGERLQHQKVLIIIDDVDDQVIL 307
Query: 304 LELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPL 363
L+G FG GSRIIV + ++H L D +YEV + + +L + +AFK+ P
Sbjct: 308 DSLVGKTQWFGNGSRIIVVTNNKHFLTAHGIDRMYEVSLPTEEHALAMLCQSAFKKKSPP 367
Query: 364 EGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYD 423
EG+ L+V+V +YA +PL L+VLGS L K++E W L +L+ N I +L++SYD
Sbjct: 368 EGFEMLVVQVARYAGSLPLVLKVLGSYLSGKDKEYWIDMLPRLQNGLNDKIERILRISYD 427
Query: 424 GLDDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISILEDRIAVHD 483
GL+ E + IF I C + V + LL + + A++G+ L D+ +I + + +H
Sbjct: 428 GLESEDQAIFRHIACIFNHMEVTTIKSLLANSIYGANVGLQNLVDKSIIHVRWGHVEMHP 487
Query: 484 LVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIEKVQLHP 543
L+ EMGR+IVR Q + P K L + +I V+ + T + I L+ +I+++ +H
Sbjct: 488 LLQEMGRKIVRTQSIGKPRKREFLVDPNDICDVLSEGIDTQKVLGISLETSKIDELCVHE 547
Query: 544 EIFKSMPNIRMLYFHKHSSFKQGQSNVI-VPDLLESLPNGLKVLHWDEFPQRSFPLDFCP 602
FK M N+R L K + G+ N + +P+ + LP LK+L W EFP R P +FCP
Sbjct: 548 SAFKRMRNLRFL---KIGTDIFGEENRLHLPESFDYLPPTLKLLCWSEFPMRCMPSNFCP 604
Query: 603 EKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWK 662
+ LV L M S+L +LW + + C L+ +DL S
Sbjct: 605 KNLVTLKMTNSKLHKLW--EGAVPLTC----------------------LKEMDLDGSVN 640
Query: 663 LVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIP-------- 714
L IPDLS + N++ + CKSL LP + L L +LN++ C+++E +P
Sbjct: 641 LKEIPDLSMATNLETLNFENCKSLVELPSFIQNLNKLLKLNMAFCNSLETLPTGFNLKSL 700
Query: 715 ------------EIKETMENLAVLVLKQTAIQELPSSLN--HLVGLE------------- 747
+ N++ L L T I+ELPS+L+ +L+ L
Sbjct: 701 NRIDFTKCSKLRTFPDFSTNISDLYLTGTNIEELPSNLHLENLIDLRISKKEIDGKQWEG 760
Query: 748 -----------------ELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFD 790
L L+ L +P S L +L LD+T C +LET P I
Sbjct: 761 VMKPLKPLLAMLSPTLTSLQLQNIPNLVELPCSFQNLIQLEVLDITNCRNLETLPTGINL 820
Query: 791 LKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPD 850
L L CSRL++FP I + + + ++L ET I+++P + L L + C
Sbjct: 821 QSLDSLSFKGCSRLRSFPEI---STNISSLNLEETGIEEVPWWIDKFSNLGLLSMDRCSR 877
Query: 851 LESLPNSICNLKLLSELDCSGCRKLT---------GIPNDIGCLSSLRTLSLQDTGVVNL 901
L+ + I LK L ++D C LT G+ + + ++ + L NL
Sbjct: 878 LKCVSLHISKLKRLGKVDFKDCGALTIVDLCGCPIGMEMEANNIDTVSKVKLDFRDCFNL 937
Query: 902 -PESIAHLSSL 911
PE++ H S+
Sbjct: 938 DPETVLHQESI 948
Score = 80.9 bits (198), Expect = 4e-15
Identities = 66/197 (33%), Positives = 102/197 (51%), Gaps = 25/197 (12%)
Query: 733 IQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLK 792
++E+P L+ LE L+ + C L +PS I L KL KL++ +C SLET P F+LK
Sbjct: 641 LKEIPD-LSMATNLETLNFENCKSLVELPSFIQNLNKLLKLNMAFCNSLETLPTG-FNLK 698
Query: 793 -LTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDL 851
L ++D CS+L+TFP + + + + L T I++LPS+L L
Sbjct: 699 SLNRIDFTKCSKLRTFPDF---STNISDLYLTGTNIEELPSNLH---------------L 740
Query: 852 ESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLS-SLRTLSLQDT-GVVNLPESIAHLS 909
E+L + + K + G K + + LS +L +L LQ+ +V LP S +L
Sbjct: 741 ENLIDLRISKKEIDGKQWEGVMK--PLKPLLAMLSPTLTSLQLQNIPNLVELPCSFQNLI 798
Query: 910 SLESLDVSYCRKLECIP 926
LE LD++ CR LE +P
Sbjct: 799 QLEVLDITNCRNLETLP 815
>At3g25510 unknown protein
Length = 1981
Score = 435 bits (1119), Expect = e-122
Identities = 309/962 (32%), Positives = 492/962 (51%), Gaps = 85/962 (8%)
Query: 3 ESSSSSSAAAAAMAPPPNPL----KYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FID- 55
++ SA+ + + PP+ L K+ VF SF G D KTF SH+ A R K + FID
Sbjct: 28 DNREDDSASLSPSSVPPSSLSRTWKHQVFPSFHGADVRKTFLSHVLEAFRGKGIDPFIDN 87
Query: 56 --DRSLETGDEICKAIEASTIYVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYR 113
+RS G E+ +AI S I +++ S NYASS C++ELV+IM+CK + + VI IFY
Sbjct: 88 SIERSKSIGPELVEAIRGSRIAIVLLSRNYASSSWCMNELVEIMKCKEDLGQIVITIFYE 147
Query: 114 VDPSTVRHQTNSYGDAFDEHQNRLVPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDR 173
VDP+ ++ QT +G F E + +RW+KAL A ++GYHS
Sbjct: 148 VDPTHIKKQTGDFGKVFKETCKGKTKEEI-KRWRKALEGVATIAGYHSS----------- 195
Query: 174 IAQDIFKKLNPPFSQGMLGIDKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYH 233
N F + ++G+ H+ +++LL+LD + VR+IGI G GIGK+T+A L
Sbjct: 196 ---------NWDF-EALIGMGAHMENMRALLRLDLDDVRMIGIWGPPGIGKTTIARFLLS 245
Query: 234 KLGTQFSSRCLIVNAQQK-----IDRYGIY-SLRKKYLSKLLGE-DIQSNGLNYAIERVK 286
++ F ++VN ++ +D Y + L+ K LSK++ + DI L A ER+K
Sbjct: 246 QVSKSFQLSTIMVNIKECYPSPCLDEYSVQLQLQNKMLSKMINQKDIMIPHLGVAQERLK 305
Query: 287 RAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQ 346
KV L+LDD+ + L FG GSRII+T+ + +L + IY+V+ S
Sbjct: 306 DKKVFLVLDDVDQLGQLDALAKETRWFGPGSRIIITTENLRLLMAHRINHIYKVEFSSTD 365
Query: 347 DSLELFRLNAFKEDYPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKL 406
++ ++F ++AF + +P G+ L EV + A G+PL L+V+GS L ++ W+ L +L
Sbjct: 366 EAFQIFCMHAFGQKHPYNGFYELSREVTELAGGLPLGLKVMGSSLRGMSKQEWKRTLPRL 425
Query: 407 KELPNVDIFDVLKLSYDGLDDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVL 466
+ + I +L SY+ L E KD+FL I CF+ + KV + L G+ VL
Sbjct: 426 RTCLDGKIESILMFSYEALSHEDKDLFLCIACFFNYQKIKKVEKHLADRFLDVRQGLYVL 485
Query: 467 KDRGLISILEDRIAVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAI 526
++ LI I +H L++++GREI Q +DP K L + +EI + +
Sbjct: 486 AEKSLIHIGTGATEMHTLLVQLGREIAHTQSTNDPRKSLFLVDEREICEALSDETMDSSR 545
Query: 527 QCIFLDMCQI----EKVQLHPEIFKSMPNIRMLYFH-----KHSS----FKQGQSNVIVP 573
+ I +D E + + + M N++ + F +HSS + +N P
Sbjct: 546 RIIGMDFDLSKNGEEVTNISEKGLQRMSNLQFIRFDGRSCARHSSNLTVVRSSDNNCAHP 605
Query: 574 DLLESLPN------GLKVLHWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQS 627
D + +L + +++LHW F + P F PE LV+L+M S LW + L+
Sbjct: 606 DTVNALQDLNYQFQEIRLLHWINFRRLCLPSTFNPEFLVELNMPSSTCHTLWEGSKALR- 664
Query: 628 LCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLT 687
NL+ +DLS S L +PDLS + N++E+IL C SL
Sbjct: 665 -----------------------NLKWMDLSYSISLKELPDLSTATNLEELILKYCVSLV 701
Query: 688 RLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQ-TAIQELPSSLNHLVGL 746
++P + KL L+ L L GC+++ +P + + L L L + +++ ELPSS+ + + L
Sbjct: 702 KVPSCVGKLGKLQVLCLHGCTSILELPSFTKNVTGLQSLDLNECSSLVELPSSIGNAINL 761
Query: 747 EELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKT 806
+ L L C +L +P SI T L K L C SL P L LDL +CS L
Sbjct: 762 QNLDLG-CLRLLKLPLSIVKFTNLKKFILNGCSSLVELPFMGNATNLQNLDLGNCSSLVE 820
Query: 807 FPAILEPAESFAHISLAE-TAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLS 865
P+ + A + ++ L+ +++ KLPS + N L+ L L++C L +P SI ++ L
Sbjct: 821 LPSSIGNAINLQNLDLSNCSSLVKLPSFIGNATNLEILDLRKCSSLVEIPTSIGHVTNLW 880
Query: 866 ELDCSGCRKLTGIPNDIGCLSSLRTLSLQD-TGVVNLPESIAHLSSLESLDVSYCRKLEC 924
LD SGC L +P+ +G +S L+ L+L + + +V LP S H ++L LD+S C L
Sbjct: 881 RLDLSGCSSLVELPSSVGNISELQVLNLHNCSNLVKLPSSFGHATNLWRLDLSGCSSLVE 940
Query: 925 IP 926
+P
Sbjct: 941 LP 942
Score = 327 bits (838), Expect = 2e-89
Identities = 225/759 (29%), Positives = 392/759 (51%), Gaps = 80/759 (10%)
Query: 38 TFTSHLHAALRRKNVF-IDDRSLETGD----EICKAIEASTIYVIVFSENYASSRCCLDE 92
+F L +RK + +D ++ G+ E+ AI S I +I+ S NYASS CLDE
Sbjct: 1264 SFNEALMKEFQRKGITPFNDNEIKRGESISPELVLAIRGSRIALILLSRNYASSSWCLDE 1323
Query: 93 LVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRLVPGVVMQ---RWKKA 149
L +I++C+ + + V+ +FY+VDPS ++ T +G F + G + RW +A
Sbjct: 1324 LAEIIKCREEFGQTVMVVFYKVDPSDIKKLTGDFGSVF----RKTCAGKTNEDTRRWIQA 1379
Query: 150 LLEAADLSGYHSIAARSESMLIDRIAQDIFKKLNPPFSQG----MLGIDKHIAQIQSLLQ 205
L + A L+GY S +E+++I++IA DI KLN ++G+ H+ +++ LL
Sbjct: 1380 LAKVATLAGYVSNNWDNEAVMIEKIATDISNKLNKSTPSRDFDELVGMGAHMERMELLLC 1439
Query: 206 LDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQKIDRYGIYS------ 259
LDS+ VR+IGI G GIGK+T+A L+ + F + N ++ + R + S
Sbjct: 1440 LDSDEVRMIGIWGPSGIGKTTIARFLFSQFSDSFELSAFMENIKELMYRKPVCSDDYSAK 1499
Query: 260 --LRKKYLSKLLGE-DIQSNGLNYAIERVKRAKVLLILDDLKISIPILELIGGHGNFGQG 316
L+ +++S+++ D++ L R+ KVL++LD++ S+ + + FG G
Sbjct: 1500 LHLQNQFMSQIINHMDVEVPHLGVVENRLNDKKVLIVLDNIDQSMQLDAIAKETRWFGHG 1559
Query: 317 SRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYANLIVEVLKY 376
SRII+T++ + +LK + IY+V S ++ ++F ++A + +P + + L +EV
Sbjct: 1560 SRIIITTQDQKLLKAHGINHIYKVDYPSTHEACQIFCMSAVGKKFPKDEFQELALEVTNL 1619
Query: 377 AKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLDDEPKDIFLDI 436
+PL L+V+GS ++ W + L +L+ + +I +LK SYD L E KD+FL I
Sbjct: 1620 LGNLPLGLRVMGSHFRGMSKQEWINALPRLRTHLDSNIQSILKFSYDALCREDKDLFLHI 1679
Query: 437 TCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISILEDRIAVHDLVLEMGREIV--R 494
C + ++ V L VL ++ LISI E I +H+L+ +GREIV
Sbjct: 1680 ACTFNNKRIENVEAHLTHKFLDTKQRFHVLAEKSLISIEEGWIKMHNLLELLGREIVCHE 1739
Query: 495 QQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQ-IEKVQLHPEIFKSMPNIR 553
+ + +PGK L + ++I V+ + G+ ++ I+ + + + ++ + F+ M N++
Sbjct: 1740 HESIREPGKRQFLVDARDICEVLTDDTGSKSVVGIYFNSAELLGELNISERAFEGMSNLK 1799
Query: 554 MLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCPEKLVKLHMRES 613
L S K + +P L+ + L++L WD FP P +FC E LV+L+MR S
Sbjct: 1800 FLRIKCDRSDK-----MYLPRGLKYISRKLRLLEWDRFPLTCLPSNFCTEYLVELNMRHS 1854
Query: 614 RLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIPDLSKSP 673
+L +LW G+L L NL+ ++L +S L +PD S +
Sbjct: 1855 KLVKLWE-----------------GNL-------SLGNLKWMNLFHSKNLKELPDFSTAT 1890
Query: 674 NIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAI 733
N++ +IL GC SL LP ++ L++L+L C T++
Sbjct: 1891 NLQTLILCGCSSLVELPYSIGSANNLQKLHLCRC-----------------------TSL 1927
Query: 734 QELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCK 772
ELP+S+ +L L+ ++LK CSKLE +P++I + + K
Sbjct: 1928 VELPASIGNLHKLQNVTLKGCSKLEVVPTNINLILDVKK 1966
Score = 124 bits (311), Expect = 3e-28
Identities = 117/396 (29%), Positives = 176/396 (43%), Gaps = 56/396 (14%)
Query: 651 NLERLDLSNSWKLVRIP-DLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSN 709
NLE LDL LV IP + N+ + LSGC SL LP ++ + L+ LNL CSN
Sbjct: 854 NLEILDLRKCSSLVEIPTSIGHVTNLWRLDLSGCSSLVELPSSVGNISELQVLNLHNCSN 913
Query: 710 VENIPEIKETMENLAVLVLKQ-TAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLT 768
+ +P NL L L +++ ELPSS+ ++ L+EL+L CS L +PSSIG L
Sbjct: 914 LVKLPSSFGHATNLWRLDLSGCSSLVELPSSIGNITNLQELNLCNCSNLVKLPSSIGNLH 973
Query: 769 KLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIK 828
L L L C+ LE P +I L +LDL DCS+ K+FP I E + L TA++
Sbjct: 974 LLFTLSLARCQKLEALPSNINLKSLERLDLTDCSQFKSFPEISTNIEC---LYLDGTAVE 1030
Query: 829 KLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSL 888
++PSS+++ L L + L+ + L +++ L+
Sbjct: 1031 EVPSSIKSWSRLTVLHMSYFEKLKEFSHV---LDIITWLEFG------------------ 1069
Query: 889 RTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAFHCPSIRSV-- 946
+ + I +S L L + CRKL +P LP + + A C S+ ++
Sbjct: 1070 -------EDIQEVAPWIKEISRLHGLRLYKCRKLLSLPQLPESLSIINAEGCESLETLDC 1122
Query: 947 MSNSRFLSDSKEGTFELYFTNNEKQDPGAQRDVVRNAEHRITDVACRFAFYCFPGSEVPN 1006
N+ + F+L N E +D Q +A PG+EVP
Sbjct: 1123 SYNNPLSLLNFAKCFKL---NQEARDFIIQIPTSNDA--------------VLPGAEVPA 1165
Query: 1007 WFHHHCKRN-SVTVDKDSLNLSNDNRIIGFAMCFVL 1041
+F H S+T+ + +S R F C VL
Sbjct: 1166 YFTHRATTGASLTIKLNERPISTSMR---FKACIVL 1198
Score = 58.2 bits (139), Expect = 3e-08
Identities = 68/278 (24%), Positives = 109/278 (38%), Gaps = 44/278 (15%)
Query: 668 DLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLA--- 724
D S N +E+ K L R+ L+F+ S + N+ ++ + N A
Sbjct: 551 DFDLSKNGEEVTNISEKGLQRMS----NLQFIRFDGRSCARHSSNLTVVRSSDNNCAHPD 606
Query: 725 -VLVLKQTAIQELPSSLNHLVGLEELSL------KFCSKLESIPSSIGTLTKLCK----- 772
V L+ Q L H + L L +F +L S+ TL + K
Sbjct: 607 TVNALQDLNYQFQEIRLLHWINFRRLCLPSTFNPEFLVELNMPSSTCHTLWEGSKALRNL 666
Query: 773 --LDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKL 830
+DL+Y SL+ P L +L L C ++ K+
Sbjct: 667 KWMDLSYSISLKELPDLSTATNLEELILKYC-----------------------VSLVKV 703
Query: 831 PSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRT 890
PS + L LQ LCL C + LP+ N+ L LD + C L +P+ IG +L+
Sbjct: 704 PSCVGKLGKLQVLCLHGCTSILELPSFTKNVTGLQSLDLNECSSLVELPSSIGNAINLQN 763
Query: 891 LSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHL 928
L L ++ LP SI ++L+ ++ C L +P +
Sbjct: 764 LDLGCLRLLKLPLSIVKFTNLKKFILNGCSSLVELPFM 801
Score = 48.1 bits (113), Expect = 3e-05
Identities = 30/83 (36%), Positives = 43/83 (51%), Gaps = 8/83 (9%)
Query: 845 LKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSL-QDTGVVNLPE 903
LKE PD + N L L GC L +P IG ++L+ L L + T +V LP
Sbjct: 1880 LKELPDFSTATN-------LQTLILCGCSSLVELPYSIGSANNLQKLHLCRCTSLVELPA 1932
Query: 904 SIAHLSSLESLDVSYCRKLECIP 926
SI +L L+++ + C KLE +P
Sbjct: 1933 SIGNLHKLQNVTLKGCSKLEVVP 1955
Score = 46.6 bits (109), Expect = 8e-05
Identities = 54/211 (25%), Positives = 90/211 (42%), Gaps = 13/211 (6%)
Query: 681 SGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLK--QTAIQELPS 738
+G KS+ + N +L L LN+S E M NL L +K ++ LP
Sbjct: 1766 TGSKSVVGIYFNSAEL--LGELNIS--------ERAFEGMSNLKFLRIKCDRSDKMYLPR 1815
Query: 739 SLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDL 798
L ++ L L +PS+ T L +L++ + + ++ + G++ L ++L
Sbjct: 1816 GLKYISRKLRLLEWDRFPLTCLPSNFCT-EYLVELNMRHSKLVKLWEGNLSLGNLKWMNL 1874
Query: 799 NDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSI 858
LK P I +++ +LP S+ + LQ L L C L LP SI
Sbjct: 1875 FHSKNLKELPDFSTATNLQTLILCGCSSLVELPYSIGSANNLQKLHLCRCTSLVELPASI 1934
Query: 859 CNLKLLSELDCSGCRKLTGIPNDIGCLSSLR 889
NL L + GC KL +P +I + ++
Sbjct: 1935 GNLHKLQNVTLKGCSKLEVVPTNINLILDVK 1965
>At5g46470 disease resistance protein-like
Length = 1160
Score = 431 bits (1107), Expect = e-120
Identities = 319/945 (33%), Positives = 486/945 (50%), Gaps = 118/945 (12%)
Query: 1 MSESSSSSSAAAAAMAPPPNPLKYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FID--- 55
M+ SSSSSS Y VF SFSGED TF SH L RK + F D
Sbjct: 1 MASSSSSSS----------RNWSYHVFPSFSGEDVRNTFLSHFLKELDRKLIISFKDNEI 50
Query: 56 DRSLETGDEICKAIEASTIYVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVD 115
+RS E+ I S I V+VFS+ YASS CL+EL++I++CK + + VIPIFY +D
Sbjct: 51 ERSQSLDPELKHGIRNSRIAVVVFSKTYASSSWCLNELLEIVKCKKEFGQLVIPIFYNLD 110
Query: 116 PSTVRHQTNSYGDAFDEH-QNRLVPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRI 174
PS VR QT +G F++ +N+ V + RWK+AL + A++ GYH + +E+ +I+ I
Sbjct: 111 PSHVRKQTGDFGKIFEKTCRNKTVDEKI--RWKEALTDVANILGYHIVTWDNEASMIEEI 168
Query: 175 AQDIFKKLNPPFS---QGMLGIDKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEAL 231
A DI K+N S + ++GI+ HI ++ SLL L+SE VR++GI G GIGK+T+A AL
Sbjct: 169 ANDILGKMNISPSNDFEDLVGIEDHITKMSSLLHLESEEVRMVGIWGPSGIGKTTIARAL 228
Query: 232 YHKLGTQFSSRCLI-----------VNAQQKIDRYGIYSLRKKYLSKLLGEDIQSNGLNY 280
+ +L QF S I + +D L++ +L+++ + +
Sbjct: 229 FSRLSCQFQSSVFIDKVFISKSMEVYSGANLVDYNMKLHLQRAFLAEIFDKKDIKIHVGA 288
Query: 281 AIERVKRAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEV 340
+ VK K L+++DDL + L FG GSRIIV + ++H L+ D IY+V
Sbjct: 289 MEKMVKHRKALIVIDDLDDQDVLDALADQTQWFGSGSRIIVVTENKHFLRANRIDHIYKV 348
Query: 341 QGMSYQDSLELFRLNAFKEDYPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWE 400
S +LE+F +AFK++ P + + L EV A +PL L VLGS L + W
Sbjct: 349 CLPSNALALEMFCRSAFKKNSPPDDFLELSSEVALRAGNLPLGLNVLGSNLRGINKGYWI 408
Query: 401 SELKKLKELPNVDIFDVLKLSYDGLDDEPKD-IFLDITCFYAGDFVDKVVELLDSCGFSA 459
L +L+ L I L++SYDGL++ + IF I C + G+ V + LL +
Sbjct: 409 DMLPRLQGLDG-KIGKTLRVSYDGLNNRKDEAIFRHIACIFNGEKVSDIKLLLANSNLDV 467
Query: 460 DIGMDVLKDRGLISILEDRIAVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRK 519
+IG+ L DR LI + + +H L+ E+G+EIVR Q + PG+ L + K+I V+
Sbjct: 468 NIGLKNLVDRSLICERFNTLEMHSLLQELGKEIVRTQS-NQPGEREFLVDLKDICDVLEH 526
Query: 520 NKGTDAIQCIFLDMCQIEKVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESL 579
N GT + I LD+ + +++ +H FK M N+ L + ++ + +P+ + L
Sbjct: 527 NTGTKKVLGITLDIDETDELHIHESSFKGMHNLLFLKIYTKKLDQKKKVRWHLPERFDYL 586
Query: 580 PNGLKVLHWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGH 639
P+ L++L +D +P + P +F PE LVKL M++S+LE+LW D V
Sbjct: 587 PSRLRLLRFDRYPSKCLPSNFHPENLVKLQMQQSKLEKLW--DGV--------------- 629
Query: 640 LIVFSLHQELPNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINL------ 693
L L +DL S L IPDLS + N++ + LS C SL LP ++
Sbjct: 630 -------HSLAGLRNMDLRGSRNLKEIPDLSMATNLETLKLSSCSSLVELPSSIQYLNKL 682
Query: 694 -----------------FKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQEL 736
LK L+RLNLSGCS +++ +I N++ L + QTA ++
Sbjct: 683 NDLDMSYCDHLETIPSGVNLKSLDRLNLSGCSRLKSFLDIP---TNISWLDIGQTA--DI 737
Query: 737 PSSLNHLVGLEELSLKFCSKLE-------------------------SIPSSIGTLTKLC 771
PS+L L L+EL L C +++ +PSSI L +L
Sbjct: 738 PSNL-RLQNLDELIL--CERVQLRTPLMTMLSPTLTRLTFSNNPSFVEVPSSIQNLYQLE 794
Query: 772 KLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLP 831
L++ C +L T P I L LDL+ CS+LKTFP I + + + ++L+ TAI+++P
Sbjct: 795 HLEIMNCRNLVTLPTGINLDSLISLDLSHCSQLKTFPDI---STNISDLNLSYTAIEEVP 851
Query: 832 SSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLT 876
S++ L L L + C +L + +I LK L D S C +LT
Sbjct: 852 LSIEKLSLLCYLDMNGCSNLLCVSPNISKLKHLERADFSDCVELT 896
Score = 43.1 bits (100), Expect = 0.001
Identities = 33/106 (31%), Positives = 54/106 (50%), Gaps = 6/106 (5%)
Query: 827 IKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLS 886
+K++P L L+TL L C L LP+SI L L++LD S C L IP+ + L
Sbjct: 646 LKEIPD-LSMATNLETLKLSSCSSLVELPSSIQYLNKLNDLDMSYCDHLETIPSGVN-LK 703
Query: 887 SLRTLSLQD----TGVVNLPESIAHLSSLESLDVSYCRKLECIPHL 928
SL L+L +++P +I+ L ++ D+ +L+ + L
Sbjct: 704 SLDRLNLSGCSRLKSFLDIPTNISWLDIGQTADIPSNLRLQNLDEL 749
>At4g11170 RPP1-WsA-like disease resistance protein
Length = 1174
Score = 427 bits (1097), Expect = e-119
Identities = 293/818 (35%), Positives = 455/818 (54%), Gaps = 61/818 (7%)
Query: 20 NPLKYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDD---RSLETGDEICKAIEASTI 74
N +YDVF SF GED F SHL K + F DD RS G E+ AI S I
Sbjct: 7 NSWRYDVFPSFRGEDVRNNFLSHLLKEFESKGIVTFRDDHIKRSHTIGHELRAAIRESKI 66
Query: 75 YVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQ 134
V++FSENYASS CLDEL++IM+CK V+P+FY+VDPS +R QT +G +F E
Sbjct: 67 SVVLFSENYASSSWCLDELIEIMKCKEEQGLKVMPVFYKVDPSDIRKQTGKFGMSFLE-- 124
Query: 135 NRLVPGVVMQR---WKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKLNPPFSQG-- 189
G +R W++AL +AA++ G H +E+ I I++D+ +KLN S+
Sbjct: 125 --TCCGKTEERQHNWRRALTDAANILGDHPQNWDNEAYKITTISKDVLEKLNATPSRDFN 182
Query: 190 -MLGIDKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNA 248
++G++ HIA+++SLL L+S+ VRI+GI G G+GK+T+A ALY++ F+ + N
Sbjct: 183 DLVGMEAHIAKMESLLCLESQGVRIVGIWGPAGVGKTTIARALYNQYHENFNLSIFMENV 242
Query: 249 QQK-----IDRYGI-YSLRKKYLSKLLGE-DIQSNGLNYAIERVKRAKVLLILDDLKISI 301
++ +D YG+ L++++LSKLL + D++ L ER+K KVL+ILDD+
Sbjct: 243 RESYGEAGLDDYGLKLHLQQRFLSKLLDQKDLRVRHLGAIEERLKSQKVLIILDDVDNIE 302
Query: 302 PILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDY 361
+ L + FG SRI+VT++++ +L + + + +Y+V S Q++L +F +AFK+
Sbjct: 303 QLKALAKENQWFGNKSRIVVTTQNKQLLVSHDINHMYQVAYPSKQEALTIFCQHAFKQSS 362
Query: 362 PLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLS 421
P + +L +E A +PLAL+VLGS + K +E WE L LK + ++ VLK+
Sbjct: 363 PSDDLKHLAIEFTTLAGHLPLALRVLGSFMRGKGKEEWEFSLPTLKSRLDGEVEKVLKVG 422
Query: 422 YDGLDDEPKDIFLDITCFYAGDFVDKVVELLDSCGFS-ADIGMDVLKDRGLISILED-RI 479
YDGL D KD+FL I C ++G + + +++ + + G+ VL D+ LI E+ RI
Sbjct: 423 YDGLHDHEKDLFLHIACIFSGQHENYLKQMIIANNDTYVSFGLQVLADKSLIQKFENGRI 482
Query: 480 AVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQI-EK 538
+H L+ ++G+E+VR+Q + +PGK L N KE V+ N GT + I LDMC+I E+
Sbjct: 483 EMHSLLRQLGKEVVRKQSIYEPGKRQFLMNAKETCGVLSNNTGTGTVLGISLDMCEIKEE 542
Query: 539 VQLHPEIFKSMPNIRMLYFHKHSSF--KQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSF 596
+ + + F+ M N+ L F+ S K + + L LP L++LHWD +P F
Sbjct: 543 LYISEKTFEEMRNLVYLKFYMSSPIDDKMKVKLQLPEEGLSYLPQ-LRLLHWDAYPLEFF 601
Query: 597 PLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLD 656
P F PE LV+L+M S+L++LW Q L++L ++ +L + E L RLD
Sbjct: 602 PSSFRPECLVELNMSHSKLKKLWSGVQPLRNL-RTMNLNSSRNLEILPNLMEATKLNRLD 660
Query: 657 LSNSWKLVRIPDLSKSPNIKEIIL---SGCKSLTRLPINLFKLKFLERLNLSGCSNVENI 713
L LV +P S N++ +IL S CK L +P N+ L LE L+ C+ ++
Sbjct: 661 LGWCESLVELP--SSIKNLQHLILLEMSCCKKLEIIPTNI-NLPSLEVLHFRYCTRLQTF 717
Query: 714 PEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSL----------------KFC--- 754
PEI N+ +L L TAI E+P S+ + ++E+ + K C
Sbjct: 718 PEIS---TNIRLLNLIGTAITEVPPSVKYWSKIDEICMERAKVKRLVHVPYVLEKLCLRE 774
Query: 755 -SKLESIPSSIGTLTKLCKLDLTYC---ESLETFPGSI 788
+LE+IP + L +L +D++YC SL PGS+
Sbjct: 775 NKELETIPRYLKYLPRLQMIDISYCINIISLPKLPGSV 812
Score = 97.8 bits (242), Expect = 3e-20
Identities = 108/452 (23%), Positives = 188/452 (40%), Gaps = 68/452 (15%)
Query: 694 FKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKF 753
F+ + L LN+S S ++ + + + NL + L + E+ +L L L L +
Sbjct: 605 FRPECLVELNMSH-SKLKKLWSGVQPLRNLRTMNLNSSRNLEILPNLMEATKLNRLDLGW 663
Query: 754 CSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEP 813
C L +PSSI L L L+++ C+ LE P +I L L C+RL+TFP I
Sbjct: 664 CESLVELPSSIKNLQHLILLEMSCCKKLEIIPTNINLPSLEVLHFRYCTRLQTFPEI--- 720
Query: 814 AESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCR 873
+ + ++L TAI ++P S++ + +C++ +
Sbjct: 721 STNIRLLNLIGTAITEVPPSVKYWSKIDEICMERAK----------------------VK 758
Query: 874 KLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMK 933
+L +P + L L+ +P + +L L+ +D+SYC + +P LP +
Sbjct: 759 RLVHVPYVLEKLCLRENKELE-----TIPRYLKYLPRLQMIDISYCINIISLPKLPGSVS 813
Query: 934 QLLAFHCPSIRSVMSNSRFLSDSKEGTFELYFTNNEKQDPGAQRDVVRNAE-HRITDVAC 992
L A +C S++ + + R + L F N K AQ + R+ H+ + +A
Sbjct: 814 ALTAVNCESLQILHGHFR------NKSIHLNFINCLKLGQRAQEKIHRSVYIHQSSYIA- 866
Query: 993 RFAFYCFPGSEVPNWFHHHCKRNSVTVDKDSLNLSNDNRIIGFAMCFVLQLEDMNYTGYR 1052
PG VP +F + +S+ + + ++LS NR F +C VL G R
Sbjct: 867 ----DVLPGEHVPAYFSYRSTGSSIMIHSNKVDLSKFNR---FKVCLVLG------AGKR 913
Query: 1053 FSNFRCRLTFESDGQTHILPIDDQLNNSLHQIDLRVGHLVRDHTFIWNYYLM-DSASNHN 1111
F + + + + L++ L L DH + + LM
Sbjct: 914 FEGCDIKFYKQFFCKPREYYVPKHLDSPL---------LKSDHLCMCEFELMPPHPPTEW 964
Query: 1112 GIIHAHNF---TFEISNGLYLKSSSLVKECGI 1140
++H + F +FE GLY VKECG+
Sbjct: 965 ELLHPNEFLEVSFESRGGLY---KCEVKECGL 993
>At3g04220 putative disease resistance protein
Length = 896
Score = 423 bits (1088), Expect = e-118
Identities = 291/916 (31%), Positives = 475/916 (51%), Gaps = 101/916 (11%)
Query: 2 SESSSSSSAAAAAMAPPPNPL----KYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FID 55
++ + SSS+ + +PP + L KYDVF SF GED K F SH+ +R+ + F+D
Sbjct: 37 NKENDSSSSTQPSPSPPLSSLSLTRKYDVFPSFRGEDVRKDFLSHIQKEFQRQGITPFVD 96
Query: 56 D---RSLETGDEICKAIEASTIYVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFY 112
+ R G E+ +AI S I +I+ S+NYASS CLDELV+I++CK + VI IFY
Sbjct: 97 NNIKRGESIGPELIRAIRGSKIAIILLSKNYASSSWCLDELVEIIKCKEEMGQTVIVIFY 156
Query: 113 RVDPSTVRHQTNSYGDAF------DEHQNRLVPGVVMQRWKKALLEAADLSGYHSIAARS 166
+VDPS V+ T +G F E +N ++RW++A + A ++GY S +
Sbjct: 157 KVDPSLVKKLTGDFGKVFRNTCKGKEREN-------IERWREAFKKVATIAGYDSRKWDN 209
Query: 167 ESMLIDRIAQDIFKKLNPPFSQ----GMLGIDKHIAQIQSLLQLDSEAVRIIGICGMGGI 222
ES +I++I DI + LN ++G+ H+ +++ LL +DS+ ++ IGI G G+
Sbjct: 210 ESGMIEKIVSDISEMLNHSTPSRDFDDLIGMGDHMEKMKPLLDIDSDEMKTIGIWGPPGV 269
Query: 223 GKSTLAEALYHKLGTQFSSRCLIVNAQQKI-------DRYGIYSLRKKYLSKLLG-EDIQ 274
GK+T+A +LY++ +F + + + D Y L++++LS++ E++Q
Sbjct: 270 GKTTIARSLYNQHSDKFQLSVFMESIKTAYTIPACSDDYYEKLQLQQRFLSQITNQENVQ 329
Query: 275 SNGLNYAIERVKRAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEA 334
L A ER+ KVL+++DD+ S+ + L + G GSRII+T++ R +L+
Sbjct: 330 IPHLGVAQERLNDKKVLVVIDDVNQSVQVDALAKENDWLGPGSRIIITTQDRGILRAHGI 389
Query: 335 DEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDK 394
+ IYEV +Y+++L++F ++AF + P +G+ L +V + +PL L+V+GS
Sbjct: 390 EHIYEVDYPNYEEALQIFCMHAFGQKSPYDGFEELAQQVTTLSGRLPLGLKVMGSYFRGM 449
Query: 395 EREVWESELKKLKELPNVDIFDVLKLSYDGLDDEPKDIFLDITCFYAGDFVDKVVELLDS 454
++ W L +++ + I +LKLSYD L D K +FL + C + D + V + L
Sbjct: 450 TKQEWTMALPRVRTHLDGKIESILKLSYDALCDVDKSLFLHLACSFHNDDTELVEQQLGK 509
Query: 455 CGFSADIGMDVLKDRGLISILEDRIAVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIY 514
G+ VL ++ LI + I +H L+ ++GREIVR+Q + +PG+ L + +I
Sbjct: 510 KFSDLRQGLHVLAEKSLIHMDLRLIRMHVLLAQLGREIVRKQSIHEPGQRQFLVDATDIR 569
Query: 515 HVVRKNKGTDAIQCIFLDMCQIEK-VQLHPEIFKSMPNIRML-----YFHKHSSFKQG-- 566
V+ + G+ ++ I D +EK + + + F+ M N++ + F +H + G
Sbjct: 570 EVLTDDTGSRSVIGIDFDFNTMEKELDISEKAFRGMSNLQFIRIYGDLFSRHGVYYFGGR 629
Query: 567 --------QSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCPEKLVKLHMRESRLEQL 618
S + P L+ LP L++LHW +FP S P +F E LVKL M S+LE+L
Sbjct: 630 GHRVSLDYDSKLHFPRGLDYLPGKLRLLHWQQFPMTSLPSEFHAEFLVKLCMPYSKLEKL 689
Query: 619 WGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIPDLSKSPNIKEI 678
W G+ Q L NLE LDL+ S L +PDLS + N++ +
Sbjct: 690 W----------EGI--------------QPLRNLEWLDLTCSRNLKELPDLSTATNLQRL 725
Query: 679 ILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQ-TAIQELP 737
+ C SL +LP ++ + L+++NL C ++ +P + NL L L++ +++ ELP
Sbjct: 726 SIERCSSLVKLPSSIGEATNLKKINLRECLSLVELPSSFGNLTNLQELDLRECSSLVELP 785
Query: 738 SSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDL-KLTKL 796
+S +L +E L CS L +PS+ G LT L L L C S+ P S +L L L
Sbjct: 786 TSFGNLANVESLEFYECSSLVKLPSTFGNLTNLRVLGLRECSSMVELPSSFGNLTNLQVL 845
Query: 797 DLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPN 856
+L CS L +LPSS NL L+ L L++C L LP+
Sbjct: 846 NLRKCSTL-----------------------VELPSSFVNLTNLENLDLRDCSSL--LPS 880
Query: 857 SICNLKLLSELDCSGC 872
S N+ L L C
Sbjct: 881 SFGNVTYLKRLKFYKC 896
Score = 101 bits (252), Expect = 2e-21
Identities = 80/237 (33%), Positives = 122/237 (50%), Gaps = 7/237 (2%)
Query: 686 LTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTA-IQELPSSLNHLV 744
+T LP F +FL +L + S +E + E + + NL L L + ++ELP L+
Sbjct: 664 MTSLPSE-FHAEFLVKLCMP-YSKLEKLWEGIQPLRNLEWLDLTCSRNLKELPD-LSTAT 720
Query: 745 GLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDL-KLTKLDLNDCSR 803
L+ LS++ CS L +PSSIG T L K++L C SL P S +L L +LDL +CS
Sbjct: 721 NLQRLSIERCSSLVKLPSSIGEATNLKKINLRECLSLVELPSSFGNLTNLQELDLRECSS 780
Query: 804 LKTFPAILEPAESFAHISLAE-TAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLK 862
L P + + E +++ KLPS+ NL L+ L L+EC + LP+S NL
Sbjct: 781 LVELPTSFGNLANVESLEFYECSSLVKLPSTFGNLTNLRVLGLRECSSMVELPSSFGNLT 840
Query: 863 LLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYC 919
L L+ C L +P+ L++L L L+D + LP S +++ L+ L C
Sbjct: 841 NLQVLNLRKCSTLVELPSSFVNLTNLENLDLRDCSSL-LPSSFGNVTYLKRLKFYKC 896
Score = 95.5 bits (236), Expect = 2e-19
Identities = 70/209 (33%), Positives = 105/209 (49%), Gaps = 10/209 (4%)
Query: 723 LAVLVLKQTAIQELPSSLNHLVGLEELSLKFC---SKLESIPSSIGTLTKLCKLDLTYCE 779
L +L +Q + LPS + E +K C SKLE + I L L LDLT
Sbjct: 654 LRLLHWQQFPMTSLPSEFH-----AEFLVKLCMPYSKLEKLWEGIQPLRNLEWLDLTCSR 708
Query: 780 SLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAET-AIKKLPSSLQNLV 838
+L+ P L +L + CS L P+ + A + I+L E ++ +LPSS NL
Sbjct: 709 NLKELPDLSTATNLQRLSIERCSSLVKLPSSIGEATNLKKINLRECLSLVELPSSFGNLT 768
Query: 839 GLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQD-TG 897
LQ L L+EC L LP S NL + L+ C L +P+ G L++LR L L++ +
Sbjct: 769 NLQELDLRECSSLVELPTSFGNLANVESLEFYECSSLVKLPSTFGNLTNLRVLGLRECSS 828
Query: 898 VVNLPESIAHLSSLESLDVSYCRKLECIP 926
+V LP S +L++L+ L++ C L +P
Sbjct: 829 MVELPSSFGNLTNLQVLNLRKCSTLVELP 857
>At5g41750 disease resistance protein-like
Length = 1068
Score = 422 bits (1084), Expect = e-118
Identities = 285/839 (33%), Positives = 452/839 (52%), Gaps = 89/839 (10%)
Query: 23 KYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FID---DRSLETGDEICKAIEASTIYVI 77
+Y VF SF G D K F SHLH+ K + F D DR G E+ + I + + ++
Sbjct: 12 RYQVFSSFHGPDVRKGFLSHLHSVFASKGITTFNDQKIDRGQTIGPELIQGIREARVSIV 71
Query: 78 VFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRL 137
V S+ YASS CLDELV+I++CK + V+ +FY VDPS V+ Q+ +G+AF++
Sbjct: 72 VLSKKYASSSWCLDELVEILKCKEALGQIVMTVFYEVDPSDVKKQSGVFGEAFEKTCQGK 131
Query: 138 VPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKLNPPFS---QGMLGID 194
V + RW+ AL A ++G HS+ +E+ +I +I D+ KLN S +GM+G++
Sbjct: 132 NEEVKI-RWRNALAHVATIAGEHSLNWDNEAKMIQKIVTDVSDKLNLTPSRDFEGMVGME 190
Query: 195 KHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQKI-- 252
H+ ++ SLL L+S+ V++IGI G GIGK+T+A L++K+ + F +C + N + I
Sbjct: 191 AHLKRLNSLLCLESDEVKMIGIWGPAGIGKTTIARTLFNKISSIFPFKCFMENLKGSIKG 250
Query: 253 --DRYGIYSLRKKYLSKLLG-EDIQSNGLNYAIERVKRAKVLLILDDLKISIPILELIGG 309
+ Y SL+K+ LS++L E+++ + L + + KVL+ILDD+ + L
Sbjct: 251 GAEHYSKLSLQKQLLSEILKQENMKIHHLGTIKQWLHDQKVLIILDDVDDLEQLEVLAED 310
Query: 310 HGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYANL 369
FG GSRIIVT+ +++LK +IY V S +++LE+ L+AFK+ +G+ L
Sbjct: 311 PSWFGSGSRIIVTTEDKNILKAHRIQDIYHVDFPSEEEALEILCLSAFKQSSIPDGFEEL 370
Query: 370 IVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLDDEP 429
+V + +PL L V+G+ L K + WE L +++ + +I ++L++ YD L E
Sbjct: 371 ANKVAELCGNLPLGLCVVGASLRRKSKNEWERLLSRIESSLDKNIDNILRIGYDRLSTED 430
Query: 430 KDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISILEDRIAV--HDLVLE 487
+ +FL I CF+ + VD + LL G ++L DR L+ I D V H L+ +
Sbjct: 431 QSLFLHIACFFNNEKVDYLTALLADRKLDVVNGFNILADRSLVRISTDGHVVMHHYLLQK 490
Query: 488 MGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIEKVQLHPEIFK 547
+GR IV +Q ++PGK L +EI V+ K GT++++ I D IE+V + F+
Sbjct: 491 LGRRIVHEQWPNEPGKRQFLIEAEEIRDVLTKGTGTESVKGISFDTSNIEEVSVGKGAFE 550
Query: 548 SMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCPEKLVK 607
M N++ L ++ S +G + +P+ +E +P +++LHW +P++S P F PE LVK
Sbjct: 551 GMRNLQFLRIYRDSFNSEG--TLQIPEDMEYIP-PVRLLHWQNYPRKSLPQRFNPEHLVK 607
Query: 608 LHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIP 667
+ M S+L++LWG G+ Q LPNL+ +D+S S+ L IP
Sbjct: 608 IRMPSSKLKKLWG----------GI--------------QPLPNLKSIDMSFSYSLKEIP 643
Query: 668 DLSKSPNIKEIILSGCKSLTRLP---INLFKLKF--------------------LERLNL 704
+LSK+ N++ + L CKSL LP +NL KL+ LERL++
Sbjct: 644 NLSKATNLEILSLEFCKSLVELPFSILNLHKLEILNVENCSMLKVIPTNINLASLERLDM 703
Query: 705 SGCSNVENIPEIKETMENLAVLVLKQTAIQELP------SSLNHLV----GLEELSLKFC 754
+GCS + P+I N+ L L T I+++P S L+HL L+ L + C
Sbjct: 704 TGCSELRTFPDIS---SNIKKLNLGDTMIEDVPPSVGCWSRLDHLYIGSRSLKRLHVPPC 760
Query: 755 --------SKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLK 805
S +ESIP SI LT+L L++ C L++ G L LD NDC LK
Sbjct: 761 ITSLVLWKSNIESIPESIIGLTRLDWLNVNSCRKLKSILG--LPSSLQDLDANDCVSLK 817
Score = 99.0 bits (245), Expect = 1e-20
Identities = 105/443 (23%), Positives = 181/443 (40%), Gaps = 89/443 (20%)
Query: 721 ENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCES 780
E+L + + + +++L + L L+ + + F L+ IP+ + T L L L +C+S
Sbjct: 603 EHLVKIRMPSSKLKKLWGGIQPLPNLKSIDMSFSYSLKEIPN-LSKATNLEILSLEFCKS 661
Query: 781 LETFPGSIFDL-KLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVG 839
L P SI +L KL L++ +CS LK P + NL
Sbjct: 662 LVELPFSILNLHKLEILNVENCSMLKVIPTNI------------------------NLAS 697
Query: 840 LQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSL----------- 888
L+ L + C +L + P+ N+K L+ G + +P +GC S L
Sbjct: 698 LERLDMTGCSELRTFPDISSNIKKLN----LGDTMIEDVPPSVGCWSRLDHLYIGSRSLK 753
Query: 889 --------RTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAFHC 940
+L L + + ++PESI L+ L+ L+V+ CRKL+ I LP ++ L A C
Sbjct: 754 RLHVPPCITSLVLWKSNIESIPESIIGLTRLDWLNVNSCRKLKSILGLPSSLQDLDANDC 813
Query: 941 PSIRSVMSNSRFLSDSKEGTFELYFTNNEKQDPGAQRDVVRNAEHRITDVACRFAFYCFP 1000
S++ V + L F N D A++ +++ + +R + C P
Sbjct: 814 VSLKRVCFSFH------NPIRALSFNNCLNLDEEARKGIIQQSVYR---------YICLP 858
Query: 1001 GSEVPNWFHHHCKRNSVTVDKDSLNLSNDNRIIGFAMCFVLQLEDMNYTGYRFSNFRCRL 1060
G ++P F H S+T+ LS +R A +L +E Y + C L
Sbjct: 859 GKKIPEEFTHKATGRSITIPLSPGTLSASSRF--KASILILPVE-----SYETDDISCSL 911
Query: 1061 TFESDGQTHI--LPIDDQLNNSLHQIDLRVGHLVRDHTFIWNYYLMDSASNHNGI-IHAH 1117
+ + H LP L + +H FI++ L + ++ + +
Sbjct: 912 RTKGGVEVHCCELPYHFLLRSR------------SEHLFIFHGDLFPQGNKYHEVDVTMS 959
Query: 1118 NFTFEISNGLYLKSSSLVKECGI 1140
TFE S + K + ECG+
Sbjct: 960 EITFEFS---HTKIGDKIIECGV 979
>At5g36930 disease resistance like protein
Length = 1164
Score = 415 bits (1066), Expect = e-116
Identities = 307/921 (33%), Positives = 469/921 (50%), Gaps = 89/921 (9%)
Query: 19 PNPLKYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGD----EICKAIEAS 72
P YDVF+SF G D K F SHL+ +LRR + F+DD L+ G+ E+ AIE S
Sbjct: 9 PERWTYDVFVSFRGADVRKNFLSHLYDSLRRCGISTFMDDVELQRGEYISPELLNAIETS 68
Query: 73 TIYVIVFSENYASSRCCLDELVKIMEC-KMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFD 131
I ++V +++YASS CLDELV IM+ K N V PIF VDPS +R Q SY +F
Sbjct: 69 KILIVVLTKDYASSAWCLDELVHIMKSHKNNPSHMVFPIFLYVDPSDIRWQQGSYAKSFS 128
Query: 132 EHQNRLVPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKLNPPF---SQ 188
+H+N P ++ W++AL + A++SG+ I R+E+ I I ++I K+L +
Sbjct: 129 KHKNSH-PLNKLKDWREALTKVANISGW-DIKNRNEAECIADITREILKRLPCQYLHVPS 186
Query: 189 GMLGIDKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNA 248
+G+ + I SLL + S+ VR+I I GMGGIGK+TLA+ +++ F + N
Sbjct: 187 YAVGLRSRLQHISSLLSIGSDGVRVIVIYGMGGIGKTTLAKVAFNEFSHLFEGSSFLENF 246
Query: 249 QQKIDR-YGIYSLRKKYLSKLLGE-DIQSNGLNYAI-ERVKRAKVLLILDDLKISIPILE 305
++ + G L+ + LS +L DI+ GL++A+ ER + +VLL++DD+ +
Sbjct: 247 REYSKKPEGRTHLQHQLLSDILRRNDIEFKGLDHAVKERFRSKRVLLVVDDVDDVHQLNS 306
Query: 306 LIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEG 365
FG GSRII+T+R+ H+LK A+ Y + + +SLELF +AF+ P +
Sbjct: 307 AAIDRDCFGHGSRIIITTRNMHLLKQLRAEGSYSPKELDGDESLELFSWHAFRTSEPPKE 366
Query: 366 YANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGL 425
+ EV+ Y G+PLA++VLG+ L ++ WES LK LK +PN +I L++S++ L
Sbjct: 367 FLQHSEEVVTYCAGLPLAVEVLGAFLIERSIREWESTLKLLKRIPNDNIQAKLQISFNAL 426
Query: 426 DDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISILEDRIAVHDLV 485
E KD+FLDI CF+ G V +LD C DI + +L +R LI+I + I +HDL+
Sbjct: 427 TIEQKDVFLDIACFFIGVDSYYVACILDGCNLYPDIVLSLLMERCLITISGNNIMMHDLL 486
Query: 486 LEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIEKVQLHPEI 545
+MGR+IVR+ G+ S LW+H ++ V++K GT+AI+ + L ++ E
Sbjct: 487 RDMGRQIVREISPKKCGERSRLWSHNDVVGVLKKKSGTNAIEGLSLKADVMDFQYFEVEA 546
Query: 546 FKSMPNIRML---YFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCP 602
F M +R+L Y + S+ E P L+ L W F FP++
Sbjct: 547 FAKMQELRLLELRYVDLNGSY-------------EHFPKDLRWLCWHGFSLECFPINLSL 593
Query: 603 EKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQE---LPNLERLDLSN 659
E L L ++ S L++ W Q + + Y+ H + + PN+E+L L N
Sbjct: 594 ESLAALDLQYSNLKRFWKAQSPPQP-ANMVKYLDLSHSVYLRETPDFSYFPNVEKLILIN 652
Query: 660 SWKLVRIPDLSKSPNI--KEII---LSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIP 714
LV + KS I K+++ LS C L LP ++KLK LE L LS CS +E +
Sbjct: 653 CKSLVLV---HKSIGILDKKLVLLNLSSCIELDVLPEEIYKLKSLESLFLSNCSKLERLD 709
Query: 715 EIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLES--------------- 759
+ +E+L L+ TA++E+PS++N L L+ LSL C L S
Sbjct: 710 DALGELESLTTLLADFTALREIPSTINQLKKLKRLSLNGCKGLLSDDIDNLYSEKSHSVS 769
Query: 760 --IPSSIGTLTKLCKLDLTYCE-SLETFPGSIFDL------------------------K 792
P S+ LT + L L YC S E P I L
Sbjct: 770 LLRPVSLSGLTYMRILSLGYCNLSDELIPEDIGSLSFLRDLDLRGNSFCNLPTDFATLPN 829
Query: 793 LTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLE 852
L +L L+DCS+L+ +IL S + + + + K + L L L +C L
Sbjct: 830 LGELLLSDCSKLQ---SILSLPRSLLFLDVGKCIMLKRTPDISKCSALFKLQLNDCISLF 886
Query: 853 SLPNSICNLKLLSELDCSGCR 873
+P I N + LS + GC+
Sbjct: 887 EIP-GIHNHEYLSFIVLDGCK 906
Score = 84.7 bits (208), Expect = 3e-16
Identities = 83/268 (30%), Positives = 122/268 (44%), Gaps = 25/268 (9%)
Query: 685 SLTRLPINLFKLKFLERLNLSGCSNVENIPEIK---ETMENLAVLVLKQTAIQELPSSLN 741
SL PINL L+ L L+L SN++ + + + + L L + +
Sbjct: 583 SLECFPINL-SLESLAALDLQ-YSNLKRFWKAQSPPQPANMVKYLDLSHSVYLRETPDFS 640
Query: 742 HLVGLEELSLKFCSKLESIPSSIGTLTK-LCKLDLTYCESLETFPGSIFDLK-LTKLDLN 799
+ +E+L L C L + SIG L K L L+L+ C L+ P I+ LK L L L+
Sbjct: 641 YFPNVEKLILINCKSLVLVHKSIGILDKKLVLLNLSSCIELDVLPEEIYKLKSLESLFLS 700
Query: 800 DCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECP-----DLESL 854
+CS+L+ L ES + TA++++PS++ L L+ L L C D+++L
Sbjct: 701 NCSKLERLDDALGELESLTTLLADFTALREIPSTINQLKKLKRLSLNGCKGLLSDDIDNL 760
Query: 855 ------------PNSICNLKLLSELDCSGCRKLTG-IPNDIGCLSSLRTLSLQDTGVVNL 901
P S+ L + L C IP DIG LS LR L L+ NL
Sbjct: 761 YSEKSHSVSLLRPVSLSGLTYMRILSLGYCNLSDELIPEDIGSLSFLRDLDLRGNSFCNL 820
Query: 902 PESIAHLSSLESLDVSYCRKLECIPHLP 929
P A L +L L +S C KL+ I LP
Sbjct: 821 PTDFATLPNLGELLLSDCSKLQSILSLP 848
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.139 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,612,822
Number of Sequences: 26719
Number of extensions: 1192192
Number of successful extensions: 9906
Number of sequences better than 10.0: 489
Number of HSP's better than 10.0 without gapping: 343
Number of HSP's successfully gapped in prelim test: 147
Number of HSP's that attempted gapping in prelim test: 3308
Number of HSP's gapped (non-prelim): 2110
length of query: 1156
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1046
effective length of database: 8,379,506
effective search space: 8764963276
effective search space used: 8764963276
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0073.8


