

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0073.3
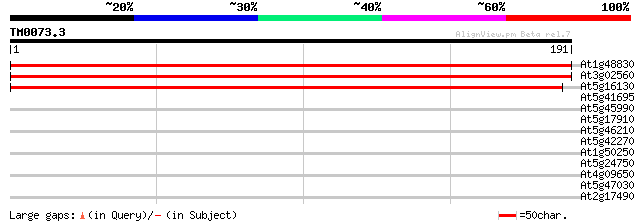
(191 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g48830 unknown protein 312 8e-86
At3g02560 putative 40S ribosomal protein 309 5e-85
At5g16130 40S ribosomal protein S7-like 305 1e-83
At5g41695 putative protein 28 2.3
At5g45990 CRN (crooked neck) protein 28 3.0
At5g17910 putative protein 28 3.0
At5g46210 cullin 28 3.9
At5g42270 cell division protein FtsH 27 5.0
At1g50250 putative chloroplast FtsH protease 27 5.0
At5g24750 sterol glucosyltransferase - like protein 27 6.6
At4g09650 H+-transporting ATP synthase-like protein 27 6.6
At5g47030 ATP synthase delta' chain, mitochondrial precursor (sp... 27 8.6
At2g17490 putative retroelement pol polyprotein 27 8.6
>At1g48830 unknown protein
Length = 191
Score = 312 bits (799), Expect = 8e-86
Identities = 150/191 (78%), Positives = 179/191 (93%)
Query: 1 MYTSRKKIHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKA 60
M++++ KIHK+K E +E +E V QA FDLENT+QELKS+LKDLY+NSAVQVD+SG RKA
Sbjct: 1 MFSAQHKIHKEKGVELSELDEQVAQAFFDLENTNQELKSELKDLYVNSAVQVDISGGRKA 60
Query: 61 VVIHVPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLT 120
+V++VPYRLRKA+RK+HV+LVRELEKKFSGKDVILIATRRI+RPPKKGSAA+RPR RTLT
Sbjct: 61 IVVNVPYRLRKAYRKIHVRLVRELEKKFSGKDVILIATRRIVRPPKKGSAAKRPRNRTLT 120
Query: 121 AVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK 180
+VHEA+L+DVVLPAEIVGKR RYR+DG+KIMKVFLDPKERNNTEYK+E F+AVY+KL+GK
Sbjct: 121 SVHEAILDDVVLPAEIVGKRTRYRLDGTKIMKVFLDPKERNNTEYKVEAFSAVYKKLTGK 180
Query: 181 DVVFEYPITEA 191
DVVFE+PITEA
Sbjct: 181 DVVFEFPITEA 191
>At3g02560 putative 40S ribosomal protein
Length = 191
Score = 309 bits (792), Expect = 5e-85
Identities = 151/191 (79%), Positives = 171/191 (89%)
Query: 1 MYTSRKKIHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKA 60
MY+ + KIHKDK PTEFEE V QALFDLENT+QELKS+LKDLYIN AVQ+D+SGNRKA
Sbjct: 1 MYSGQNKIHKDKGVAPTEFEEQVTQALFDLENTNQELKSELKDLYINQAVQMDISGNRKA 60
Query: 61 VVIHVPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLT 120
VVI+VP+RLRKAFRK+H++LVRELEKKFSGKDVI +ATRRI+RPPKKGSA QRPR RTLT
Sbjct: 61 VVIYVPFRLRKAFRKIHLRLVRELEKKFSGKDVIFVATRRIMRPPKKGSAVQRPRNRTLT 120
Query: 121 AVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK 180
+VHEAMLEDV PAEIVGKR RYR+DG+KIMKVFLD K +N+TEYKLET VYRKL+GK
Sbjct: 121 SVHEAMLEDVAYPAEIVGKRTRYRLDGTKIMKVFLDSKLKNDTEYKLETMVGVYRKLTGK 180
Query: 181 DVVFEYPITEA 191
DVVFEYP+ EA
Sbjct: 181 DVVFEYPVIEA 191
>At5g16130 40S ribosomal protein S7-like
Length = 190
Score = 305 bits (780), Expect = 1e-83
Identities = 148/188 (78%), Positives = 169/188 (89%)
Query: 1 MYTSRKKIHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKA 60
M++++ KI KDK+AEPTE EE V QALFDLENT+QELKS+LKDLYIN AV +D+SGNRKA
Sbjct: 1 MFSAQNKIKKDKNAEPTECEEQVAQALFDLENTNQELKSELKDLYINQAVHMDISGNRKA 60
Query: 61 VVIHVPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLT 120
VVI+VP+RLRKAFRK+H +LVRELEKKFSGKDVI + TRRI+RPPKKG+A QRPR RTLT
Sbjct: 61 VVIYVPFRLRKAFRKIHPRLVRELEKKFSGKDVIFVTTRRIMRPPKKGAAVQRPRNRTLT 120
Query: 121 AVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK 180
+VHEAMLEDV PAEIVGKR RYR+DGSKIMKVFLD KE+NNTEYKLET VYRKL+GK
Sbjct: 121 SVHEAMLEDVAFPAEIVGKRTRYRLDGSKIMKVFLDAKEKNNTEYKLETMVGVYRKLTGK 180
Query: 181 DVVFEYPI 188
DVVFEYP+
Sbjct: 181 DVVFEYPV 188
>At5g41695 putative protein
Length = 548
Score = 28.5 bits (62), Expect = 2.3
Identities = 29/114 (25%), Positives = 47/114 (40%), Gaps = 9/114 (7%)
Query: 2 YTSRKKIHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKAV 61
Y R+ + D+D P EF E VG L H K+++ + IN V + + +
Sbjct: 261 YLRRESLPIDEDETPPEFVEAVGVRKKTLFVAHLSRKTEITHI-INFFKDVGEVVHVRLI 319
Query: 62 VIHVPYRLRKAFRKV----HVKLVRELEKK----FSGKDVILIATRRILRPPKK 107
+ H + AF + K+VR LE K + + L + + PP K
Sbjct: 320 LNHTGKHVGCAFVEFGSANEAKMVRALETKNGEYLNDCKIFLEVAKMVPYPPPK 373
>At5g45990 CRN (crooked neck) protein
Length = 673
Score = 28.1 bits (61), Expect = 3.0
Identities = 16/55 (29%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query: 132 LPAEIVGKRVRYRVDGSKIMKVFLD---PKERNNTEYKLETFAAVYRKLSGKDVV 183
LP ++ +++ R DGS + + D P+E T K+ A ++KL G++ V
Sbjct: 619 LPKKLKKRKMTSREDGSTEYEEYFDYLFPEESGTTNLKILEAAYKWKKLKGEECV 673
>At5g17910 putative protein
Length = 1342
Score = 28.1 bits (61), Expect = 3.0
Identities = 22/91 (24%), Positives = 37/91 (40%), Gaps = 5/91 (5%)
Query: 86 KKFSGKDVILIATRRILRPPK-----KGSAAQRPRTRTLTAVHEAMLEDVVLPAEIVGKR 140
K+ + DV+L+ TR + PK KG A + + A +V L E+V ++
Sbjct: 1247 KEITKSDVLLVETRALEEFPKPSELKKGMAMEVISEGVVIPTKAAGPSNVTLSDEVVTEK 1306
Query: 141 VRYRVDGSKIMKVFLDPKERNNTEYKLETFA 171
+ S P+ + E +ET A
Sbjct: 1307 AKAETTASNTDANVQSPESKETPENSVETIA 1337
>At5g46210 cullin
Length = 792
Score = 27.7 bits (60), Expect = 3.9
Identities = 21/83 (25%), Positives = 40/83 (47%), Gaps = 13/83 (15%)
Query: 7 KIHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKAVVIHVP 66
K++K K PT FE ENT ++L+S ++ +++ + D+ +AV
Sbjct: 79 KLNKAKPTLPTNFE----------ENTWEKLQSAIRAIFLKKKISFDLESLYQAV---DN 125
Query: 67 YRLRKAFRKVHVKLVRELEKKFS 89
L K K++ ++ +E E+ S
Sbjct: 126 LCLHKLDGKLYDQIEKECEEHIS 148
>At5g42270 cell division protein FtsH
Length = 704
Score = 27.3 bits (59), Expect = 5.0
Identities = 17/46 (36%), Positives = 21/46 (44%)
Query: 80 LVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLTAVHEA 125
L E K KD I A RI+ P+K +A + L A HEA
Sbjct: 469 LAARRELKEISKDEISDALERIIAGPEKKNAVVSEEKKRLVAYHEA 514
>At1g50250 putative chloroplast FtsH protease
Length = 716
Score = 27.3 bits (59), Expect = 5.0
Identities = 17/46 (36%), Positives = 21/46 (44%)
Query: 80 LVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLTAVHEA 125
L E K KD I A RI+ P+K +A + L A HEA
Sbjct: 481 LAARRELKEISKDEISDALERIIAGPEKKNAVVSEEKKRLVAYHEA 526
>At5g24750 sterol glucosyltransferase - like protein
Length = 517
Score = 26.9 bits (58), Expect = 6.6
Identities = 14/36 (38%), Positives = 22/36 (60%), Gaps = 1/36 (2%)
Query: 150 IMKVFLDPKERNNTEYKLETFAAVYRKLSGKDVVFE 185
+ K+FL+ KER E++ E +A +R + GKD E
Sbjct: 73 LRKMFLEEKERIKREHRQECHSA-FRTIFGKDPCME 107
>At4g09650 H+-transporting ATP synthase-like protein
Length = 234
Score = 26.9 bits (58), Expect = 6.6
Identities = 31/138 (22%), Positives = 55/138 (39%), Gaps = 20/138 (14%)
Query: 14 AEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKAVVIHVPYRLRKAF 73
A PT E Q + D+ +KS + ++ + V V NR I++ + K F
Sbjct: 93 ANPTITVEKKRQVIDDI------VKSSSLQSHTSNFLNVLVDANR----INIVTEIVKEF 142
Query: 74 RKVHVKLVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLTAVHEAMLEDVVLP 133
V+ KL D L R +++ A + + LT ++ V+
Sbjct: 143 ELVYNKLT----------DTQLAEVRSVVKLEAPQLAQIAKQVQKLTGAKNVRVKTVIDA 192
Query: 134 AEIVGKRVRYRVDGSKIM 151
+ + G +RY GSK++
Sbjct: 193 SLVAGFTIRYGESGSKLI 210
>At5g47030 ATP synthase delta' chain, mitochondrial precursor
(sp|Q96252)
Length = 203
Score = 26.6 bits (57), Expect = 8.6
Identities = 14/40 (35%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query: 104 PPKKGSAAQRPRTRTLTAVHEAMLEDVVLP--AEIVGKRV 141
PP+ SA +PR T +++ + + VLP +E+ GK V
Sbjct: 51 PPQTPSAFMKPRPSTPSSIPTKLTVNFVLPYTSELTGKEV 90
>At2g17490 putative retroelement pol polyprotein
Length = 822
Score = 26.6 bits (57), Expect = 8.6
Identities = 16/55 (29%), Positives = 29/55 (52%), Gaps = 2/55 (3%)
Query: 127 LEDVVLPAEIVGKRV--RYRVDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSG 179
LE + L E +++ ++ ++ SKIM L P+ ++ E + + VYR L G
Sbjct: 702 LEGIFLSQECYARKLLKKFNMEDSKIMSTPLLPQRKDQEEDESLADSKVYRSLVG 756
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,100,578
Number of Sequences: 26719
Number of extensions: 162857
Number of successful extensions: 465
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 458
Number of HSP's gapped (non-prelim): 13
length of query: 191
length of database: 11,318,596
effective HSP length: 94
effective length of query: 97
effective length of database: 8,807,010
effective search space: 854279970
effective search space used: 854279970
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0073.3


