

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0071.7
(393 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
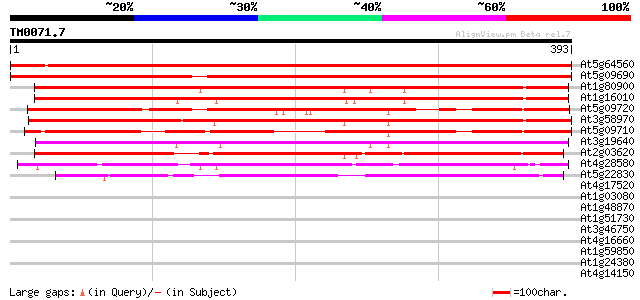
Score E
Sequences producing significant alignments: (bits) Value
At5g64560 unknown protein 598 e-171
At5g09690 unknown protein 544 e-155
At1g80900 unknown protein 330 8e-91
At1g16010 unknown protein 330 8e-91
At5g09720 putative protein 329 2e-90
At3g58970 unknown protein 328 4e-90
At5g09710 putative protein 305 2e-83
At3g19640 unknown protein 295 2e-80
At2g03620 unknown protein 287 6e-78
At4g28580 MRS2-6 244 5e-65
At5g22830 magnesium transporter protein (GMN10) 91 1e-18
At4g17520 nuclear RNA binding protein A-like protein 39 0.004
At1g03080 unknown protein 35 0.093
At1g48870 hypothetical protein 34 0.12
At1g51730 unknown protein 33 0.27
At3g46750 hypothetical protein 32 0.60
At4g16660 HSP like protein 31 1.0
At1g59850 hypothetical protein 31 1.3
At1g24380 hypothetical protein 31 1.3
At4g14150 kinesin like protein 30 1.8
>At5g64560 unknown protein
Length = 394
Score = 598 bits (1542), Expect = e-171
Identities = 312/395 (78%), Positives = 342/395 (85%), Gaps = 3/395 (0%)
Query: 1 MARDGSVLPADPQAVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRI 60
MA++G ++PADP AV VKKKT + SW L DA GQ LDVDKY IMHRVQIHARDLRI
Sbjct: 1 MAQNGYLVPADPSAVVTVKKKTPQA-SWALIDATGQSEPLDVDKYEIMHRVQIHARDLRI 59
Query: 61 LDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSS 120
LDP LSYPSTILGRE+AIVLNLEHIKAIIT+EEVLLRDP+DENV+PVVEEL+RRLP ++
Sbjct: 60 LDPNLSYPSTILGRERAIVLNLEHIKAIITSEEVLLRDPSDENVIPVVEELRRRLPVGNA 119
Query: 121 LHQQ-QGDGQEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPAL 179
H QGDG+E +Q+D + +EDESPFEFRALEVALEAICSFLAART ELE AAYPAL
Sbjct: 120 SHNGGQGDGKEIAGAQNDGDTGDEDESPFEFRALEVALEAICSFLAARTAELETAAYPAL 179
Query: 180 DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRKAGS-SS 238
DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK S SS
Sbjct: 180 DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRKLSSASS 239
Query: 239 PVSGSGAANWFGGSPTIGSKISRASRASLATVRFDENDVEELEMLLEAYFMQIDGTFNKL 298
P+S G NW+ SPTIGSKISRASRASLATV DENDVEELEMLLEAYFMQID T N+L
Sbjct: 240 PISSIGEPNWYTTSPTIGSKISRASRASLATVHGDENDVEELEMLLEAYFMQIDSTLNRL 299
Query: 299 TTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDN 358
TTLREYIDDTEDYINIQLDNHRNQLIQLEL LSSGTVCLS YSLVA IFGMNIPY+WND
Sbjct: 300 TTLREYIDDTEDYINIQLDNHRNQLIQLELVLSSGTVCLSMYSLVAGIFGMNIPYTWNDG 359
Query: 359 HGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLVGS 393
HG+MFK+VV ++G ++F++I+ YAR KGLVGS
Sbjct: 360 HGYMFKYVVGLTGTLCVVVFVIIMSYARYKGLVGS 394
>At5g09690 unknown protein
Length = 397
Score = 544 bits (1401), Expect = e-155
Identities = 288/396 (72%), Positives = 323/396 (80%), Gaps = 13/396 (3%)
Query: 1 MARDGSVLPADPQAVAVVKKKT-QSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLR 59
M+ DG ++P D AV K+KT Q SRSWI DA GQ ++LDVDK+ IMHRVQIHARDLR
Sbjct: 12 MSPDGELVPVDSSAVVTAKRKTSQLSRSWISIDATGQKTVLDVDKHVIMHRVQIHARDLR 71
Query: 60 ILDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVS 119
ILDP L YPS ILGRE+AIVLNLEHIKAIITAEEVL+RD +DENV+PV+EE QRRLP +
Sbjct: 72 ILDPNLFYPSAILGRERAIVLNLEHIKAIITAEEVLIRDSSDENVIPVLEEFQRRLPVGN 131
Query: 120 SLHQQQGDGQEYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPAL 179
H GDG + EEDESPFEFRALEVALEAICSFLAARTTELE AYPAL
Sbjct: 132 EAHGVHGDG----------DLGEEDESPFEFRALEVALEAICSFLAARTTELEKFAYPAL 181
Query: 180 DELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK-AGSSS 238
DELT KISSRNL+RVRKLKSAMTRLTARVQKVRDELEQLLDDD DMADLYL+RK G+SS
Sbjct: 182 DELTLKISSRNLERVRKLKSAMTRLTARVQKVRDELEQLLDDDGDMADLYLTRKLVGASS 241
Query: 239 PVSGSGAANWFGGSPTIGSKISRASRASLATVRFD-ENDVEELEMLLEAYFMQIDGTFNK 297
VS S W+ SPTIGS ISRASR SL TVR D E DVEELEMLLEAYFMQID T NK
Sbjct: 242 SVSVSDEPIWYPTSPTIGSMISRASRVSLVTVRGDDETDVEELEMLLEAYFMQIDSTLNK 301
Query: 298 LTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWND 357
LT LREYIDDTEDYINIQLDNHRNQLIQLEL LS+GTVC+S YS++A IFGMNIP +WN
Sbjct: 302 LTELREYIDDTEDYINIQLDNHRNQLIQLELMLSAGTVCVSVYSMIAGIFGMNIPNTWNH 361
Query: 358 NHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLVGS 393
+HG++FKWVV ++G F ++F++I+ YAR +GL+GS
Sbjct: 362 DHGYIFKWVVSLTGTFCIVLFVIILSYARFRGLIGS 397
>At1g80900 unknown protein
Length = 443
Score = 330 bits (846), Expect = 8e-91
Identities = 192/400 (48%), Positives = 252/400 (63%), Gaps = 27/400 (6%)
Query: 18 VKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREKA 77
+KK+ Q +SWI D +++VDK+ +M R + ARDLR+LDPL YPSTILGREKA
Sbjct: 43 LKKRGQGLKSWIRVDTSANSQVIEVDKFTMMRRCDLPARDLRLLDPLFVYPSTILGREKA 102
Query: 78 IVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYV----- 132
IV+NLE I+ IITA+EVLL + D V+ V ELQ+RL S D E
Sbjct: 103 IVVNLEQIRCIITADEVLLLNSLDNYVLRYVVELQQRLKASSVTEVWNQDSLELSRRRSR 162
Query: 133 SSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLD 192
S + + + D PFEFRALEVALEA C+FL ++ +ELE+ AYP LDELTSKIS+ NL+
Sbjct: 163 SLDNVLQNSSPDYLPFEFRALEVALEAACTFLDSQASELEIEAYPLLDELTSKISTLNLE 222
Query: 193 RVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK---------AGSSSPVSGS 243
R R+LKS + LT RVQKVRDE+EQL+DDD DMA++YL+ K S PV +
Sbjct: 223 RARRLKSRLVALTRRVQKVRDEIEQLMDDDGDMAEMYLTEKKKRMEGSLYGDQSLPVYRT 282
Query: 244 GAANWFGG--SPTIGSKISRASRASLATVRFDEN----------DVEELEMLLEAYFMQI 291
SP SR SL+ VR + ++EELEMLLEAYF+ I
Sbjct: 283 NDCFSLSAPVSPVSSPPESRRLEKSLSIVRSRHDSARSSEDATENIEELEMLLEAYFVVI 342
Query: 292 DGTFNKLTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNI 351
D T NKLT+L+EYIDDTED+INIQLDN RNQLIQ EL L++ T ++ + +VA IFGMN
Sbjct: 343 DSTLNKLTSLKEYIDDTEDFINIQLDNVRNQLIQFELLLTTATFVVAIFGVVAGIFGMNF 402
Query: 352 PYSWNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLV 391
+ + G FKWV+ ++GV ++FL + Y +++ L+
Sbjct: 403 EIDFFEKPG-AFKWVLAITGVCGLVVFLAFLWYYKRRRLM 441
>At1g16010 unknown protein
Length = 442
Score = 330 bits (846), Expect = 8e-91
Identities = 194/399 (48%), Positives = 256/399 (63%), Gaps = 26/399 (6%)
Query: 18 VKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREKA 77
+KK+ Q RSWI D G +++VDK+ +M R + ARDLR+LDPL YPSTILGREKA
Sbjct: 43 LKKRGQGLRSWIRVDTSGNTQVMEVDKFTMMRRCDLPARDLRLLDPLFVYPSTILGREKA 102
Query: 78 IVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLP--KVSSLHQQQGDGQEYVSSQ 135
IV+NLE I+ IITA+EVLL + D V+ V ELQ+RL V + QQ+ S+
Sbjct: 103 IVVNLEQIRCIITADEVLLLNSLDNYVLRYVVELQQRLKTSSVGEMWQQENSQLSRRRSR 162
Query: 136 HDTEAAEE---DESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLD 192
A E D PFEFRALE+ALEA C+FL ++ +ELE+ AYP LDELTSKIS+ NL+
Sbjct: 163 SFDNAFENSSPDYLPFEFRALEIALEAACTFLDSQASELEIEAYPLLDELTSKISTLNLE 222
Query: 193 RVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRKA--------GSSSPV---S 241
RVR+LKS + LT RVQKVRDE+EQL+DDD DMA++YL+ K G S + S
Sbjct: 223 RVRRLKSRLVALTRRVQKVRDEIEQLMDDDGDMAEMYLTEKKRRMEGSMYGDQSLLGYRS 282
Query: 242 GSGAANWFGGSPTIGSKISRASRASLATVRFDEN---------DVEELEMLLEAYFMQID 292
G + SP SR SL+ R + ++EELEMLLEAYF+ ID
Sbjct: 283 NDGLSVSAPVSPVSSPPDSRRLDKSLSIARSRHDSARSSEGAENIEELEMLLEAYFVVID 342
Query: 293 GTFNKLTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIP 352
T NKLT+L+EYIDDTED+INIQLDN RNQLIQ EL L++ T ++ + +VA IFGMN
Sbjct: 343 STLNKLTSLKEYIDDTEDFINIQLDNVRNQLIQFELLLTTATFVVAIFGVVAGIFGMNFE 402
Query: 353 YSWNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLV 391
+ + G F+WV+I++GV ++F + + + + L+
Sbjct: 403 IDFFNQPG-AFRWVLIITGVCGFVIFSAFVWFFKYRRLM 440
>At5g09720 putative protein
Length = 397
Score = 329 bits (843), Expect = 2e-90
Identities = 211/431 (48%), Positives = 263/431 (60%), Gaps = 91/431 (21%)
Query: 13 QAVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTIL 72
+ V V + QSS SWI DA G+ ++LDVDKY IMHRVQIHARDLRILDP L YPS IL
Sbjct: 6 ELVPVKRITPQSSWSWISIDATGKKTVLDVDKYVIMHRVQIHARDLRILDPNLFYPSAIL 65
Query: 73 GREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYV 132
GRE+AIVLNLEHIKAIITA+E D +DEN++P +EE Q RL + H Q DG
Sbjct: 66 GRERAIVLNLEHIKAIITAKE----DSSDENLIPTLEEFQTRLSVGNKAHGGQLDG---- 117
Query: 133 SSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSK----ISS 188
+ EEDES FEFRALEVALEAICSFLAART ELE +AYPALDELT K I S
Sbjct: 118 ------DVVEEDESAFEFRALEVALEAICSFLAARTIELEKSAYPALDELTLKFHDPIDS 171
Query: 189 RN----------------------LDRVRKLKSAMTRLTA---------RVQ-------- 209
+ ++++ K ++ + RV+
Sbjct: 172 SGPKGEQESLGTGSMFRKEIYLGVMHKIKEFKDSLKEPSKLEYMLLGLYRVKVGSKSEYD 231
Query: 210 ---KVRDELEQLLDDDDDMADLYLSRK-AGSSSPVSGSGA-ANWFGGSPTIGSKISRASR 264
+++DELEQLL+DD+DMA+LYLSRK AG+SSP SG NW+ SPTIG+KISRA
Sbjct: 232 VDLQIKDELEQLLEDDEDMAELYLSRKLAGASSPAIDSGEHINWYPTSPTIGAKISRAKS 291
Query: 265 --ASLATVRFDE-NDVEELEMLLEAYFMQIDGTFNKLTTLREYIDDTEDYINIQLDNHRN 321
ATVR D+ NDVEE+EMLLE LREY+D+TED++N
Sbjct: 292 HLVRSATVRGDDKNDVEEVEMLLE---------------LREYVDETEDFLN-------- 328
Query: 322 QLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDNHGFMFKWVVIVSGVFSAIMFLLI 381
IQ E+ L++G++C+S YS+V I GMNIP+ WN MFKWVV + AI+F++I
Sbjct: 329 --IQFEIILTAGSICVSVYSVVVGILGMNIPFPWNIKK-HMFKWVVSGTATVCAILFVII 385
Query: 382 IVYARKKGLVG 392
+ +AR K L G
Sbjct: 386 MSFARYKKLFG 396
>At3g58970 unknown protein
Length = 436
Score = 328 bits (840), Expect = 4e-90
Identities = 189/388 (48%), Positives = 252/388 (64%), Gaps = 10/388 (2%)
Query: 14 AVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILG 73
A KKKT +R W+ FD G +++ DK I+ R + ARDLRIL P+ S+ S IL
Sbjct: 51 ATGKAKKKTGGARLWMRFDRTGAMEVVECDKSTIIKRASVPARDLRILGPVFSHSSNILA 110
Query: 74 REKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVS 133
REKAIV+NLE IKAI+TAEEVLL DP V+P VE L+++ P+ + ++ V
Sbjct: 111 REKAIVVNLEVIKAIVTAEEVLLLDPLRPEVLPFVERLKQQFPQRNG-NENALQASANVQ 169
Query: 134 SQHDTEAAE--EDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNL 191
S D EAAE + E PFEF+ LE+ALE +CSF+ LE A+P LDELT +S+ NL
Sbjct: 170 SPLDPEAAEGLQSELPFEFQVLEIALEVVCSFVDKSVAALETEAWPVLDELTKNVSTENL 229
Query: 192 DRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK---AGSSSPVSGSGAANW 248
+ VR LKS +TRL ARVQKVRDELE LLDD++DMADLYL+RK + + A+N
Sbjct: 230 EYVRSLKSNLTRLLARVQKVRDELEHLLDDNEDMADLYLTRKWIQNQQTEAILAGTASNS 289
Query: 249 FGGSPTIGSKISRASR---ASLATVRFDENDVEELEMLLEAYFMQIDGTFNKLTTLREYI 305
S + R + AS+ T +E+DVE+LEMLLEAYFMQ+DG NK+ T+REYI
Sbjct: 290 IALPAHNTSNLHRLTSNRSASMVTSNTEEDDVEDLEMLLEAYFMQLDGMRNKILTVREYI 349
Query: 306 DDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDNHGFMFKW 365
DDTEDY+NIQLDN RN+LIQL+L L+ + ++ +L+A++FGMNIP HG +F +
Sbjct: 350 DDTEDYVNIQLDNQRNELIQLQLTLTIASFAIAAETLLASLFGMNIPCPLYSIHG-VFGY 408
Query: 366 VVIVSGVFSAIMFLLIIVYARKKGLVGS 393
V ++F++ + YAR K L+GS
Sbjct: 409 FVWSVTALCIVLFMVTLGYARWKKLLGS 436
>At5g09710 putative protein
Length = 328
Score = 305 bits (782), Expect = 2e-83
Identities = 194/388 (50%), Positives = 236/388 (60%), Gaps = 72/388 (18%)
Query: 11 DPQAVAVVKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPST 70
DP V+ K+K SRSW+L DA G ++L+VD YAI+ RV I+ARDLR+ + +S P +
Sbjct: 8 DPSEVSTAKRKP--SRSWLLIDAAGNSTMLNVDSYAIIRRVHIYARDLRVFESSISSPLS 65
Query: 71 ILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQE 130
I RE AIVLNLEHIK IITA+ EE +RRL + + Q DG+E
Sbjct: 66 IRTREGAIVLNLEHIKVIITAD----------------EEFERRLGVENRERRGQPDGKE 109
Query: 131 YVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRN 190
++ D AE+DESPFEFRALEVALEAICSFLAARTTELE + YPAL+EL SK
Sbjct: 110 DSGAEVD---AEKDESPFEFRALEVALEAICSFLAARTTELEKSGYPALNELASK----- 161
Query: 191 LDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK-AGSSSPVSGSG-AANW 248
DDDD+ DL LSRK A +SSPVS S N
Sbjct: 162 ------------------------------DDDDLGDLCLSRKIATTSSPVSDSDEQINS 191
Query: 249 FGGSPTIGSKISRASR--ASLATVR-FDENDVEELEMLLEAYFMQIDGTFNKLTTLREYI 305
+ SPTIG+KISRA ATVR D+NDVEE+EMLLEA++MQID T NKL LREY+
Sbjct: 192 YPTSPTIGAKISRAKSHLVRSATVRGDDQNDVEEVEMLLEAHYMQIDRTLNKLAELREYL 251
Query: 306 DDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDNHGFMFKW 365
DDTEDYIN Q E+ +++G+VC+S YSLV I NIP+SWN MFKW
Sbjct: 252 DDTEDYIN----------FQFEVIITAGSVCISVYSLVVGILSTNIPFSWNTKE-HMFKW 300
Query: 366 VVIVSGVFSAIMFLLIIVYARKKGLVGS 393
VV + AI F++II YAR K LVG+
Sbjct: 301 VVSATATLCAIFFVIIISYARYKKLVGN 328
>At3g19640 unknown protein
Length = 484
Score = 295 bits (756), Expect = 2e-80
Identities = 183/446 (41%), Positives = 253/446 (56%), Gaps = 73/446 (16%)
Query: 19 KKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREKAI 78
+KK R+W++ ++ GQ + K++IM R + ARDLRILDPLLSYPST+LGRE+AI
Sbjct: 38 RKKGVGVRTWLVLNSSGQSEPKEEGKHSIMRRTGLPARDLRILDPLLSYPSTVLGRERAI 97
Query: 79 VLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRL----------------------- 115
V+NLEHIKAIITA+EVLL + D +V P ++ELQRR+
Sbjct: 98 VINLEHIKAIITAQEVLLLNSKDPSVSPFIDELQRRILCHHHATKPQEEQNSGGEPHTRV 157
Query: 116 -PKVSSLHQQQGDGQEYVSSQHDTEAAEEDES-----PFEFRALEVALEAICSFLAARTT 169
P +Q G + ++ D + + E++ PFEF ALE LEA S L
Sbjct: 158 DPAQGEAGTEQSSGDQGSEAKKDAKQSLENQDGSKVLPFEFVALEACLEAASSSLEHEAL 217
Query: 170 ELEMAAYPALDELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLY 229
LE+ A+PALD+LTSKIS+ NL+RVR++KS + +T RVQKVRDELE LLDDD+DMA++Y
Sbjct: 218 RLELEAHPALDKLTSKISTLNLERVRQIKSRLVAITGRVQKVRDELEHLLDDDEDMAEMY 277
Query: 230 LSRKAGSSSPVSGSGAANWFGG--------------SPTIGSKISRASR----------- 264
L+ K S + + N P S+ +R R
Sbjct: 278 LTEKLAQKLEDSSNSSMNESDTFEVDLPQGDEDDRLPPEFASEANRDGRYLQANDAHELL 337
Query: 265 ----------------ASLATVRFDENDVEELEMLLEAYFMQIDGTFNKLTTLREYIDDT 308
+S + ++ DVEELEMLLEAYF+QIDG NKL+TLREY+DDT
Sbjct: 338 MSTQSALSRNSRGTHTSSTRSAMTNKLDVEELEMLLEAYFVQIDGILNKLSTLREYVDDT 397
Query: 309 EDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYS-WNDN-HG-FMFKW 365
EDYINI LD+ +N L+Q+ + L++ T+ +S + VA +FGMNI + DN HG F W
Sbjct: 398 EDYINIMLDDKQNHLLQMGVMLTTATLVMSAFIAVAGVFGMNITIELFTDNKHGPSRFIW 457
Query: 366 VVIVSGVFSAIMFLLIIVYARKKGLV 391
VI + S +++ I + + K L+
Sbjct: 458 TVIGGSIGSICLYVGAIGWCKYKRLL 483
>At2g03620 unknown protein
Length = 421
Score = 287 bits (735), Expect = 6e-78
Identities = 169/398 (42%), Positives = 243/398 (60%), Gaps = 49/398 (12%)
Query: 18 VKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREKA 77
+KK+ QSSRSW+ D DG ++L++DK IM R + +RDLR+LDPL YPS+ILGRE+A
Sbjct: 41 LKKRGQSSRSWVKIDQDGNSAVLELDKATIMKRCSLPSRDLRLLDPLFIYPSSILGRERA 100
Query: 78 IVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVSSQHD 137
IV++LE I+ IITAEEV+L + D +VV EL +RL S H+
Sbjct: 101 IVVSLEKIRCIITAEEVILMNARDASVVQYQSELCKRL-----------------QSNHN 143
Query: 138 TEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLDRVRKL 197
+D+ PFEF+ALE+ LE C L A+ ELEM YP LDEL + IS+ NL+ VR+L
Sbjct: 144 LNV--KDDLPFEFKALELVLELSCLSLDAQVNELEMEVYPVLDELATNISTLNLEHVRRL 201
Query: 198 KSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK------------------------ 233
K + LT +VQKV DE+E L+DDDDDMA++YL+ K
Sbjct: 202 KGRLLTLTQKVQKVCDEIEHLMDDDDDMAEMYLTEKKERAEAHASEELEDNIGEDFESSG 261
Query: 234 -AGSSSPVS--GSGAANWFGGSPTIGSKISRASRASLATVRFDENDVEELEMLLEAYFMQ 290
S+PVS GS + N FG S I + ++ L++ EN +++LEMLLEAYF+
Sbjct: 262 IVSKSAPVSPVGSTSGN-FGKLQRAFSSIVGSHKSLLSSSSIGEN-IDQLEMLLEAYFVV 319
Query: 291 IDGTFNKLTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMN 350
+D T +KL++L+EYIDDTED INI+L N +NQLIQ +L L++ T + ++ V A+FGMN
Sbjct: 320 VDNTLSKLSSLKEYIDDTEDLINIKLGNVQNQLIQFQLLLTAATFVAAIFAAVTAVFGMN 379
Query: 351 IPYSWNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARKK 388
+ S N F++V++++G+ ++ ++Y + K
Sbjct: 380 LQDSVFQN-PTTFQYVLLITGIGCGFLYFGFVLYFKHK 416
>At4g28580 MRS2-6
Length = 408
Score = 244 bits (624), Expect = 5e-65
Identities = 155/401 (38%), Positives = 225/401 (55%), Gaps = 35/401 (8%)
Query: 6 SVLPADPQAVAVV------KKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLR 59
S+ P P A+V KK+ W FD G + DK I+ R + A+DLR
Sbjct: 27 SLSPPTPPCSAIVGGTGKSKKRRGGVCLWTRFDRTGFMEVAGCDKSTIIERSSVSAKDLR 86
Query: 60 ILDPLLSYPSTILGREKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVS 119
S+ S IL REKAIVLNLE IKA+IT+E+V+L D V+ + + L+ P+
Sbjct: 87 TA---FSHSSKILAREKAIVLNLEVIKAVITSEQVMLLDSLRPEVLTLTDRLKHHFPRK- 142
Query: 120 SLHQQQGDGQEYV---SSQHDTEAAEE---DESPFEFRALEVALEAICSFLAARTTELEM 173
DG E + SS E EE + PFEFR LE+A E CSF+ + +LE
Sbjct: 143 -------DGPENILQASSHGHQEGGEEGLKSKLPFEFRVLEIAFEVFCSFVDSNVVDLET 195
Query: 174 AAYPALDELTSKISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRK 233
A+ LDELT K+S+ NL +R LK+++T L ARVQKVRDE+E LDD +DM DLYL+RK
Sbjct: 196 QAWSILDELTKKVSNENLKDLRSLKTSLTHLLARVQKVRDEIEHFLDDKEDMEDLYLTRK 255
Query: 234 AGSSSPVSGSGAANWFGGSPTIGSKISRASRASLATVRFDENDVEELEMLLEAYFMQIDG 293
+ A+N P + S S+ T +E+D++++EMLLEAYFMQ++G
Sbjct: 256 WIQNQQT--EAASNSIVSQPNLQRHTSNRISTSMVT---EEDDIDDMEMLLEAYFMQLEG 310
Query: 294 TFNKLTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIP- 352
NK+ ++E+ID TE Y+ I ++ RN LI L + ++ G ++ ++V +FGMNIP
Sbjct: 311 MRNKILLMKEHIDSTEAYVKILQNSRRNGLIHLMMLVNIGNYAITAGTVVVNLFGMNIPI 370
Query: 353 --YSWNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLV 391
YS D G++ W V+ ++F++ + YA+ K L+
Sbjct: 371 GLYSTPDIFGYVV-WAVV---ALCIVLFIVTVGYAKWKKLL 407
>At5g22830 magnesium transporter protein (GMN10)
Length = 459
Score = 90.5 bits (223), Expect = 1e-18
Identities = 89/361 (24%), Positives = 154/361 (42%), Gaps = 46/361 (12%)
Query: 33 ADGQGSLLDVDKYAIMHRVQIHARDLRILDPLL----SYPSTILGREKAIVLNLEHIKAI 88
A G S +++ ++ + RD+R +DP L S PS +L RE AI+LNL ++AI
Sbjct: 137 ATGAISTRKINRRQLLKSSGLRPRDIRSVDPSLFMTNSVPS-LLVREHAILLNLGSLRAI 195
Query: 89 ITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVSSQHDTEAAEEDESPF 148
+ VL+ D V+ L +P+++ G PF
Sbjct: 196 AMRDRVLIFDYNRRGGRAFVDTL---MPRLNPRSMNGGPSM-----------------PF 235
Query: 149 EFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLDRVRKLKSAMTRLTARV 208
E A+E AL + L R ++E L+ L +++++ L+ +R K + L +R
Sbjct: 236 ELEAVESALISRIQRLEQRLMDIEPRVQALLEVLPNRLTADILEELRISKQRLVELGSRA 295
Query: 209 QKVRDELEQLLDDDDDMADLYLSRKAGSSSPVSGSGAANWFGGSPTIGSKISRASRASLA 268
+R L LL+D ++ + + G + T+ +
Sbjct: 296 GALRQMLLDLLEDPHEIRRICI------------------MGRNCTLRRGDDDLECTLPS 337
Query: 269 TVRFDENDVEELEMLLEAYFMQIDGTFNKLTTLREYIDDTEDYINIQLDNHRNQLIQLEL 328
E + EE+EMLLE Y + + + L + + ED I + L + R ++ + EL
Sbjct: 338 DKLIAEEEEEEIEMLLENYLQRCESCHGQAERLLDSAKEMEDSIAVNLSSRRLEVSRFEL 397
Query: 329 FLSSGTVCLSFYSLVAAIFGMNI-PYSWNDNHGFMFKWVVIVSGVFSAIMFLLIIVYARK 387
L GT C++ +L+A IFGMN+ Y F I+ G +A+ F L+ Y +
Sbjct: 398 LLQVGTFCVAVGALIAGIFGMNLRSYLEEQASAFWLTTGGIIIG--AAVAFFLMYSYLSR 455
Query: 388 K 388
+
Sbjct: 456 R 456
>At4g17520 nuclear RNA binding protein A-like protein
Length = 360
Score = 39.3 bits (90), Expect = 0.004
Identities = 32/123 (26%), Positives = 58/123 (47%), Gaps = 8/123 (6%)
Query: 100 TDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVSSQHDTEAAEEDESP----FEFRALEV 155
T +++ PV EE + K ++ +Q G+G E ++++T A + +E P E
Sbjct: 165 TQDDITPVTEESTAVVDKNLTVEKQDGEG-EATDAKNETPAEKAEEKPEDKEMTLEEYEK 223
Query: 156 ALEAICSFLAARTTE---LEMAAYPALDELTSKISSRNLDRVRKLKSAMTRLTARVQKVR 212
LE L A E ++ A+ A+ +L+SK S+ + ++ R+T R +K R
Sbjct: 224 VLEEKKKALQATKVEERKVDTKAFEAMQQLSSKKSNNDEVFIKLGTEKDKRITEREEKTR 283
Query: 213 DEL 215
L
Sbjct: 284 KSL 286
>At1g03080 unknown protein
Length = 1744
Score = 34.7 bits (78), Expect = 0.093
Identities = 38/146 (26%), Positives = 63/146 (43%), Gaps = 12/146 (8%)
Query: 98 DPT-DENVVPVVEELQRRLPKVSSLHQQQGDGQEYVSSQHDTEAAEEDESPF---EFRAL 153
DP D N V+E L L K+S+LH D + V ++ E +E+E +
Sbjct: 1559 DPNKDANKRKVLERLNSDLQKLSNLHVAVEDLKIKVETEEKDEKGKENEYETIKGQINEA 1618
Query: 154 EVALEAICSFLAARTTELE--------MAAYPALDELTSKISSRNLDRVRKLKSAMTRLT 205
E ALE + S T+++ + LDE S R ++ R+ + RL
Sbjct: 1619 EEALEKLLSINRKLVTKVQNGFERSDGSKSSMDLDENESSRRRRISEQARRGSEKIGRLQ 1678
Query: 206 ARVQKVRDELEQLLDDDDDMADLYLS 231
+Q+++ L +L D +D A +S
Sbjct: 1679 LEIQRLQFLLLKLEGDREDRAKAKIS 1704
>At1g48870 hypothetical protein
Length = 593
Score = 34.3 bits (77), Expect = 0.12
Identities = 41/183 (22%), Positives = 69/183 (37%), Gaps = 10/183 (5%)
Query: 128 GQEYVSSQHDTEAAEEDE---SPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTS 184
G++ V + + E AE P V L R E ++ Y DE+TS
Sbjct: 25 GEQVVVEEEEVEEAEFQVWSGEPISVEERRVRFLKKMGLLEERCLERMVSDYS--DEVTS 82
Query: 185 KISSRNL-DRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRKAGSSSPVSGS 243
S +L D + + +ELE + DDDDD ++ + S S
Sbjct: 83 SSSDSSLCDSGLQCCVREENYGSTTSISDEELEGVDDDDDDSEEISSNASTSPSRSSSKK 142
Query: 244 GAANWFGGSPTIGSKIS----RASRASLATVRFDENDVEELEMLLEAYFMQIDGTFNKLT 299
A W + G K ++S +++ V+ N +E+ +I+G K+
Sbjct: 143 SAKKWLFNCFSAGVKDKDFKYKSSEETMSKVKVKTNKKSHVELSAAYMVQKINGHKGKIW 202
Query: 300 TLR 302
TL+
Sbjct: 203 TLK 205
>At1g51730 unknown protein
Length = 252
Score = 33.1 bits (74), Expect = 0.27
Identities = 42/182 (23%), Positives = 77/182 (42%), Gaps = 23/182 (12%)
Query: 75 EKAIVLNLEHIKAIITAEEVLLRDPTDENV-----VPVVEELQRRLPKVSSLHQQQGDGQ 129
++A +L+++ I+ I ++ +L++ ++ + ++ L S H Q D
Sbjct: 73 DEAPLLDVKSIRGIHVSDLTILKEKLEQEASENLGMAMIYTLVSSAKDWLSEHYGQDDAA 132
Query: 130 EYVSSQHDTEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDE--LTSKIS 187
E+ + EAA+EDE V LE ++ EL + + E LT+
Sbjct: 133 EFA----EVEAAKEDEVIVP-HGEPVTLETFLAWRERYEAELALERAKLMPESALTAP-K 186
Query: 188 SRNLDRVRKLKSAMTRLTARVQKVRDELEQ----------LLDDDDDMADLYLSRKAGSS 237
+ L + +S R T + DE E DD++DM + YL+ K+ SS
Sbjct: 187 EKKLTGRQWFESGRGRGTVVIADEEDEEEDEEDIDFEDEDFEDDEEDMLEHYLAEKSDSS 246
Query: 238 SP 239
+P
Sbjct: 247 AP 248
>At3g46750 hypothetical protein
Length = 415
Score = 32.0 bits (71), Expect = 0.60
Identities = 45/231 (19%), Positives = 89/231 (38%), Gaps = 30/231 (12%)
Query: 93 EVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVSSQHDTEAAEEDESPFEFRA 152
+V P D + +V E + K + Q+ + V TE E +P +
Sbjct: 204 DVKQEKPADSDTTTIVTESSEKTRKECTSQQEPISPSKTV-----TETVTEKLAP-GYAK 257
Query: 153 LEVALEAICSFLAARTTELEMAAYPA---LDELTSKISSRNLDRVRKLKSAMTRLTARVQ 209
+ A +AI T +++ A+P +E + ++ N + T++ +
Sbjct: 258 VSDATQAI-------TKKIQDMAFPEPTEREEEVNDVAEINTAGTNQPTGFNTKVWDKGV 310
Query: 210 KVRDELEQLLDDDDDMADLYLSRKAGSS-SPVSGSGAANWFGGSPTIGSKISRASRASLA 268
+++ + Q + +D D LSR + SP GS + FG + + + A +
Sbjct: 311 SMKEYISQKFEPSED--DRELSRVISKAISPRKGSSQTSSFGAATNMAPASNSADNKAPL 368
Query: 269 TVRFDENDVEELEMLLEAYFMQIDGTFNKLTTLREYIDDTEDYINIQLDNH 319
+E++ F+ T N LT L+ Y D + ++ +NH
Sbjct: 369 VANTNESE-----------FLFYSSTINNLTHLQLYSDAFDSAAVVEAENH 408
>At4g16660 HSP like protein
Length = 331
Score = 31.2 bits (69), Expect = 1.0
Identities = 17/61 (27%), Positives = 33/61 (53%)
Query: 86 KAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVSSQHDTEAAEEDE 145
K + T+ EV + T ++ V V ++ + PK+ + + + +E S+ EAA+E+E
Sbjct: 267 KPVFTSTEVYAKVFTLQDKVTKVNKIPKPKPKIEKVTKTENTTKEEEQSKSSDEAAKEEE 326
Query: 146 S 146
S
Sbjct: 327 S 327
>At1g59850 hypothetical protein
Length = 498
Score = 30.8 bits (68), Expect = 1.3
Identities = 51/207 (24%), Positives = 81/207 (38%), Gaps = 49/207 (23%)
Query: 98 DPTDENVVPVVEELQRRLPKVSSLHQQQGDGQE------------YVSSQHDTEA----- 140
D DE P VE+LQ+ LPK+ L + +G + V ++ +A
Sbjct: 169 DAADE---PDVEQLQKALPKIGKLLKSEGFKAKAELLGAIGTVIGAVGGRNSEKAVLDWL 225
Query: 141 ---AEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKI-SSRNLDRVRK 196
E S ++RA + A EA+ AR +E P + I SR D+V+
Sbjct: 226 LPNVSEFLSSDDWRARKAAAEAM-----ARVAMVEEELAPLYKKTCLGILESRRFDKVKL 280
Query: 197 LKSAMTRLTARVQKVRDELEQLLDDDDDMADLYLSRKAGSS--SPVSGSGAANWFGGSPT 254
++ M R +QL D ++++ S K+ SS S SG + G
Sbjct: 281 VRETMNRTLGL-------WKQLEGDSTEVSESSSSSKSASSGLSATSGKRSNTLKGKDRN 333
Query: 255 IGSKISRASRASLATVRFDENDVEELE 281
+ + +S S NDVE L+
Sbjct: 334 LNTPLSSKS-----------NDVEPLD 349
>At1g24380 hypothetical protein
Length = 273
Score = 30.8 bits (68), Expect = 1.3
Identities = 22/83 (26%), Positives = 42/83 (50%), Gaps = 6/83 (7%)
Query: 74 REKAIVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPK--VSSLHQQQGDGQEY 131
R+K V ++ I+ A+ + DP+ + E +R L K ++ H++ G+G +Y
Sbjct: 162 RQKIEVARMKEENKILFADLNSISDPSSSAYIE--NERKRILEKRAQTNQHEEDGEGSQY 219
Query: 132 VSSQHDTEAAEEDESPFEFRALE 154
SQ+ A+ ESPF + ++
Sbjct: 220 HGSQY--RASHYQESPFHGKQVQ 240
>At4g14150 kinesin like protein
Length = 1662
Score = 30.4 bits (67), Expect = 1.8
Identities = 20/78 (25%), Positives = 38/78 (48%), Gaps = 2/78 (2%)
Query: 154 EVALEAICSFLAARTTELE--MAAYPALDELTSKISSRNLDRVRKLKSAMTRLTARVQKV 211
E+ALE C+ A+ T+L + Y E + I D++ +L+S M + ++ +
Sbjct: 854 EMALEEFCTKQASEITQLNRLVQQYKHERECNAIIGQTREDKIIRLESLMDGVLSKEDFL 913
Query: 212 RDELEQLLDDDDDMADLY 229
+E LL + + D+Y
Sbjct: 914 DEEFASLLHEHKLLKDMY 931
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,461,011
Number of Sequences: 26719
Number of extensions: 356329
Number of successful extensions: 1444
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 1382
Number of HSP's gapped (non-prelim): 50
length of query: 393
length of database: 11,318,596
effective HSP length: 101
effective length of query: 292
effective length of database: 8,619,977
effective search space: 2517033284
effective search space used: 2517033284
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0071.7


