

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0069.6
(167 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
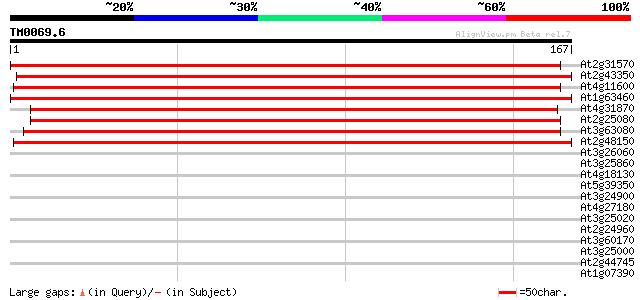
Score E
Sequences producing significant alignments: (bits) Value
At2g31570 putative glutathione peroxidase 256 5e-69
At2g43350 putative glutathione peroxidase 249 5e-67
At4g11600 phospholipid hydroperoxide glutathione peroxidase 248 8e-67
At1g63460 glutathione peroxidase like protein 234 2e-62
At4g31870 glutathione peroxidase - like protein 228 9e-61
At2g25080 putative glutathione peroxidase 228 2e-60
At3g63080 glutathione peroxidase -like protein 209 4e-55
At2g48150 glutathione peroxidase like protein 201 2e-52
At3g26060 putative peroxiredoxin 34 0.032
At3g25860 dihydrolipoamide S-acetyltransferase 29 1.3
At4g18130 phytochrome E 28 1.8
At5g39350 putative protein 28 2.3
At3g24900 leucine-rich repeat disease resistance protein, putative 28 3.0
At4g27180 heavy chain polypeptide of kinesin-like protein (katB) 27 3.9
At3g25020 disease resistance protein, putative 27 5.1
At2g24960 hypothetical protein 27 5.1
At3g60170 putative protein 27 6.7
At3g25000 disease resistance protein, putative, 3' partial 27 6.7
At2g44745 WRKY transcription factor 12 (WRKY12) 27 6.7
At1g07390 disease resistance protein, putative 27 6.7
>At2g31570 putative glutathione peroxidase
Length = 169
Score = 256 bits (653), Expect = 5e-69
Identities = 115/164 (70%), Positives = 141/164 (85%)
Query: 1 MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKS 60
MA+++ KS+YDFTVKDI GNDVSL QY GK L++VNVAS+CGLT NYKELN+LYEKYK
Sbjct: 1 MADESPKSIYDFTVKDIGGNDVSLDQYKGKTLLVVNVASKCGLTDANYKELNVLYEKYKE 60
Query: 61 KGLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQK 120
+GLEILAFPCNQF GQEPG N+EIQ VCTRFK+EFP+FDKV+VNGKN PL+K+LK +K
Sbjct: 61 QGLEILAFPCNQFLGQEPGNNEEIQQTVCTRFKAEFPIFDKVDVNGKNTAPLYKYLKAEK 120
Query: 121 GGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
GG+ D IKWNFTKFLV+ +GKV++RY+P TSP++ EKD++ L
Sbjct: 121 GGLLIDAIKWNFTKFLVSPDGKVLQRYSPRTSPLQFEKDIQTAL 164
>At2g43350 putative glutathione peroxidase
Length = 206
Score = 249 bits (636), Expect = 5e-67
Identities = 114/165 (69%), Positives = 143/165 (86%)
Query: 3 EQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKG 62
EQ+S S+Y+ +VKDI G DVSLS+++GKVL+IVNVAS+CGLT NYKE+NILY KYK++G
Sbjct: 42 EQSSTSIYNISVKDIEGKDVSLSKFTGKVLLIVNVASKCGLTHGNYKEMNILYAKYKTQG 101
Query: 63 LEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGG 122
EILAFPCNQF QEPG+N EI++ VC FK+EFP+FDK+EVNGKN PL+ FLK+QKGG
Sbjct: 102 FEILAFPCNQFGSQEPGSNMEIKETVCNIFKAEFPIFDKIEVNGKNTCPLYNFLKEQKGG 161
Query: 123 IFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQSS 167
+FGD IKWNF KFLV+++G VV+RYAPTTSP++IEKD+ KLL S+
Sbjct: 162 LFGDAIKWNFAKFLVDRQGNVVDRYAPTTSPLEIEKDIVKLLASA 206
>At4g11600 phospholipid hydroperoxide glutathione peroxidase
Length = 232
Score = 248 bits (634), Expect = 8e-67
Identities = 116/163 (71%), Positives = 134/163 (82%)
Query: 2 AEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSK 61
A KSLYDFTVKD +GNDV LS Y GKVL+IVNVASQCGLT +NY EL LYEKYK
Sbjct: 66 ASSEPKSLYDFTVKDAKGNDVDLSIYKGKVLLIVNVASQCGLTNSNYTELAQLYEKYKGH 125
Query: 62 GLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKG 121
G EILAFPCNQF QEPGTN+EI CTRFK+E+P+FDKV+VNG A P++KFLK KG
Sbjct: 126 GFEILAFPCNQFGNQEPGTNEEIVQFACTRFKAEYPIFDKVDVNGDKAAPVYKFLKSSKG 185
Query: 122 GIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
G+FGDGIKWNF KFLV+K+G VV+R+APTTSP+ IEKD++KLL
Sbjct: 186 GLFGDGIKWNFAKFLVDKDGNVVDRFAPTTSPLSIEKDVKKLL 228
>At1g63460 glutathione peroxidase like protein
Length = 167
Score = 234 bits (596), Expect = 2e-62
Identities = 106/167 (63%), Positives = 137/167 (81%)
Query: 1 MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKS 60
MA + +S+Y+ +++D +GN+++LSQY KVL+IVNVAS+CG+T +NY ELN LY +YK
Sbjct: 1 MATKEPESVYELSIEDAKGNNLALSQYKDKVLLIVNVASKCGMTNSNYTELNELYNRYKD 60
Query: 61 KGLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQK 120
KGLEILAFPCNQF +EPGTND+I D VCTRFKSEFP+F+K+EVNG+NA PL+KFLK K
Sbjct: 61 KGLEILAFPCNQFGDEEPGTNDQITDFVCTRFKSEFPIFNKIEVNGENASPLYKFLKKGK 120
Query: 121 GGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQSS 167
GIFGD I+WNF KFLV+K G+ V+RY PTTSP+ +E D++ LL S
Sbjct: 121 WGIFGDDIQWNFAKFLVDKNGQAVQRYYPTTSPLTLEHDIKNLLNIS 167
>At4g31870 glutathione peroxidase - like protein
Length = 230
Score = 228 bits (582), Expect = 9e-61
Identities = 107/157 (68%), Positives = 128/157 (81%)
Query: 7 KSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLEIL 66
KS++DFTVKDI GNDVSL ++ GK L+IVNVAS+CGLT +NY EL+ LYEKYK++G EIL
Sbjct: 74 KSVHDFTVKDIDGNDVSLDKFKGKPLLIVNVASRCGLTSSNYSELSQLYEKYKNQGFEIL 133
Query: 67 AFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIFGD 126
AFPCNQF GQEPG+N EI+ CTRFK+EFP+FDKV+VNG + P++KFLK GG GD
Sbjct: 134 AFPCNQFGGQEPGSNPEIKQFACTRFKAEFPIFDKVDVNGPSTAPIYKFLKSNAGGFLGD 193
Query: 127 GIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKL 163
IKWNF KFLV+K+GKVVERY PTTSP +IE KL
Sbjct: 194 IIKWNFEKFLVDKKGKVVERYPPTTSPFQIEVPNSKL 230
>At2g25080 putative glutathione peroxidase
Length = 236
Score = 228 bits (580), Expect = 2e-60
Identities = 103/158 (65%), Positives = 132/158 (83%)
Query: 7 KSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLEIL 66
K+++DFTVKDI G DV+L+++ GKV++IVNVAS+CGLT +NY EL+ LYEKYK++G EIL
Sbjct: 77 KTVHDFTVKDIDGKDVALNKFKGKVMLIVNVASRCGLTSSNYSELSHLYEKYKTQGFEIL 136
Query: 67 AFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIFGD 126
AFPCNQF QEPG+N EI+ CTRFK+EFP+FDKV+VNG + P+++FLK GG G
Sbjct: 137 AFPCNQFGFQEPGSNSEIKQFACTRFKAEFPIFDKVDVNGPSTAPIYEFLKSNAGGFLGG 196
Query: 127 GIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
IKWNF KFL++K+GKVVERY PTTSP +IEKD++KLL
Sbjct: 197 LIKWNFEKFLIDKKGKVVERYPPTTSPFQIEKDIQKLL 234
>At3g63080 glutathione peroxidase -like protein
Length = 173
Score = 209 bits (533), Expect = 4e-55
Identities = 94/160 (58%), Positives = 121/160 (74%)
Query: 5 TSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLE 64
+ KS++ FTVKD G +V LS Y GKVL++VNVAS+CG T++NY +L LY KYK +G
Sbjct: 10 SEKSIHQFTVKDSSGKEVDLSVYQGKVLLVVNVASKCGFTESNYTQLTELYRKYKDQGFV 69
Query: 65 ILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIF 124
+LAFPCNQF QEPGT++E CTRFK+E+PVF KV VNG+NA P++KFLK +K
Sbjct: 70 VLAFPCNQFLSQEPGTSEEAHQFACTRFKAEYPVFQKVRVNGQNAAPVYKFLKSKKPSFL 129
Query: 125 GDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
G IKWNFTKFLV K+G+V++RY T SP+ I+KD+EK L
Sbjct: 130 GSRIKWNFTKFLVGKDGQVIDRYGTTVSPLSIQKDIEKAL 169
>At2g48150 glutathione peroxidase like protein
Length = 170
Score = 201 bits (511), Expect = 2e-52
Identities = 93/166 (56%), Positives = 120/166 (72%)
Query: 2 AEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSK 61
A +S++ FTVKD G D+++S Y GKVL+IVNVAS+CG T+TNY +L LY KYK +
Sbjct: 5 ASVPERSVHQFTVKDSSGKDLNMSIYQGKVLLIVNVASKCGFTETNYTQLTELYRKYKDQ 64
Query: 62 GLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKG 121
EILAFPCNQF QEPGT+ E + C RFK+E+PVF KV VNG+NA P++KFLK K
Sbjct: 65 DFEILAFPCNQFLYQEPGTSQEAHEFACERFKAEYPVFQKVRVNGQNAAPIYKFLKASKP 124
Query: 122 GIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQSS 167
G IKWNFTKFLV K+G V++RY +P+ IEKD++K L+ +
Sbjct: 125 TFLGSRIKWNFTKFLVGKDGLVIDRYGTMVTPLSIEKDIKKALEDA 170
>At3g26060 putative peroxiredoxin
Length = 216
Score = 34.3 bits (77), Expect = 0.032
Identities = 19/60 (31%), Positives = 31/60 (51%), Gaps = 7/60 (11%)
Query: 11 DFTVKDIRGNDVSLSQYSGKVLII----VNVASQCGLTQTNYKELNILYEKYKSKGLEIL 66
DFT+KD G VSL +Y GK +++ + C +++ YEK+K G E++
Sbjct: 77 DFTLKDQNGKPVSLKKYKGKPVVLYFYPADETPGCTKQACAFRD---SYEKFKKAGAEVI 133
>At3g25860 dihydrolipoamide S-acetyltransferase
Length = 480
Score = 28.9 bits (63), Expect = 1.3
Identities = 24/84 (28%), Positives = 35/84 (41%), Gaps = 10/84 (11%)
Query: 47 NYKELNILYEKYKSKGL---EILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVE 103
N L+ LYEK K KG+ +LA Q P N +D + S + V
Sbjct: 291 NTDALDALYEKVKPKGVTMTALLAKAAGMALAQHPVVNASCKDGKSFSYNSSINIAVAVA 350
Query: 104 VNG-------KNAEPLFKFLKDQK 120
+NG ++A+ L +L QK
Sbjct: 351 INGGLITPVLQDADKLDLYLLSQK 374
>At4g18130 phytochrome E
Length = 1112
Score = 28.5 bits (62), Expect = 1.8
Identities = 22/62 (35%), Positives = 26/62 (41%), Gaps = 4/62 (6%)
Query: 96 FPVFDKVEVNGKNAEPLFKFLKDQKGGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMK 155
F VF KV K + L KFL GI GD + + NKEGK +E
Sbjct: 784 FGVFCKV----KCQDSLTKFLISLYQGIAGDNVPESSLVEFFNKEGKYIEASLTANKSTN 839
Query: 156 IE 157
IE
Sbjct: 840 IE 841
>At5g39350 putative protein
Length = 677
Score = 28.1 bits (61), Expect = 2.3
Identities = 12/28 (42%), Positives = 20/28 (70%)
Query: 34 IVNVASQCGLTQTNYKELNILYEKYKSK 61
+V+V S+CG ++ +K N + EK+KSK
Sbjct: 461 LVHVYSKCGTLESAHKIFNGIQEKHKSK 488
>At3g24900 leucine-rich repeat disease resistance protein, putative
Length = 884
Score = 27.7 bits (60), Expect = 3.0
Identities = 19/74 (25%), Positives = 31/74 (41%)
Query: 7 KSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLEIL 66
++L T+ D+ N S + L ++ + L N+ ++ YE LE+L
Sbjct: 168 RNLRKLTILDVSHNHFSGTLNPNSSLFELHNLAYLDLGSNNFTSSSLPYEFGNLNKLELL 227
Query: 67 AFPCNQFAGQEPGT 80
N F GQ P T
Sbjct: 228 DVSSNSFFGQVPPT 241
>At4g27180 heavy chain polypeptide of kinesin-like protein (katB)
Length = 745
Score = 27.3 bits (59), Expect = 3.9
Identities = 24/72 (33%), Positives = 36/72 (49%), Gaps = 6/72 (8%)
Query: 1 MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVA-----SQCGLTQTNYKELNILY 55
M EQ+S+S + FT+K I G + S Q VL ++++A S+ G T KE +
Sbjct: 596 MNEQSSRSHFVFTLK-ISGFNESTEQQVQGVLNLIDLAGSERLSKSGSTGDRLKETQAIN 654
Query: 56 EKYKSKGLEILA 67
+ S G I A
Sbjct: 655 KSLSSLGDVIFA 666
>At3g25020 disease resistance protein, putative
Length = 890
Score = 26.9 bits (58), Expect = 5.1
Identities = 22/83 (26%), Positives = 35/83 (41%), Gaps = 5/83 (6%)
Query: 3 EQTSKSLYDFTVKDIRGNDVSLSQYSGKV-----LIIVNVASQCGLTQTNYKELNILYEK 57
E T + ++ +R DVS + +SG + L ++ L N+ ++ YE
Sbjct: 158 ELTGSLSFVRNLRKLRVLDVSYNHFSGILNPNSSLFELHHLIYLNLRYNNFTSSSLPYEF 217
Query: 58 YKSKGLEILAFPCNQFAGQEPGT 80
LE+L N F GQ P T
Sbjct: 218 GNLNKLEVLDVSSNSFFGQVPPT 240
>At2g24960 hypothetical protein
Length = 797
Score = 26.9 bits (58), Expect = 5.1
Identities = 10/36 (27%), Positives = 20/36 (54%)
Query: 112 LFKFLKDQKGGIFGDGIKWNFTKFLVNKEGKVVERY 147
L K+ KD + + DG W+ T+ +++ + V + Y
Sbjct: 230 LLKYYKDMEAILKEDGFSWDETRLMISADDAVWDSY 265
>At3g60170 putative protein
Length = 1339
Score = 26.6 bits (57), Expect = 6.7
Identities = 10/26 (38%), Positives = 18/26 (68%)
Query: 50 ELNILYEKYKSKGLEILAFPCNQFAG 75
EL I Y + K++ L+++AF + +AG
Sbjct: 1166 ELGIFYRRRKNRSLKLMAFTDSDYAG 1191
>At3g25000 disease resistance protein, putative, 3' partial
Length = 667
Score = 26.6 bits (57), Expect = 6.7
Identities = 20/72 (27%), Positives = 32/72 (43%), Gaps = 5/72 (6%)
Query: 14 VKDIRGNDVSLSQYSGKV-----LIIVNVASQCGLTQTNYKELNILYEKYKSKGLEILAF 68
++ +R DVS + +SG + L ++ L N+ ++ YE LE+L
Sbjct: 200 LRKLRVLDVSYNHFSGILNPNSSLFELHHIIYLNLRYNNFTSSSLPYEFGNLNKLEVLDV 259
Query: 69 PCNQFAGQEPGT 80
N F GQ P T
Sbjct: 260 SSNSFFGQVPPT 271
>At2g44745 WRKY transcription factor 12 (WRKY12)
Length = 218
Score = 26.6 bits (57), Expect = 6.7
Identities = 17/65 (26%), Positives = 30/65 (46%), Gaps = 1/65 (1%)
Query: 100 DKVEVNGKNAEPLFKFLKDQKGGIFGDGIKW-NFTKFLVNKEGKVVERYAPTTSPMKIEK 158
+KV++ K EP F F + DG KW + + +V Y T + +++K
Sbjct: 120 NKVKIRRKLREPRFCFQTKSDVDVLDDGYKWRKYGQKVVKNSLHPRSYYRCTHNNCRVKK 179
Query: 159 DLEKL 163
+E+L
Sbjct: 180 RVERL 184
>At1g07390 disease resistance protein, putative
Length = 976
Score = 26.6 bits (57), Expect = 6.7
Identities = 19/64 (29%), Positives = 32/64 (49%), Gaps = 1/64 (1%)
Query: 21 DVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEK-YKSKGLEILAFPCNQFAGQEPG 79
D+S + +SG + VN S L N + ++ +K+ GLE+L N F+G+
Sbjct: 628 DISHNSFSGSIPRNVNFPSLRELRLQNNEFTGLVPGNLFKAAGLEVLDLRNNNFSGKILN 687
Query: 80 TNDE 83
T D+
Sbjct: 688 TIDQ 691
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.135 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,800,834
Number of Sequences: 26719
Number of extensions: 163614
Number of successful extensions: 288
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 274
Number of HSP's gapped (non-prelim): 23
length of query: 167
length of database: 11,318,596
effective HSP length: 92
effective length of query: 75
effective length of database: 8,860,448
effective search space: 664533600
effective search space used: 664533600
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0069.6


