

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
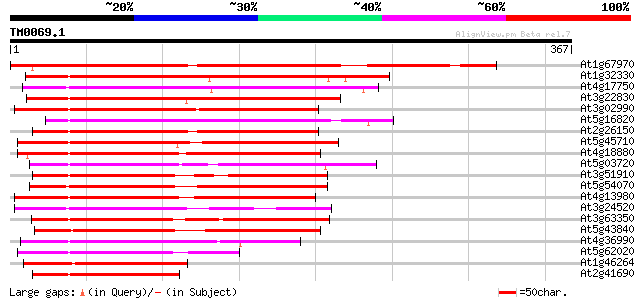
Query= TM0069.1
(367 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g67970 putative heat shock transcription factor 277 6e-75
At1g32330 heat shock transcription factor HSF8 like protein 210 9e-55
At4g17750 heat shock transcription factor HSF1 202 2e-52
At3g22830 putative heat shock protein 195 3e-50
At3g02990 putative heat shock transcription factor 190 1e-48
At5g16820 Heat Shock Factor 3 187 6e-48
At2g26150 heat shock transcription factor like protein 175 3e-44
At5g45710 heat shock transcription factor 172 2e-43
At4g18880 heat shock transcription factor 21 (AtHSF21) 171 4e-43
At5g03720 heat shock transcription factor -like protein 163 2e-40
At3g51910 putative heat shock transcription factor 163 2e-40
At5g54070 putative protein 157 7e-39
At4g13980 unknown protein 155 3e-38
At3g24520 heat shock transcription factor HSF1, putative 154 6e-38
At3g63350 heat shock transcription factor-like protein 154 7e-38
At5g43840 heat shock transcription factor-like protein 150 1e-36
At4g36990 heat shock transcription factor HSF4 126 2e-29
At5g62020 heat shock factor 6 120 1e-27
At1g46264 heat shock transcription factor, putative 120 1e-27
At2g41690 putative heat shock transcription factor 114 6e-26
>At1g67970 putative heat shock transcription factor
Length = 374
Score = 277 bits (709), Expect = 6e-75
Identities = 157/323 (48%), Positives = 204/323 (62%), Gaps = 32/323 (9%)
Query: 1 MVKSKENASGSSS---VAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLL 57
MVKS + GSSS VAPFL KCYDMV+D +TDSIISW+ + ++FVI D T FSV LL
Sbjct: 1 MVKSTDGGGGSSSSSSVAPFLRKCYDMVDDSTTDSIISWSPSADNSFVILDTTVFSVQLL 60
Query: 58 PTYFKHNNFASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQK 117
P YFKH+NF+SFIRQLNIYGFRKVD DRWEFAN+ FVRGQK LLKN+ RRK+ ++Q K
Sbjct: 61 PKYFKHSNFSSFIRQLNIYGFRKVDADRWEFANDGFVRGQKDLLKNVIRRKNVQSSEQSK 120
Query: 118 ALPEHNNSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQ 177
+ S + GL KEV+ LK D+ L QEL+ + Q+ E ++KML L DR+Q
Sbjct: 121 -----HESTSTTYAQEKSGLWKEVDILKGDKQVLAQELIKVRQYQEVTDTKMLHLEDRVQ 175
Query: 178 GMEKHQQQMLSFLVMVVQSPGFMVQLLHPKE-NSWRLAEAGNMFDPGKEDDKPVASDGMI 236
GME+ QQ+MLSFLVMV+++P +VQLL PKE N+WR A G I
Sbjct: 176 GMEESQQEMLSFLVMVMKNPSLLVQLLQPKEKNTWRKAGEG----------------AKI 219
Query: 237 VQYKPPVGEKRKHVIP-IPLSPGFDRQPEPELSADRLKDLCISSEFLKVLLDEKVSPLDN 295
V+ GE + +P + P D + +++ + D +++ LK LDE N
Sbjct: 220 VEEVTDEGESNSYGLPLVTYQPPSDNNGTAKSNSNDVNDFLRNADMLKFCLDE------N 273
Query: 296 HSPFLLPDLPDDGSWEQLFLGSP 318
H P ++PDL DDG+WE+L L SP
Sbjct: 274 HVPLIIPDLYDDGAWEKLLLLSP 296
>At1g32330 heat shock transcription factor HSF8 like protein
Length = 482
Score = 210 bits (535), Expect = 9e-55
Identities = 115/250 (46%), Positives = 158/250 (63%), Gaps = 13/250 (5%)
Query: 11 SSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFI 70
S++ PFL+K YDMV+D +TDSI+SW+ + ++F++ F+ LLP FKHNNF+SF+
Sbjct: 32 SNAPPPFLSKTYDMVDDHNTDSIVSWSA-NNNSFIVWKPPEFARDLLPKNFKHNNFSSFV 90
Query: 71 RQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPS- 129
RQLN YGFRKVD DRWEFANE F+RGQKHLL++I RRK H Q +H+N S
Sbjct: 91 RQLNTYGFRKVDPDRWEFANEGFLRGQKHLLQSITRRKPAHGQGQGHQRSQHSNGQNSSV 150
Query: 130 ---REAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQM 186
E GL +EVE LK D+N LMQELV L Q +S ++++ + RLQGME QQQ+
Sbjct: 151 SACVEVGKFGLEEEVERLKRDKNVLMQELVRLRQQQQSTDNQLQTMVQRLQGMENRQQQL 210
Query: 187 LSFLVMVVQSPGFMVQLLHPK----ENSWRLAEAGN----MFDPGKEDDKPVASDGMIVQ 238
+SFL VQSP F+ Q L + E++ R+++ D ++ DG IV+
Sbjct: 211 MSFLAKAVQSPHFLSQFLQQQNQQNESNRRISDTSKKRRFKRDGIVRNNDSATPDGQIVK 270
Query: 239 YKPPVGEKRK 248
Y+PP+ E+ K
Sbjct: 271 YQPPMHEQAK 280
>At4g17750 heat shock transcription factor HSF1
Length = 495
Score = 202 bits (514), Expect = 2e-52
Identities = 114/253 (45%), Positives = 150/253 (59%), Gaps = 21/253 (8%)
Query: 9 SGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFAS 68
+ +S PFL+K YDMVED +TD+I+SW+ P+ ++F++ D FS LLP YFKHNNF+S
Sbjct: 45 NANSLPPPFLSKTYDMVEDPATDAIVSWS-PTNNSFIVWDPPEFSRDLLPKYFKHNNFSS 103
Query: 69 FIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEP 128
F+RQLN YGFRKVD DRWEFANE F+RGQKHLLK I RRK + P+ +
Sbjct: 104 FVRQLNTYGFRKVDPDRWEFANEGFLRGQKHLLKKISRRKSVQGHGSSSSNPQSQQLSQG 163
Query: 129 SR---------EAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGM 179
E GL +EVE LK D+N LMQELV L Q ++ ++K+ VL LQ M
Sbjct: 164 QGSMAALSSCVEVGKFGLEEEVEQLKRDKNVLMQELVKLRQQQQTTDNKLQVLVKHLQVM 223
Query: 180 EKHQQQMLSFLVMVVQSPGFMVQLLHPKENSWRLAEAGNMFDPGKEDDKPV--------- 230
E+ QQQ++SFL VQ+P F+ Q + + +S N +ED
Sbjct: 224 EQRQQQIMSFLAKAVQNPTFLSQFIQKQTDSNMHVTEANKKRRLREDSTAATESNSHSHS 283
Query: 231 --ASDGMIVQYKP 241
ASDG IV+Y+P
Sbjct: 284 LEASDGQIVKYQP 296
>At3g22830 putative heat shock protein
Length = 406
Score = 195 bits (496), Expect = 3e-50
Identities = 100/207 (48%), Positives = 140/207 (67%), Gaps = 3/207 (1%)
Query: 12 SSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIR 71
S PFL K YD+VED T+ ++SW++ S ++F++ D AFSVTLLP +FKHNNF+SF+R
Sbjct: 57 SGPPPFLTKTYDLVEDSRTNHVVSWSK-SNNSFIVWDPQAFSVTLLPRFFKHNNFSSFVR 115
Query: 72 QLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTD--QQKALPEHNNSDEPS 129
QLN YGFRKV+ DRWEFANE F+RGQKHLLKNIRRRK + ++ QQ E + D
Sbjct: 116 QLNTYGFRKVNPDRWEFANEGFLRGQKHLLKNIRRRKTSNNSNQMQQPQSSEQQSLDNFC 175
Query: 130 REAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQMLSF 189
E +GL E+++L+ D+ LM ELV L Q +S + + ++ ++L+ E Q+QM+SF
Sbjct: 176 IEVGRYGLDGEMDSLRRDKQVLMMELVRLRQQQQSTKMYLTLIEEKLKKTESKQKQMMSF 235
Query: 190 LVMVVQSPGFMVQLLHPKENSWRLAEA 216
L +Q+P F+ QL+ KE + EA
Sbjct: 236 LARAMQNPDFIQQLVEQKEKRKEIEEA 262
>At3g02990 putative heat shock transcription factor
Length = 468
Score = 190 bits (482), Expect = 1e-48
Identities = 98/199 (49%), Positives = 132/199 (66%), Gaps = 2/199 (1%)
Query: 4 SKENASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKH 63
+K + + SS+ PFL+K YDMV+D TD ++SW+ ++FV+ ++ F+ LP YFKH
Sbjct: 11 AKSSTAVMSSIPPFLSKTYDMVDDPLTDDVVSWSS-GNNSFVVWNVPEFAKQFLPKYFKH 69
Query: 64 NNFASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHN 123
NNF+SF+RQLN YGFRKVD DRWEFANE F+RGQK +LK+I RRK V Q+ +H
Sbjct: 70 NNFSSFVRQLNTYGFRKVDPDRWEFANEGFLRGQKQILKSIVRRKPAQVQPPQQPQVQH- 128
Query: 124 NSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQ 183
+S E GL +EVE L+ D+N LMQELV L Q + E + + ++ ME+ Q
Sbjct: 129 SSVGACVEVGKFGLEEEVERLQRDKNVLMQELVRLRQQQQVTEHHLQNVGQKVHVMEQRQ 188
Query: 184 QQMLSFLVMVVQSPGFMVQ 202
QQM+SFL VQSPGF+ Q
Sbjct: 189 QQMMSFLAKAVQSPGFLNQ 207
>At5g16820 Heat Shock Factor 3
Length = 447
Score = 187 bits (476), Expect = 6e-48
Identities = 103/242 (42%), Positives = 143/242 (58%), Gaps = 21/242 (8%)
Query: 24 MVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNIYGFRKVDT 83
MV+D T+ ++SW+ ++FV+ FS LLP YFKHNNF+SF+RQLN YGFRKVD
Sbjct: 1 MVDDPLTNEVVSWSS-GNNSFVVWSAPEFSKVLLPKYFKHNNFSSFVRQLNTYGFRKVDP 59
Query: 84 DRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSREAPNHGLRKEVEN 143
DRWEFANE F+RG+K LLK+I RRK HV Q+ ++S E G+ +EVE
Sbjct: 60 DRWEFANEGFLRGRKQLLKSIVRRKPSHVQQNQQQTQVQSSSVGACVEVGKFGIEEEVER 119
Query: 144 LKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQMLSFLVMVVQSPGFMVQL 203
LK D+N LMQELV L Q ++ E+++ + ++Q ME+ QQQM+SFL VQSPGF+ QL
Sbjct: 120 LKRDKNVLMQELVRLRQQQQATENQLQNVGQKVQVMEQRQQQMMSFLAKAVQSPGFLNQL 179
Query: 204 LHPKENSWRLAEAGNMFDPGKEDDKPVASD--------------GMIVQYKPPVGEKRKH 249
+ N GN PG + + D IV+Y+P + E ++
Sbjct: 180 VQQNNND------GNRQIPGSNKKRRLPVDEQENRGDNVANGLNRQIVRYQPSINEAAQN 233
Query: 250 VI 251
++
Sbjct: 234 ML 235
>At2g26150 heat shock transcription factor like protein
Length = 345
Score = 175 bits (444), Expect = 3e-44
Identities = 90/187 (48%), Positives = 122/187 (65%), Gaps = 6/187 (3%)
Query: 16 PFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNI 75
PFL K Y+MVED +TD+++SW+ ++FV+ D FS TLLP YFKH+NF+SFIRQLN
Sbjct: 44 PFLTKTYEMVEDPATDTVVSWSN-GRNSFVVWDSHKFSTTLLPRYFKHSNFSSFIRQLNT 102
Query: 76 YGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSREAPNH 135
YGFRK+D DRWEFANE F+ GQKHLLKNI+RR++ + + + S E +
Sbjct: 103 YGFRKIDPDRWEFANEGFLAGQKHLLKNIKRRRNMGLQNVNQ-----QGSGMSCVEVGQY 157
Query: 136 GLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQMLSFLVMVVQ 195
G EVE LK D L+ E+V L Q S++S++ + RL EK QQQM++FL +
Sbjct: 158 GFDGEVERLKRDHGVLVAEVVRLRQQQHSSKSQVAAMEQRLLVTEKRQQQMMTFLAKALN 217
Query: 196 SPGFMVQ 202
+P F+ Q
Sbjct: 218 NPNFVQQ 224
>At5g45710 heat shock transcription factor
Length = 345
Score = 172 bits (437), Expect = 2e-43
Identities = 87/212 (41%), Positives = 133/212 (62%), Gaps = 10/212 (4%)
Query: 6 ENASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNN 65
EN GSSS+ PFL K Y+MV+D S+DS+++W+E + +F++ + FS LLP +FKH N
Sbjct: 3 ENNGGSSSLPPFLTKTYEMVDDSSSDSVVAWSE-NNKSFIVKNPAEFSRDLLPRFFKHKN 61
Query: 66 FASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRK--HPHVTDQQKALPEHN 123
F+SFIRQLN YGFRKVD ++WEF N++FVRG+ +L+KNI RRK H H +A
Sbjct: 62 FSSFIRQLNTYGFRKVDPEKWEFLNDDFVRGRPYLMKNIHRRKPVHSHSLVNLQA----- 116
Query: 124 NSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQ 183
P E+ + ++E LK+++ L+ EL + Q + E ++ L DRLQ ME+HQ
Sbjct: 117 --QNPLTESERRSMEDQIERLKNEKEGLLAELQNQEQERKEFELQVTTLKDRLQHMEQHQ 174
Query: 184 QQMLSFLVMVVQSPGFMVQLLHPKENSWRLAE 215
+ +++++ V+ PG + L + + R E
Sbjct: 175 KSIVAYVSQVLGKPGLSLNLENHERRKRRFQE 206
>At4g18880 heat shock transcription factor 21 (AtHSF21)
Length = 401
Score = 171 bits (434), Expect = 4e-43
Identities = 90/200 (45%), Positives = 128/200 (64%), Gaps = 7/200 (3%)
Query: 6 ENASG--SSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKH 63
EN G SSS+ PFL K Y+MV+D S+DSI+SW++ S +F++ + FS LLP +FKH
Sbjct: 3 ENNHGVSSSSLPPFLTKTYEMVDDSSSDSIVSWSQ-SNKSFIVWNPPEFSRDLLPRFFKH 61
Query: 64 NNFASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHN 123
NNF+SFIRQLN YGFRK D ++WEFAN++FVRGQ HL+KNI RRK H +LP
Sbjct: 62 NNFSSFIRQLNTYGFRKADPEQWEFANDDFVRGQPHLMKNIHRRKPVH----SHSLPNLQ 117
Query: 124 NSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQ 183
P ++ + ++E L ++ L++EL + E E ++ L +RLQ MEK Q
Sbjct: 118 AQLNPLTDSERVRMNNQIERLTKEKEGLLEELHKQDEEREVFEMQVKELKERLQHMEKRQ 177
Query: 184 QQMLSFLVMVVQSPGFMVQL 203
+ M+SF+ V++ PG + L
Sbjct: 178 KTMVSFVSQVLEKPGLALNL 197
>At5g03720 heat shock transcription factor -like protein
Length = 476
Score = 163 bits (412), Expect = 2e-40
Identities = 89/231 (38%), Positives = 139/231 (59%), Gaps = 12/231 (5%)
Query: 14 VAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQL 73
+ PFL+K +D+V+D + D +ISW +G +FV+ D F+ +LP FKHNNF+SF+RQL
Sbjct: 117 IPPFLSKTFDLVDDPTLDPVISWGL-TGASFVVWDPLEFARIILPRNFKHNNFSSFVRQL 175
Query: 74 NIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSREAP 133
N YGFRK+DTD+WEFANE F+RG+KHLLKNI RR+ P ++Q + P+
Sbjct: 176 NTYGFRKIDTDKWEFANEAFLRGKKHLLKNIHRRRSPQ-SNQTCCSSTSQSQGSPTE--- 231
Query: 134 NHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQMLSFLVMV 193
+ E+E L+ +R +LM+E+V L Q + ++ RL+ E+ Q+Q+LSFL +
Sbjct: 232 ---VGGEIEKLRKERRALMEEMVELQQQSRGTARHVDTVNQRLKAAEQRQKQLLSFLAKL 288
Query: 194 VQSPGFMVQLLH----PKENSWRLAEAGNMFDPGKEDDKPVASDGMIVQYK 240
Q+ GF+ +L + K + L +A F + + + G +V+Y+
Sbjct: 289 FQNRGFLERLKNFKGKEKGGALGLEKARKKFIKHHQQPQDSPTGGEVVKYE 339
>At3g51910 putative heat shock transcription factor
Length = 272
Score = 163 bits (412), Expect = 2e-40
Identities = 84/193 (43%), Positives = 121/193 (62%), Gaps = 21/193 (10%)
Query: 16 PFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNI 75
PFL K ++MV+D +TD I+SW G +FV+ D+ +FS LLP +FKH+NF+SFIRQLN
Sbjct: 29 PFLTKTFEMVDDPNTDHIVSWNR-GGTSFVVWDLHSFSTILLPRHFKHSNFSSFIRQLNT 87
Query: 76 YGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSREAPNH 135
YGFRK++ +RWEFANE F+ GQ+ LLKNI+RR S PS +A N
Sbjct: 88 YGFRKIEAERWEFANEEFLLGQRQLLKNIKRRN------------PFTPSSSPSHDACN- 134
Query: 136 GLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQMLSFLVMVVQ 195
L+ ++ LM E+V L Q ++ +S + + R++G E+ Q+QM+SFL +Q
Sbjct: 135 -------ELRREKQVLMMEIVSLRQQQQTTKSYIKAMEQRIEGTERKQRQMMSFLARAMQ 187
Query: 196 SPGFMVQLLHPKE 208
SP F+ QLL ++
Sbjct: 188 SPSFLHQLLKQRD 200
>At5g54070 putative protein
Length = 331
Score = 157 bits (398), Expect = 7e-39
Identities = 79/195 (40%), Positives = 122/195 (62%), Gaps = 15/195 (7%)
Query: 14 VAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQL 73
+ PFL K +++V+D TD ++SW+ P+ +F+I D FS LLP YFKH NF+SFIRQL
Sbjct: 69 ITPFLRKTFEIVDDKVTDPVVSWS-PTRKSFIIWDSYEFSENLLPKYFKHKNFSSFIRQL 127
Query: 74 NIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSREAP 133
N YGF+KVD+DRWEFANE F G+KHLLKNI+RR N+ ++EA
Sbjct: 128 NSYGFKKVDSDRWEFANEGFQGGKKHLLKNIKRRS--------------KNTKCCNKEAS 173
Query: 134 NHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQMLSFLVMV 193
EVE+LK +++ + E++ L Q E ++ +M+ + +++ G++ QQ MLSF +
Sbjct: 174 TTTTETEVESLKEEQSPMRLEMLKLKQQQEESQHQMVTVQEKIHGVDTEQQHMLSFFAKL 233
Query: 194 VQSPGFMVQLLHPKE 208
+ F+ +L+ ++
Sbjct: 234 AKDQRFVERLVKKRK 248
>At4g13980 unknown protein
Length = 466
Score = 155 bits (393), Expect = 3e-38
Identities = 77/197 (39%), Positives = 123/197 (62%), Gaps = 10/197 (5%)
Query: 4 SKENASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKH 63
S G+ APFL K Y+MV+D STD I+SW+ + ++F++ + FS LLPTYFKH
Sbjct: 11 SVSGGEGAGGPAPFLVKTYEMVDDSSTDQIVSWSA-NNNSFIVWNHAEFSRLLLPTYFKH 69
Query: 64 NNFASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHN 123
NNF+SFIRQLN YGFRK+D +RWEF N++F++ QKHLLKNI RRK H H+
Sbjct: 70 NNFSSFIRQLNTYGFRKIDPERWEFLNDDFIKDQKHLLKNIHRRKPIH---------SHS 120
Query: 124 NSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQ 183
+ S + L+++++ L ++ ++ +L+ Q A+ + +++ + ME Q
Sbjct: 121 HPPASSTDQERAVLQEQMDKLSREKAAIEAKLLKFKQQKVVAKHQFEEMTEHVDDMENRQ 180
Query: 184 QQMLSFLVMVVQSPGFM 200
+++L+FL +++P F+
Sbjct: 181 KKLLNFLETAIRNPTFV 197
>At3g24520 heat shock transcription factor HSF1, putative
Length = 330
Score = 154 bits (390), Expect = 6e-38
Identities = 82/207 (39%), Positives = 122/207 (58%), Gaps = 29/207 (14%)
Query: 4 SKENASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKH 63
+ N + ++ +APF+ K Y MV D STD +I+W P+ ++F++ D FS +LP YFKH
Sbjct: 5 NSNNNNNNNVIAPFIVKTYQMVNDPSTDWLITWG-PAHNSFIVVDPLDFSQRILPAYFKH 63
Query: 64 NNFASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHN 123
NNF+SF+RQLN YGFRKVD DRWEFANE+F+RGQKHLL NI RRKH Q
Sbjct: 64 NNFSSFVRQLNTYGFRKVDPDRWEFANEHFLRGQKHLLNNIARRKHARGMYGQ------- 116
Query: 124 NSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQ 183
+ + + +E+E LK ++ L E+ +++ R++ EK
Sbjct: 117 -------DLEDGEIVREIERLKEEQRELEAEIQRMNR--------------RIEATEKRP 155
Query: 184 QQMLSFLVMVVQSPGFMVQLLHPKENS 210
+QM++FL VV+ P + +++ KE +
Sbjct: 156 EQMMAFLYKVVEDPDLLPRMMLEKERT 182
>At3g63350 heat shock transcription factor-like protein
Length = 282
Score = 154 bits (389), Expect = 7e-38
Identities = 80/195 (41%), Positives = 119/195 (61%), Gaps = 10/195 (5%)
Query: 15 APFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLN 74
+PFL K ++MV D +T+ I+SW G +FV+ D +FS T+LP YFKHNNF+SF+RQLN
Sbjct: 27 SPFLTKTFEMVGDPNTNHIVSWNR-GGISFVVWDPHSFSATILPLYFKHNNFSSFVRQLN 85
Query: 75 IYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSREAPN 134
YGFRK++ +RWEF NE F+ GQ+ LLK+I+RR + P N + EA +
Sbjct: 86 TYGFRKIEAERWEFMNEGFLMGQRDLLKSIKRR-------TSSSSPPSLNYSQSQPEAHD 138
Query: 135 HGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQMLSFLVMVV 194
G+ E+ L+ +R+ LM E+ L Q + A + + R+ G EK Q+ M+SFL V
Sbjct: 139 PGV--ELPQLREERHVLMMEISTLRQEEQRARGYVQAMEQRINGAEKKQRHMMSFLRRAV 196
Query: 195 QSPGFMVQLLHPKEN 209
++P + Q+ K +
Sbjct: 197 ENPSLLQQIFEQKRD 211
>At5g43840 heat shock transcription factor-like protein
Length = 251
Score = 150 bits (379), Expect = 1e-36
Identities = 74/187 (39%), Positives = 119/187 (63%), Gaps = 20/187 (10%)
Query: 17 FLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNIY 76
FL K Y++VED ST++I+SW+ + ++F++ + F++ LP FKHNNF+SF+RQLN Y
Sbjct: 20 FLTKTYNIVEDSSTNNIVSWSRDN-NSFIVWEPETFALICLPRCFKHNNFSSFVRQLNTY 78
Query: 77 GFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSREAPNHG 136
GF+K+DT+RWEFANE+F++G++HLLKNI+RRK S +
Sbjct: 79 GFKKIDTERWEFANEHFLKGERHLLKNIKRRK-------------------TSSQTQTQS 119
Query: 137 LRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQMLSFLVMVVQS 196
L E+ L+ DR +L ELV L + ES ++ + ++ ++L+ E Q+ M++FL+ ++
Sbjct: 120 LEGEIHELRRDRMALEVELVRLRRKQESVKTYLHLMEEKLKVTEVKQEMMMNFLLKKIKK 179
Query: 197 PGFMVQL 203
P F+ L
Sbjct: 180 PSFLQSL 186
>At4g36990 heat shock transcription factor HSF4
Length = 284
Score = 126 bits (317), Expect = 2e-29
Identities = 75/185 (40%), Positives = 105/185 (56%), Gaps = 4/185 (2%)
Query: 8 ASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFA 67
A+ S APFL+K Y +V+D STD ++SW E G FV+ F+ LLP YFKHNNF+
Sbjct: 6 AAQRSVPAPFLSKTYQLVDDHSTDDVVSWNE-EGTAFVVWKTAEFAKDLLPQYFKHNNFS 64
Query: 68 SFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDE 127
SFIRQLN YGFRK D+WEFAN+ F RG + LL +IRRRK + K + + S+
Sbjct: 65 SFIRQLNTYGFRKTVPDKWEFANDYFRRGGEDLLTDIRRRKSVIASTAGKCVVVGSPSES 124
Query: 128 PSREAPNHGLRKEVENLKSDRN--SLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQ 185
S +HG + S +N S+ + LS E + + LS L +K + +
Sbjct: 125 NSGGGDDHG-SSSTSSPGSSKNPGSVENMVADLSGENEKLKRENNNLSSELAAAKKQRDE 183
Query: 186 MLSFL 190
+++FL
Sbjct: 184 LVTFL 188
>At5g62020 heat shock factor 6
Length = 299
Score = 120 bits (301), Expect = 1e-27
Identities = 64/145 (44%), Positives = 86/145 (59%), Gaps = 10/145 (6%)
Query: 6 ENASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNN 65
E++S S PFL K +++VED S D +ISW E G +F++ + T F+ LLP +FKHNN
Sbjct: 13 ESSSQRSIPTPFLTKTFNLVEDSSIDDVISWNE-DGSSFIVWNPTDFAKDLLPKHFKHNN 71
Query: 66 FASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNS 125
F+SF+RQLN YGF+KV DRWEF+N+ F RG+K LL+ I+RR K H
Sbjct: 72 FSSFVRQLNTYGFKKVVPDRWEFSNDFFKRGEKRLLREIQRR---------KITTTHQTV 122
Query: 126 DEPSREAPNHGLRKEVENLKSDRNS 150
PS E N + N D N+
Sbjct: 123 VAPSSEQRNQTMVVSPSNSGEDNNN 147
>At1g46264 heat shock transcription factor, putative
Length = 348
Score = 120 bits (301), Expect = 1e-27
Identities = 59/107 (55%), Positives = 73/107 (68%), Gaps = 1/107 (0%)
Query: 10 GSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASF 69
G + APFL K Y +V+D +TD ++SW + TFV+ F+ LLP YFKHNNF+SF
Sbjct: 28 GKAVPAPFLTKTYQLVDDPATDHVVSWGDDDT-TFVVWRPPEFARDLLPNYFKHNNFSSF 86
Query: 70 IRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQ 116
+RQLN YGFRK+ DRWEFANE F RG+KHLL I RRK + QQ
Sbjct: 87 VRQLNTYGFRKIVPDRWEFANEFFKRGEKHLLCEIHRRKTSQMIPQQ 133
>At2g41690 putative heat shock transcription factor
Length = 244
Score = 114 bits (286), Expect = 6e-26
Identities = 57/96 (59%), Positives = 68/96 (70%), Gaps = 1/96 (1%)
Query: 16 PFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNI 75
PFL K Y +VED +TD +ISW E G FV+ F+ LLPT FKH NF+SF+RQLN
Sbjct: 40 PFLVKTYKVVEDPTTDGVISWNE-YGTGFVVWQPAEFARDLLPTLFKHCNFSSFVRQLNT 98
Query: 76 YGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPH 111
YGFRKV T RWEF+NE F +GQ+ L+ NIRRRK H
Sbjct: 99 YGFRKVTTIRWEFSNEMFRKGQRELMSNIRRRKSQH 134
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.133 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,060,471
Number of Sequences: 26719
Number of extensions: 420745
Number of successful extensions: 1095
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 35
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 1008
Number of HSP's gapped (non-prelim): 62
length of query: 367
length of database: 11,318,596
effective HSP length: 101
effective length of query: 266
effective length of database: 8,619,977
effective search space: 2292913882
effective search space used: 2292913882
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0069.1


