

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0067b.6
(404 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
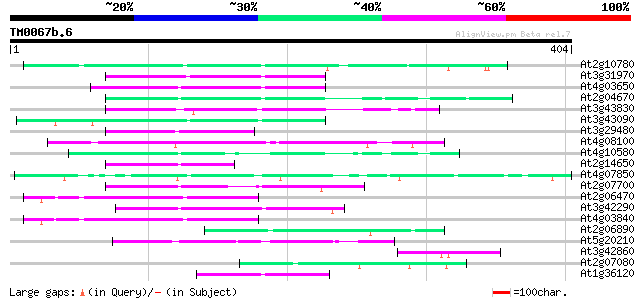
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 96 4e-20
At3g31970 hypothetical protein 95 6e-20
At4g03650 putative reverse transcriptase 94 1e-19
At2g04670 putative retroelement pol polyprotein 92 4e-19
At3g43830 putative protein 80 3e-15
At3g43090 putative protein 79 4e-15
At3g29480 hypothetical protein 77 1e-14
At4g08100 putative polyprotein 76 4e-14
At4g10580 putative reverse-transcriptase -like protein 73 3e-13
At2g14650 putative retroelement pol polyprotein 73 3e-13
At4g07850 putative polyprotein 65 9e-11
At2g07700 putative retroelement pol polyprotein 64 1e-10
At2g06470 putative retroelement pol polyprotein 62 4e-10
At3g42290 putative protein 61 1e-09
At4g03840 putative transposon protein 59 5e-09
At2g06890 putative retroelement integrase 55 9e-08
At5g20210 putative protein 51 1e-06
At3g42860 unknown protein (At3g42860) 46 3e-05
At2g07080 putative gag-protease polyprotein 45 5e-05
At1g36120 putative reverse transcriptase gb|AAD22339.1 45 9e-05
>At2g10780 pseudogene
Length = 1611
Score = 95.9 bits (237), Expect = 4e-20
Identities = 96/372 (25%), Positives = 143/372 (37%), Gaps = 38/372 (10%)
Query: 11 MATMAQAVTAQAQAVTAQANDNAMRRATE-EAREQHQRQREITLDQNKGLNDFRRQNPPK 69
+AT+ +AV A + + E + H R+R D L R
Sbjct: 125 LATLTKAVIADPVVLKVDNQKDDDSEVVEVNPQPNHGRKRG---DYLSLLEHVSRLGTRH 181
Query: 70 FSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGTRGIMEDNHEE 129
F G TDP AD W +++ F + E + +A + L GDA WW D E
Sbjct: 182 FMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVEKRRGD--EV 239
Query: 130 ISWNSFRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAKHFQFFHDHVNE 189
S+ F F +KYFP A D E +L L QG +V E+ + L ++ + E
Sbjct: 240 RSFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRYVG--RELEEE 297
Query: 190 RYMCKRFVNGLRPDIEDSVRPLRIMRFQSLVEKATEVE--LMKNRRLNRAGTGGPMRSGS 247
+ +RF+ GLR +I + LVE+A +E + + R LNR ++
Sbjct: 298 QAQLRRFIRGLRIEIRNHCLVRTFNSVSELVERAAMIEEGIEEERYLNRE------KAPI 351
Query: 248 QNFQSREKFQSRGPYQRPAGTGFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPGHYAN 307
+N QS + + + + T + + +T SG K C RCG H
Sbjct: 352 RNNQSTKPADKKRKFDKVDNTKSDAKTGECVTCGKNHSG-TCWKAIGACGRCGSKDHAIQ 410
Query: 308 ACPDTRP-----------KCFNCNRMGHTAGQCRAPKTEPTVNTA--RG--------KRP 346
+CP P CF C + GH +C E RG KR
Sbjct: 411 SCPKMEPGQSKVLGEETRTCFYCGKTGHLKRECPKLTAEKQAGQRDNRGGNGLPPPPKRQ 470
Query: 347 AAKARVYTMDGE 358
A +RVY + E
Sbjct: 471 AVASRVYELSEE 482
>At3g31970 hypothetical protein
Length = 1329
Score = 95.1 bits (235), Expect = 6e-20
Identities = 57/158 (36%), Positives = 78/158 (49%), Gaps = 4/158 (2%)
Query: 70 FSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGTRGIMEDNHEE 129
F GGT P++AD W VE FG + +V +A + L GDA WW+ H
Sbjct: 158 FFGGTSPEEADSWKSRVEHNFGSSRCPAEYRVDLAVHFLEGDAHLWWRSVTARRRQAH-- 215
Query: 130 ISWNSFRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAKHFQFFHDHVNE 189
+SW F F KYFP A D E++FL L QG SV E+ + L + ++
Sbjct: 216 MSWADFVAEFNAKYFPQEALDRMEARFLELTQGERSVREYEREFNRLLVYAG--RGMQDD 273
Query: 190 RYMCKRFVNGLRPDIEDSVRPLRIMRFQSLVEKATEVE 227
+ +RF+ GLRPD+ R L+ +LVE A EVE
Sbjct: 274 QAQMRRFLRGLRPDLRVRCRVLQYATKAALVETAAEVE 311
>At4g03650 putative reverse transcriptase
Length = 839
Score = 94.4 bits (233), Expect = 1e-19
Identities = 56/169 (33%), Positives = 82/169 (48%), Gaps = 4/169 (2%)
Query: 59 LNDFRRQNPPKFSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKG 118
+ +R FSGGT P++AD W +VE+ FG + +V + + L GDA WW+
Sbjct: 145 MKQLQRIGTEYFSGGTSPEEADSWRSQVERNFGSSRCPAEYRVDLTVHFLEGDAHLWWRS 204
Query: 119 TRGIMEDNHEEISWNSFRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAK 178
++SW F F KYFP A D E++FL L QG SV E+ K L
Sbjct: 205 VTA--RRRQADMSWADFMAEFNAKYFPREALDRMEARFLELTQGVRSVREYDRKFNRLLV 262
Query: 179 HFQFFHDHVNERYMCKRFVNGLRPDIEDSVRPLRIMRFQSLVEKATEVE 227
+ +++ +RF+ GLRP++ R + +LVE A EVE
Sbjct: 263 YAG--RGMEDDQAQMRRFLRGLRPNLRVRCRVSQYATKAALVETAAEVE 309
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 92.4 bits (228), Expect = 4e-19
Identities = 80/293 (27%), Positives = 118/293 (39%), Gaps = 41/293 (13%)
Query: 70 FSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGTRGIMEDNHEE 129
FS GT P++AD W V++ FG + +V +A + L GDA WW+ +
Sbjct: 138 FSSGTSPEEADSWRSRVQRNFGSSRCPAEYRVDLAVHFLEGDAHLWWRSVTA--RRRQAD 195
Query: 130 ISWNSFRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAKHFQFFHDHVNE 189
+SW F F KYFP A D E++FL L QG SV E+ + L + ++
Sbjct: 196 MSWADFVAEFNAKYFPQEALDRMEARFLELTQGEWSVREYDREFNRLLAYAG--RGMEDD 253
Query: 190 RYMCKRFVNGLRPDIEDSVRPLRIMRFQSLVEKATEVELMKNRRLNRAGTGGPMRSGSQN 249
+ +RF+ GLRPD+ R + +LVE A EVE
Sbjct: 254 QAQMRRFLRGLRPDLRVQCRVSQYATKAALVETAAEVE---------------------- 291
Query: 250 FQSREKFQSRGPYQRPAGTGFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPGHYANAC 309
E Q + PA T + + +T + GG Q K++ H + A
Sbjct: 292 ----EDLQRQVVGVSPAVQ--TKNTQQQVTPSKGGKPAQGQKRKW--------DHPSRAG 337
Query: 310 PDTRPKCFNCNRMGHTAGQCRAPKTEPTVNTARGKRPAAKARVYTMDGERAEE 362
R F+C + HT C + V A G+ A R +D + A E
Sbjct: 338 QGGRAGYFSCGSLDHTVADC-TERGHLHVKVAGGQFLAVLGRAKGVDIQIAGE 389
>At3g43830 putative protein
Length = 329
Score = 79.7 bits (195), Expect = 3e-15
Identities = 66/244 (27%), Positives = 100/244 (40%), Gaps = 44/244 (18%)
Query: 70 FSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGTRGIMEDNHEE 129
F+GGTDP KA W +E F ++ +A+ L G+A+ WW+ + E+
Sbjct: 116 FNGGTDPVKAYNWRNMLECKFKSMRCPVEFWRDLASCYLRGEAQEWWERVK-----QREQ 170
Query: 130 IS----WNSFRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAKHFQFFHD 185
+ W+ F+ F +Y P D+ E +FL L+QG +V ++ + SL + F
Sbjct: 171 VGCVDQWSFFKEEFTRRYLPEETIDDLEMKFLRLQQGTKTVRKYEKEFHSLER---FERR 227
Query: 186 HVNERYMCKRFVNGLRPDIEDSVRPLRIMRFQSLVEKATEVELMKNRRLNRAGTGGPMRS 245
E + +F++GLR DI LVEKA +E+
Sbjct: 228 KRGEHELIHKFISGLRVDIRLCCHVRDFDNMIDLVEKAASLEI----------------- 270
Query: 246 GSQNFQSREKFQSRGPYQRPAGTGFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPGHY 305
+ Y+R A GSY G G S + +E TC+RCG GH
Sbjct: 271 ---------GLEEEARYKRIAQEKEAMGSY----GQTGHSKRRC--QEVTCYRCGVAGHI 315
Query: 306 ANAC 309
A C
Sbjct: 316 ARDC 319
>At3g43090 putative protein
Length = 357
Score = 79.0 bits (193), Expect = 4e-15
Identities = 65/242 (26%), Positives = 95/242 (38%), Gaps = 23/242 (9%)
Query: 6 QLAEMMATMAQAVTAQAQAVTAQAND--NAMRRATEEAREQHQRQREITLDQNKG----- 58
++A A + ++ Q Q + + N +R+AT A Q Q + +Q +
Sbjct: 47 RVANQRAAINASLLGQIQGMRGMFEELMNRIRQATPAAAGTQQPQENVLPEQQQPPPPPP 106
Query: 59 -------------LNDFRRQNPPKFSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMAT 105
+ R F+GG DP KAD W + +E F + + +
Sbjct: 107 PPPPPPPPAQMEIMKVMRTMGTEYFTGGIDPVKADDWRKLLENNFENARCPVEFQKDIIV 166
Query: 106 YLLLGDAEYWWKGTRGIMEDNHEEISWNSFRTAFLEKYFPTSARDERESQFLTLRQGGMS 165
+ L +A +WW G G H I W FR F K+FP A D E F LRQ
Sbjct: 167 HFLREEASHWWDGVLGNTPVQHV-IFWEDFREEFNRKFFPQEAMDSLEDDFEELRQDTKK 225
Query: 166 VPEFASKLESLAKHFQFFHDHVNERYMCKRFVNGLRPDIEDSVRPLRIMRFQSLVEKATE 225
V E +L L++ E+ M +R + GLRPDI LVEKA
Sbjct: 226 VRENERELSHLSRF--SVRAGRGEQSMIRRLMRGLRPDILTRCASREFFSMIDLVEKAAR 283
Query: 226 VE 227
+E
Sbjct: 284 IE 285
>At3g29480 hypothetical protein
Length = 718
Score = 77.4 bits (189), Expect = 1e-14
Identities = 42/107 (39%), Positives = 53/107 (49%), Gaps = 2/107 (1%)
Query: 70 FSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGTRGIMEDNHEE 129
FSGGT P++AD W VE+ FG + +V +A + L GDA WWK
Sbjct: 51 FSGGTSPEEADSWRSRVERNFGSSRCPVEYRVDLAVHFLEGDAHLWWKSV--TTRRRQAN 108
Query: 130 ISWNSFRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESL 176
+SW F F KYFP A D E+ FL L QG SV E+ + L
Sbjct: 109 MSWADFVAEFNAKYFPQEALDRMEAHFLELTQGERSVREYDREFNRL 155
>At4g08100 putative polyprotein
Length = 1054
Score = 75.9 bits (185), Expect = 4e-14
Identities = 67/294 (22%), Positives = 122/294 (40%), Gaps = 25/294 (8%)
Query: 28 QANDNAMRRATEEAREQHQRQREITLDQNKGLNDFRRQNPPKFSGGTDPDKADLWIQEVE 87
+A + ++ ++ +H+R+R D GL + P F G +PD W Q++E
Sbjct: 61 EARNYYSHASSHSSQRRHRRERPPPRDPLGGL----KLKIPAFHGTDNPDTYLEWEQKIE 116
Query: 88 KIFGVLQTAEGAKVRMATYLLLGDAEYWWKG--TRGIMEDNHEEISWNSFRTAFLEKYFP 145
+F + + KV++A A WW T ++ +WN + +++ P
Sbjct: 117 LVFLCQECLQSNKVKIAATKFYNYALSWWDQLVTSRRRTRDYPIKTWNQLKFVMRKRFVP 176
Query: 146 TSARDERESQFLTLRQGGMSVPEFASKLESLAKHFQFFHDHVNERYMCKRFVNGLRPDIE 205
+ E + L QG +V E+ ++E+L D E M RF+ GL +I+
Sbjct: 177 SYYHRELHQRLRNLVQGSKTVEEYFLEMETLMLRADLQED--GEAVM-SRFMGGLNREIQ 233
Query: 206 DSVRPLRIMRFQSLVEKATEVE-LMKNRRLNRAGTGGPMRSGSQNFQSREKF---QSRGP 261
D + + + ++ KA E +K + + T SG ++Q EKF P
Sbjct: 234 DRLETQHYVELEEMLHKAVMFEQQIKRKNARSSHTKTNYSSGKPSYQKEEKFGYQNDSKP 293
Query: 262 YQRPAGTGFTSGSYRPMTGAAGGSGDQ--TLKKETTCFRCGEPGHYANACPDTR 313
+ +P +P+ G G + T ++ CF+ HYA+ C + R
Sbjct: 294 FVKP----------KPVDLDPKGKGKEVITRARDIRCFKSQGLRHYASKCSNKR 337
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 72.8 bits (177), Expect = 3e-13
Identities = 69/282 (24%), Positives = 105/282 (36%), Gaps = 67/282 (23%)
Query: 43 EQHQRQREITLDQNKGLNDFRRQNPPKFSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVR 102
+Q E L + + +R + FSGGT P++AD W V + FG + +V
Sbjct: 125 QQRAAVAEEVLSYLRMMEQLQRIDTGYFSGGTSPEEADSWRSRVGRNFGSSRCPAEYRVD 184
Query: 103 MATYLLLGDAEYWWKGTRGIMEDNHEEISWNSFRTAFLEKYFPTSARDERESQFLTLRQG 162
+A + L GDA WW+ ++SW F F KYFP A D Q
Sbjct: 185 LAVHFLEGDAHLWWRSVTA--RRRQTDMSWADFVAEFKAKYFPQEALDPYAGQ------- 235
Query: 163 GMSVPEFASKLESLAKHFQFFHDHVNERYMCKRFVNGLRPDIEDSVRPLRIMRFQSLVEK 222
GM +++ +RF+ GLRPD+ R + +LVE
Sbjct: 236 GME----------------------DDQAQMRRFLRGLRPDLRVRCRVSQYATKAALVET 273
Query: 223 ATEVELMKNRRLNRAGTGGPMRSGSQNFQSREKFQSRGPYQRPAGTGFTSGSYRPMTGAA 282
A EVE ++FQ + P +P T + +T +
Sbjct: 274 AAEVE--------------------EDFQ--RQVVGVSPVVQPKKT------QQQVTPSK 305
Query: 283 GGSGDQTLKKETTCFRCGEPGHYANACPDTRPKCFNCNRMGH 324
GG Q K++ H + A R +CF+C + H
Sbjct: 306 GGKPAQGQKRKW--------DHPSRAGQGGRARCFSCGSLDH 339
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 72.8 bits (177), Expect = 3e-13
Identities = 37/93 (39%), Positives = 47/93 (49%), Gaps = 2/93 (2%)
Query: 70 FSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGTRGIMEDNHEE 129
FSGGT P+ AD W VE+ FG + ++ +A + L GDA WW+ +
Sbjct: 156 FSGGTSPEVADSWRSRVERNFGSSRCPAEYRIDLAVHFLEGDAHLWWRSVTA--RRRQAD 213
Query: 130 ISWNSFRTAFLEKYFPTSARDERESQFLTLRQG 162
ISW F F KYFP A D E+ FL L QG
Sbjct: 214 ISWADFVAEFNAKYFPQEALDRMEAHFLELTQG 246
>At4g07850 putative polyprotein
Length = 1138
Score = 64.7 bits (156), Expect = 9e-11
Identities = 93/422 (22%), Positives = 159/422 (37%), Gaps = 73/422 (17%)
Query: 4 TNQLAEMMATMAQAVTAQAQAVTAQANDNAMRRAT------EEAREQHQRQREITLDQNK 57
T +A+MM +A + + +N+ R AT R H+RQR +
Sbjct: 13 TTNMAKMMDERFEAARSNQERKNRTMRNNSDREATLSYYSQSSTRTSHRRQR-------R 65
Query: 58 GLNDFRRQNPPKFSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWK 117
G + ++PP+ + A+L + +I + E KV++A A WW
Sbjct: 66 GQEE---RHPPR------DNLANLKL----RIPPFHEYTEANKVKVAATEFYDYALSWWD 112
Query: 118 GT----RGIMEDNHEEISWNSFRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKL 173
R + +D+ E +WN + ++ P+ E + L QG +V E+ ++
Sbjct: 113 QVVTTKRRLGDDSIE--TWNQLKNIMKRRFVPSHYHRELHQRLRNLVQGNRTVEEYFKEM 170
Query: 174 ESLAKHFQFFHDHVNERYMC--KRFVNGLRPDIEDSVRPLRIMRFQSLVEKATEVELMKN 231
E+L V E RF+ GL DI D + + L KA E
Sbjct: 171 ETL-----MLRADVQEECEATMSRFMGGLNRDILDRFEVIHYENLEELFHKAVMFEKQIK 225
Query: 232 RRLNRAGTGGPMRSGSQNFQSREKFQSRGPYQRPAGTGFTSGSYRPMT-------GAAGG 284
RR S ++ S S+ YQR +GF Y+P + G
Sbjct: 226 RR-----------SAKPSYNS-----SKPSYQREEKSGFQK-EYKPFVKPKVEEISSKGK 268
Query: 285 SGDQTLKKETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQCRAPKTEPTVNTARGK 344
+ T ++ CF+C GHYA+ C + R + G + P+ +G+
Sbjct: 269 EKEVTRTRDLKCFKCHGLGHYASECSNKRIMIIRDS--GEVESEDEKPEESDVEEAPKGE 326
Query: 345 RPAAKARVYTMDGERAEEFARGERKNDGNFLTILSYSSITCSITL--VACANTSNLLILS 402
+ ++ +AEE A+ E F T CS+ + +C N ++ ++
Sbjct: 327 LLVTMRVLSVLN--KAEEQAQRENL----FHTRCLIKGKVCSLIIDGGSCTNVASETMVQ 380
Query: 403 KL 404
KL
Sbjct: 381 KL 382
>At2g07700 putative retroelement pol polyprotein
Length = 543
Score = 64.3 bits (155), Expect = 1e-10
Identities = 48/188 (25%), Positives = 82/188 (43%), Gaps = 25/188 (13%)
Query: 70 FSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGTRGIMEDNHEE 129
F G + AD W++++E F + + + K ++A L DA W + + +
Sbjct: 53 FKGEHNTVVADKWLRDLEMNFEISRCPDNFKRQIAVNFLDKDARIWRESVTA--RNQGQM 110
Query: 130 ISWNSFRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAKHFQFFHDHVNE 189
ISW +FR F +KYFP ARD E QF+ K + ++ +E
Sbjct: 111 ISWEAFRREFEKKYFPPEARDRLEQQFM--------------------KRY-LYNGQDDE 149
Query: 190 RYMCKRFVNGLRPDIEDSVRPLRIMRFQSLVEKA--TEVELMKNRRLNRAGTGGPMRSGS 247
M +RF+ GLR +I ++ ++ L E+A E E + + N GP+ G
Sbjct: 150 ALMVRRFLRGLRLEIWGRLQAVKYDSVNELTERADNVEEESIPELKTNLTYPEGPLEIGE 209
Query: 248 QNFQSREK 255
+ + +K
Sbjct: 210 RRIRKFKK 217
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 62.4 bits (150), Expect = 4e-10
Identities = 51/171 (29%), Positives = 74/171 (42%), Gaps = 8/171 (4%)
Query: 11 MATMAQAVTAQ--AQAVTAQANDNAMRRATEEAREQHQRQREITLDQNKGLNDFRRQNPP 68
+AT+ +AV A Q V Q +D++ + H R+R D L R
Sbjct: 90 LATLTKAVVADPVVQRVDNQKDDDS-EVVEVNPQPNHGRKRG---DYLSLLEHVSRLGTR 145
Query: 69 KFSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGTRGIMEDNHE 128
F G TDP AD W +++ F + E + +A + L GDA WW D E
Sbjct: 146 HFMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVEKRRGD--E 203
Query: 129 EISWNSFRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAKH 179
S+ F F +KYFP A D E +L L QG +V E+ + L ++
Sbjct: 204 VRSFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRY 254
>At3g42290 putative protein
Length = 462
Score = 61.2 bits (147), Expect = 1e-09
Identities = 50/176 (28%), Positives = 73/176 (41%), Gaps = 15/176 (8%)
Query: 77 DKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGTRGIMEDNHEEISWNSFR 136
+KAD W V++ F + +V +A + L DA WWK ++SW F
Sbjct: 161 EKADSWRSRVKRNFSSSRCLMEYRVDLAVHFLEEDAHLWWKSVTA--RRRQADMSWADFV 218
Query: 137 TAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAKHFQFFHDHVNERYMCKRF 196
F KYF D E +FL L SV E+ + + + + + + +RF
Sbjct: 219 VEFNAKYFLQDELDRMEVRFLELTLVERSVWEYDREFNRVLVYAGW--GMKDGQAELRRF 276
Query: 197 VNGLRPDIEDSVRPLRIMRFQSLVEKATEVELMKN-----------RRLNRAGTGG 241
+ GLRPD+ R + +LVE A EV K L+RAG GG
Sbjct: 277 MRGLRPDLRVRCRMSQYALKAALVETAAEVAPRKGGKPMQGKKRSWNHLSRAGQGG 332
>At4g03840 putative transposon protein
Length = 973
Score = 58.9 bits (141), Expect = 5e-09
Identities = 49/171 (28%), Positives = 72/171 (41%), Gaps = 8/171 (4%)
Query: 11 MATMAQAVTAQ--AQAVTAQANDNAMRRATEEAREQHQRQREITLDQNKGLNDFRRQNPP 68
+AT+ +A+ A Q V Q +D + + H R+R D L R
Sbjct: 90 LATLTKAIVANPVVQRVDNQKDDE-LEVVEVNPQPNHGRKRG---DYLSLLEHVSRLGTR 145
Query: 69 KFSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGTRGIMEDNHE 128
F G TDP AD W +++ F + E + +A + L DA WW D E
Sbjct: 146 HFMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLERDAHNWWLTVEKRRGD--E 203
Query: 129 EISWNSFRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAKH 179
S+ F F +KYFP A D E +L L QG +V E+ + L ++
Sbjct: 204 VRSFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRY 254
>At2g06890 putative retroelement integrase
Length = 1215
Score = 54.7 bits (130), Expect = 9e-08
Identities = 40/175 (22%), Positives = 70/175 (39%), Gaps = 7/175 (4%)
Query: 141 EKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAKHFQFFHDHVNERYMCKRFVNGL 200
+++ P+ E + L QG SV E+ ++E+L D RF+ L
Sbjct: 7 KRFVPSHYHRELHLKLRNLTQGNRSVEEYYKEMETLMLRADISEDR---EATLSRFLGDL 63
Query: 201 RPDIEDSVRPLRIMRFQSLVEKATEVELMKNRRLNRAGTGGPMRSGSQNFQSREKFQS-- 258
DI+D + ++ + ++ KA E R+ + + G +Q E+ S
Sbjct: 64 NRDIQDRLETQYYVQIEEMLHKAILFEQQVKRKSSSRSSYGSGTIAKPTYQREERTSSYH 123
Query: 259 RGPYQRPAGTGFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPGHYANACPDTR 313
P P + + G A S + ++ C++C GHYAN CP+ R
Sbjct: 124 NKPIVSPRAESKPYAAVQDHKGKAEISTSRV--RDVRCYKCQGKGHYANECPNKR 176
>At5g20210 putative protein
Length = 256
Score = 50.8 bits (120), Expect = 1e-06
Identities = 50/204 (24%), Positives = 83/204 (40%), Gaps = 26/204 (12%)
Query: 75 DPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDAEYWWKGTRGIMEDNHEEISWNS 134
D D +W++ +E F + E KV A + GDA W + + + ++W+
Sbjct: 71 DGDHLFIWLEIIEVYFTLQLAPEVCKVDTAFRSMEGDALLWLQ----CLRQENPTLTWSQ 126
Query: 135 FRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAKHFQFFHDHVNERYMCK 194
+T +E++ E Q +LRQ G SV E+A++ + A N+ + K
Sbjct: 127 LKTELIEEFGGGDITASPEEQLASLRQTG-SVDEYANEFRARAAQIG------NKDQLVK 179
Query: 195 -RFVNGLRPDIEDSVRPLRIMRFQSLVEKATEVELMKNRRLNRAGTGGPMRSGSQNFQSR 253
F+NGL+ +I RP + + A +E N NRA + R
Sbjct: 180 GMFLNGLKEEIRVMFRPNDAEGPEMAIRTAKAIERELNFN-NRA-------------KGR 225
Query: 254 EKFQSRGPYQRPAGTGFTSGSYRP 277
++ S Y GTG Y+P
Sbjct: 226 RQYSSHQEYFDMEGTGLCYPGYQP 249
>At3g42860 unknown protein (At3g42860)
Length = 372
Score = 46.2 bits (108), Expect = 3e-05
Identities = 25/85 (29%), Positives = 35/85 (40%), Gaps = 11/85 (12%)
Query: 280 GAAGGSGDQTLKKETTCFRCGEPGHYANAC---PDTRP--------KCFNCNRMGHTAGQ 328
G + G Q K T C++CG+ GH+A C DT P CF C + GH +
Sbjct: 223 GDSDTRGYQNAKTGTPCYKCGKEGHWARDCTVQSDTGPVKSTSAAGDCFKCGKPGHWSRD 282
Query: 329 CRAPKTEPTVNTARGKRPAAKARVY 353
C A P + K ++ Y
Sbjct: 283 CTAQSGNPKYEPGQMKSSSSSGECY 307
Score = 38.1 bits (87), Expect = 0.009
Identities = 17/74 (22%), Positives = 30/74 (39%), Gaps = 16/74 (21%)
Query: 296 CFRCGEPGHYANAC------PDTRP----------KCFNCNRMGHTAGQCRAPKTEPTVN 339
CF+CG+PGH++ C P P +C+ C + GH + C +
Sbjct: 270 CFKCGKPGHWSRDCTAQSGNPKYEPGQMKSSSSSGECYKCGKQGHWSRDCTGQSSNQQFQ 329
Query: 340 TARGKRPAAKARVY 353
+ + K ++ Y
Sbjct: 330 SGQAKSTSSTGDCY 343
Score = 33.5 bits (75), Expect = 0.21
Identities = 18/66 (27%), Positives = 28/66 (42%), Gaps = 19/66 (28%)
Query: 296 CFRCGEPGHYANAC----------------PDTRPKCFNCNRMGHTAGQCRAPKTEPTVN 339
C++CG+ GH++ C + C+ C + GH + C +P T N
Sbjct: 306 CYKCGKQGHWSRDCTGQSSNQQFQSGQAKSTSSTGDCYKCGKAGHWSRDCTSP--AQTTN 363
Query: 340 TARGKR 345
T GKR
Sbjct: 364 TP-GKR 368
>At2g07080 putative gag-protease polyprotein
Length = 627
Score = 45.4 bits (106), Expect = 5e-05
Identities = 42/180 (23%), Positives = 65/180 (35%), Gaps = 19/180 (10%)
Query: 166 VPEFASKLESLAKHFQFFHDHVNERYMCKRFVNGLRPDIEDSVRPLRIMRFQSLVEKATE 225
+ +F+SK+ +LA + ++ + K+ + L P +MR +K +
Sbjct: 124 IVKFSSKISALANEAEVMGKTYKDQKLVKKLLRCLPPKF---AAHKAVMRVAGNTDKISF 180
Query: 226 VELMKNRRLNRAGTGGPMRSGSQNF-----QSREKFQSRGPYQRPAGTGFTSGSYRPMTG 280
V+L+ +L S+N Q E+FQ F R
Sbjct: 181 VDLVGMLKLEEMKADQDKVKPSKNIAFNADQGSEQFQEIKDGMALLARNFGKALKRVEID 240
Query: 281 AAGGSG--------DQTLKKETTCFRCGEPGHYANACPDTR---PKCFNCNRMGHTAGQC 329
G D KKE C+ CG GH CP T+ KC C +GHT +C
Sbjct: 241 GERSRGRFSRSENDDLRKKKEIQCYECGGFGHIKPECPITKRKEMKCLKCKGVGHTKFEC 300
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 44.7 bits (104), Expect = 9e-05
Identities = 28/96 (29%), Positives = 46/96 (47%), Gaps = 2/96 (2%)
Query: 135 FRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAKHFQFFHDHVNERYMCK 194
F F KYFP A D E++FL L +G +SV E+ + + L + +++ +
Sbjct: 157 FVAEFNAKYFPQEALDRMEARFLELTKGELSVREYGREFKRLLVYAG--RGMEDDQAQMR 214
Query: 195 RFVNGLRPDIEDSVRPLRIMRFQSLVEKATEVELMK 230
RF+ GLRPD+ R + +L E +L +
Sbjct: 215 RFLRGLRPDLRVRCRVSQYATKAALTAAEVEEDLQR 250
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,970,084
Number of Sequences: 26719
Number of extensions: 387171
Number of successful extensions: 1438
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 70
Number of HSP's successfully gapped in prelim test: 37
Number of HSP's that attempted gapping in prelim test: 1176
Number of HSP's gapped (non-prelim): 211
length of query: 404
length of database: 11,318,596
effective HSP length: 102
effective length of query: 302
effective length of database: 8,593,258
effective search space: 2595163916
effective search space used: 2595163916
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0067b.6


