

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0066.6
(137 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
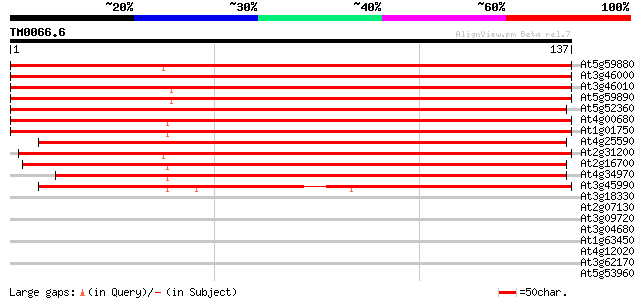
Score E
Sequences producing significant alignments: (bits) Value
At5g59880 actin depolymerizing factor 3 - like protein 236 2e-63
At3g46000 actin depolymerizing factor 2 (ADF2) 236 2e-63
At3g46010 actin depolymerizing factor 1 (ADF1) 224 1e-59
At5g59890 actin depolymerizing factor 4 - like protein 224 1e-59
At5g52360 actin depolymerizing factor-like 213 3e-56
At4g00680 putative actin-depolymerizing factor 210 2e-55
At1g01750 actin depolymerizing factor like protein 208 6e-55
At4g25590 actin depolymerizing factor - like protein 203 3e-53
At2g31200 actin depolymerizing factor 6 (ADF6) 166 5e-42
At2g16700 actin depolymerizing factor 5 (ADF5) 158 1e-39
At4g34970 actin depolymerizing factor - like protein 157 2e-39
At3g45990 actin depolymerising like protein 153 3e-38
At3g18330 hypothetical protein 31 0.18
At2g07130 hypothetical protein 30 0.30
At3g09720 RNA helicase like protein 30 0.39
At3g04680 unknown protein 29 0.67
At1g63450 unknown protein 27 4.3
At4g12020 putative disease resistance protein 26 5.6
At3g62170 PECTINESTERASE-like protein 26 5.6
At5g53960 unknown protein 26 7.4
>At5g59880 actin depolymerizing factor 3 - like protein
Length = 139
Score = 236 bits (603), Expect = 2e-63
Identities = 113/139 (81%), Positives = 130/139 (93%), Gaps = 2/139 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIED--KQVIVEKLGEPGQGYEDFTANL 58
MANAASGMAVHDDCKL+FMELKTKRT+RFI+YKIE+ KQVIVEK+GEPGQ +ED A+L
Sbjct: 1 MANAASGMAVHDDCKLKFMELKTKRTHRFIIYKIEELQKQVIVEKIGEPGQTHEDLAASL 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
PADECRYA++DF++++ VP+SRIFF+AWSPDT+RVRSKMIYASSKDRFKRELDGIQ+E
Sbjct: 61 PADECRYAIFDFDFVSSEGVPRSRIFFVAWSPDTARVRSKMIYASSKDRFKRELDGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQATDPTEM LDVFKSRAN
Sbjct: 121 LQATDPTEMDLDVFKSRAN 139
>At3g46000 actin depolymerizing factor 2 (ADF2)
Length = 137
Score = 236 bits (603), Expect = 2e-63
Identities = 112/137 (81%), Positives = 125/137 (90%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQVIVEKLGEPGQGYEDFTANLPA 60
MANAASGMAVHDDCKL+FMELK KRT+R IVYKIEDKQVIVEKLGEP Q Y+DF A+LPA
Sbjct: 1 MANAASGMAVHDDCKLKFMELKAKRTFRTIVYKIEDKQVIVEKLGEPEQSYDDFAASLPA 60
Query: 61 DECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQ 120
D+CRY +YDF+++T N KS+IFFIAWSPDT++VR KMIYASSKDRFKRELDGIQ+ELQ
Sbjct: 61 DDCRYCIYDFDFVTAENCQKSKIFFIAWSPDTAKVRDKMIYASSKDRFKRELDGIQVELQ 120
Query: 121 ATDPTEMGLDVFKSRAN 137
ATDPTEMGLDVFKSR N
Sbjct: 121 ATDPTEMGLDVFKSRTN 137
>At3g46010 actin depolymerizing factor 1 (ADF1)
Length = 139
Score = 224 bits (571), Expect = 1e-59
Identities = 109/139 (78%), Positives = 125/139 (89%), Gaps = 2/139 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQ--VIVEKLGEPGQGYEDFTANL 58
MANAASGMAVHDDCKLRF+ELK KRT+RFIVYKIE+KQ V+VEK+G+P Q YE+F A L
Sbjct: 1 MANAASGMAVHDDCKLRFLELKAKRTHRFIVYKIEEKQKQVVVEKVGQPIQTYEEFAACL 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
PADECRYA+YDF+++T N KS+IFFIAW PD ++VRSKMIYASSKDRFKRELDGIQ+E
Sbjct: 61 PADECRYAIYDFDFVTAENCQKSKIFFIAWCPDIAKVRSKMIYASSKDRFKRELDGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQATDPTEM LDVF+SRAN
Sbjct: 121 LQATDPTEMDLDVFRSRAN 139
>At5g59890 actin depolymerizing factor 4 - like protein
Length = 139
Score = 224 bits (570), Expect = 1e-59
Identities = 110/139 (79%), Positives = 123/139 (88%), Gaps = 2/139 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQ--VIVEKLGEPGQGYEDFTANL 58
MANAASGMAVHDDCKLRF+ELK KRT+RFIVYKIE+KQ VIVEK+GEP YEDF A+L
Sbjct: 1 MANAASGMAVHDDCKLRFLELKAKRTHRFIVYKIEEKQKQVIVEKVGEPILTYEDFAASL 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
PADECRYA+YDF+++T N KS+IFFIAW PD ++VRSKMIYASSKDRFKRELDGIQ+E
Sbjct: 61 PADECRYAIYDFDFVTAENCQKSKIFFIAWCPDVAKVRSKMIYASSKDRFKRELDGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQATDPTEM LDV KSR N
Sbjct: 121 LQATDPTEMDLDVLKSRVN 139
>At5g52360 actin depolymerizing factor-like
Length = 137
Score = 213 bits (541), Expect = 3e-56
Identities = 98/136 (72%), Positives = 119/136 (87%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQVIVEKLGEPGQGYEDFTANLPA 60
MANAASGMAV D+CKL+F+ELK KR YRFI+++I+ +QV+VEKLG P + Y+DFT LP
Sbjct: 1 MANAASGMAVEDECKLKFLELKAKRNYRFIIFRIDGQQVVVEKLGSPQENYDDFTNYLPP 60
Query: 61 DECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQ 120
+ECRYAVYDF++ T N+ KS+IFFIAWSPD+SRVR KM+YASSKDRFKRELDGIQ+ELQ
Sbjct: 61 NECRYAVYDFDFTTAENIQKSKIFFIAWSPDSSRVRMKMVYASSKDRFKRELDGIQVELQ 120
Query: 121 ATDPTEMGLDVFKSRA 136
ATDP+EM LD+ KSRA
Sbjct: 121 ATDPSEMSLDIIKSRA 136
>At4g00680 putative actin-depolymerizing factor
Length = 140
Score = 210 bits (535), Expect = 2e-55
Identities = 98/139 (70%), Positives = 122/139 (87%), Gaps = 2/139 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANL 58
MAN+ASGM V+D+CK++F+ELK KRTYRFIV+KI++K QV +EKLG P + Y+DFT+++
Sbjct: 1 MANSASGMHVNDECKIKFLELKAKRTYRFIVFKIDEKAQQVQIEKLGNPEETYDDFTSSI 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
P DECRYAVYDF++ TE N KS+IFFIAWSPDTSRVRSKM+YASSKDRFKRE++GIQ+E
Sbjct: 61 PDDECRYAVYDFDFTTEDNCQKSKIFFIAWSPDTSRVRSKMLYASSKDRFKREMEGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQATDP+EM LD+ K R N
Sbjct: 121 LQATDPSEMSLDIIKGRLN 139
>At1g01750 actin depolymerizing factor like protein
Length = 140
Score = 208 bits (530), Expect = 6e-55
Identities = 98/139 (70%), Positives = 119/139 (85%), Gaps = 2/139 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANL 58
MAN+ASGM V D+CKL+F+ELK KR YRFIV+KI++K QV+++KLG P + YEDFT ++
Sbjct: 1 MANSASGMHVSDECKLKFLELKAKRNYRFIVFKIDEKAQQVMIDKLGNPEETYEDFTRSI 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
P DECRYAVYD+++ T N KS+IFFIAWSPDTSRVRSKM+YASSKDRFKRELDGIQ+E
Sbjct: 61 PEDECRYAVYDYDFTTPENCQKSKIFFIAWSPDTSRVRSKMLYASSKDRFKRELDGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQATDP+EM LD+ K R N
Sbjct: 121 LQATDPSEMSLDIIKGRVN 139
>At4g25590 actin depolymerizing factor - like protein
Length = 130
Score = 203 bits (516), Expect = 3e-53
Identities = 91/129 (70%), Positives = 116/129 (89%)
Query: 8 MAVHDDCKLRFMELKTKRTYRFIVYKIEDKQVIVEKLGEPGQGYEDFTANLPADECRYAV 67
MAV D+CKL+F+ELK+KR YRFI+++I+ +QV+VEKLG P + Y+DFTA+LPA+ECRYAV
Sbjct: 1 MAVEDECKLKFLELKSKRNYRFIIFRIDGQQVVVEKLGNPDETYDDFTASLPANECRYAV 60
Query: 68 YDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQATDPTEM 127
+DF+++T+ N KS+IFFIAWSPD+SRVR KM+YASSKDRFKRELDGIQ+ELQATDP+EM
Sbjct: 61 FDFDFITDENCQKSKIFFIAWSPDSSRVRMKMVYASSKDRFKRELDGIQVELQATDPSEM 120
Query: 128 GLDVFKSRA 136
D+ KSRA
Sbjct: 121 SFDIIKSRA 129
>At2g31200 actin depolymerizing factor 6 (ADF6)
Length = 146
Score = 166 bits (419), Expect = 5e-42
Identities = 75/137 (54%), Positives = 107/137 (77%), Gaps = 2/137 (1%)
Query: 3 NAASGMAVHDDCKLRFMELKTKRTYRFIVYKIED--KQVIVEKLGEPGQGYEDFTANLPA 60
NA SGM V D+ K F+EL+ K+T+R++V+KI++ K+V+VEK G P + Y+DF A+LP
Sbjct: 10 NAISGMGVADESKTTFLELQRKKTHRYVVFKIDESKKEVVVEKTGNPTESYDDFLASLPD 69
Query: 61 DECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQ 120
++CRYAVYDF+++T N KS+IFF AWSP TS +R+K++Y++SKD+ REL GI E+Q
Sbjct: 70 NDCRYAVYDFDFVTSENCQKSKIFFFAWSPSTSGIRAKVLYSTSKDQLSRELQGIHYEIQ 129
Query: 121 ATDPTEMGLDVFKSRAN 137
ATDPTE+ L+V + RAN
Sbjct: 130 ATDPTEVDLEVLRERAN 146
>At2g16700 actin depolymerizing factor 5 (ADF5)
Length = 143
Score = 158 bits (399), Expect = 1e-39
Identities = 72/135 (53%), Positives = 103/135 (75%), Gaps = 2/135 (1%)
Query: 4 AASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANLPAD 61
A +GM V D+C FM++K K+ +R+IV+KIE+K +V V+K+G G+ Y D +LP D
Sbjct: 8 ATTGMRVTDECTSSFMDMKWKKVHRYIVFKIEEKSRKVTVDKVGGAGESYHDLEDSLPVD 67
Query: 62 ECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQA 121
+CRYAV+DF+++T N KS+IFFIAWSP+ S++R+K++YA+SKD +R L+GI ELQA
Sbjct: 68 DCRYAVFDFDFVTVDNCRKSKIFFIAWSPEASKIRAKILYATSKDGLRRVLEGIHYELQA 127
Query: 122 TDPTEMGLDVFKSRA 136
TDPTEMG D+ + RA
Sbjct: 128 TDPTEMGFDIIQDRA 142
>At4g34970 actin depolymerizing factor - like protein
Length = 130
Score = 157 bits (396), Expect = 2e-39
Identities = 71/127 (55%), Positives = 96/127 (74%), Gaps = 2/127 (1%)
Query: 12 DDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANLPADECRYAVYD 69
DDCK FME+K K+ +R++VYK+E+K +V V+K+G G+ Y+D A+LP D+CRYAV+D
Sbjct: 3 DDCKKSFMEMKWKKVHRYVVYKLEEKSRKVTVDKVGAAGESYDDLAASLPEDDCRYAVFD 62
Query: 70 FEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQATDPTEMGL 129
F+Y+T N S+IFFI WSP+ SR+R KM+YA+SK +R LDG+ ELQATDPTEMG
Sbjct: 63 FDYVTVDNCRMSKIFFITWSPEASRIREKMMYATSKSGLRRVLDGVHYELQATDPTEMGF 122
Query: 130 DVFKSRA 136
D + RA
Sbjct: 123 DKIQDRA 129
>At3g45990 actin depolymerising like protein
Length = 133
Score = 153 bits (386), Expect = 3e-38
Identities = 82/138 (59%), Positives = 99/138 (71%), Gaps = 13/138 (9%)
Query: 8 MAVHDDCKLRFMELKTKRTYRFIVYKIEDK-QVIVEKL------GEPGQGYEDFTANLPA 60
M +HDDCKL F+ELK +RT+R IVYKIED QVIVEK GE Q YE+F +LPA
Sbjct: 1 MVLHDDCKLTFLELKERRTFRSIVYKIEDNMQVIVEKHHYKKMHGEREQSYEEFANSLPA 60
Query: 61 DECRYAVYDFEYLTEGNVPKSR-IFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIEL 119
DECRYA+ D E+ VP R I FIAWSP T+++R KMIY+S+KDRFKRELDGIQ+E
Sbjct: 61 DECRYAILDIEF-----VPGERKICFIAWSPSTAKMRKKMIYSSTKDRFKRELDGIQVEF 115
Query: 120 QATDPTEMGLDVFKSRAN 137
ATD T++ LD + R N
Sbjct: 116 HATDLTDISLDAIRRRIN 133
>At3g18330 hypothetical protein
Length = 376
Score = 31.2 bits (69), Expect = 0.18
Identities = 21/94 (22%), Positives = 41/94 (43%), Gaps = 3/94 (3%)
Query: 40 IVEKLGEPGQGYEDFTANLPADECR--YAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRS 97
+ K+ + +G +T L D R +A++ +L + K I ++W D +S
Sbjct: 278 VTNKIDDTTEGAVSWTKVLELDLSRELHALFTSNFLVDEE-KKVFICCVSWKEDEDENKS 336
Query: 98 KMIYASSKDRFKRELDGIQIELQATDPTEMGLDV 131
+Y +D +E+D + +PT + L V
Sbjct: 337 NKVYIVGEDNIVKEIDSGEDATSGCEPTILSLYV 370
>At2g07130 hypothetical protein
Length = 209
Score = 30.4 bits (67), Expect = 0.30
Identities = 22/94 (23%), Positives = 41/94 (43%), Gaps = 3/94 (3%)
Query: 40 IVEKLGEPGQGYEDFTANLPADECR--YAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRS 97
+ K+ + +G +T L D R A++ +L + K I ++W D +S
Sbjct: 111 VTNKIDDTTEGAVSWTKVLVLDLSRDLQALFTSNFLVDEE-KKVFICCVSWKEDEDENKS 169
Query: 98 KMIYASSKDRFKRELDGIQIELQATDPTEMGLDV 131
K +Y +D +E+D + +PT + L V
Sbjct: 170 KKVYIVGEDNRVKEIDSGEDATSGCEPTILSLYV 203
>At3g09720 RNA helicase like protein
Length = 541
Score = 30.0 bits (66), Expect = 0.39
Identities = 19/66 (28%), Positives = 32/66 (47%), Gaps = 10/66 (15%)
Query: 70 FEYLTEGNVPKSRI-FFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQATDPTEMG 128
F+ TE + ++ FF+ DT + ++ + SSK R +R + + +E G
Sbjct: 26 FKNSTEDDDSNKKVNFFVEEEEDTEQPEAEKVIVSSKKRKRRSSNSVPVE---------G 76
Query: 129 LDVFKS 134
DVFKS
Sbjct: 77 FDVFKS 82
>At3g04680 unknown protein
Length = 444
Score = 29.3 bits (64), Expect = 0.67
Identities = 17/38 (44%), Positives = 24/38 (62%), Gaps = 2/38 (5%)
Query: 67 VYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASS 104
V F Y+TE NV K +I ++A SP T + SK++ A S
Sbjct: 402 VSGFVYVTEVNVQKKKITYLAPSPGT--LPSKLLVAGS 437
>At1g63450 unknown protein
Length = 641
Score = 26.6 bits (57), Expect = 4.3
Identities = 23/89 (25%), Positives = 36/89 (39%), Gaps = 17/89 (19%)
Query: 46 EPGQGYEDFTANLPADECRYAVYDFEYLTEGNVPKSRIFFI-AWSPDTSRVRSKM----- 99
+P Y +T +LP D RY+V Y+ + +V R+ I T R R M
Sbjct: 548 DPYSAYYQYTWHLPEDHRRYSV----YINKEDVKLKRVNVIEKLMSKTLREREDMRSYIV 603
Query: 100 -------IYASSKDRFKRELDGIQIELQA 121
+Y S +F+R D I + +
Sbjct: 604 HELLPGLVYGDSNAKFERFRDAFDITMDS 632
Score = 25.4 bits (54), Expect = 9.6
Identities = 12/37 (32%), Positives = 20/37 (53%), Gaps = 3/37 (8%)
Query: 73 LTEGNVPKSRI---FFIAWSPDTSRVRSKMIYASSKD 106
L + ++PK+ I F I W P+T R + A ++D
Sbjct: 189 LDQDSLPKNEIDQDFIIDWDPETGEERYRYFKAKTED 225
>At4g12020 putative disease resistance protein
Length = 1895
Score = 26.2 bits (56), Expect = 5.6
Identities = 15/48 (31%), Positives = 24/48 (49%), Gaps = 1/48 (2%)
Query: 40 IVEKLGEPGQGYEDFTANLPADECRYAVYDFEYLTEGN-VPKSRIFFI 86
+V + G EDF ++L A CR + +E E + +PK R+ I
Sbjct: 670 VVIRYGRADISNEDFISHLRASLCRRGISVYEKFNEVDALPKCRVLII 717
>At3g62170 PECTINESTERASE-like protein
Length = 588
Score = 26.2 bits (56), Expect = 5.6
Identities = 15/42 (35%), Positives = 22/42 (51%), Gaps = 12/42 (28%)
Query: 5 ASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQVIVEKLGE 46
A+ AV D CK R ++Y +ED + IVE++GE
Sbjct: 108 ATSKAVLDYCK------------RVLMYALEDLETIVEEMGE 137
>At5g53960 unknown protein
Length = 189
Score = 25.8 bits (55), Expect = 7.4
Identities = 17/81 (20%), Positives = 35/81 (42%), Gaps = 5/81 (6%)
Query: 41 VEKLGEPGQGYEDFTANLPADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMI 100
+EKL + + E+ N Y +Y F E + DT + +K++
Sbjct: 44 IEKLKKVVRSLEEINRNSTMASLTYKLYRFILEKEDEAAAELN-----AVDTDSIETKLV 98
Query: 101 YASSKDRFKRELDGIQIELQA 121
+K R+ RE++ I++++
Sbjct: 99 KLENKQRYIREVEDNPIDIRS 119
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,950,070
Number of Sequences: 26719
Number of extensions: 115163
Number of successful extensions: 294
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 265
Number of HSP's gapped (non-prelim): 23
length of query: 137
length of database: 11,318,596
effective HSP length: 89
effective length of query: 48
effective length of database: 8,940,605
effective search space: 429149040
effective search space used: 429149040
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0066.6


