

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0066.5
(667 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
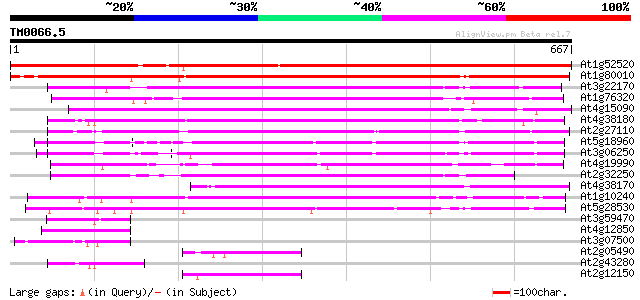
Score E
Sequences producing significant alignments: (bits) Value
At1g52520 F6D8.26 899 0.0
At1g80010 hypothetical protein 577 e-164
At3g22170 far-red impaired response protein, putative 372 e-103
At1g76320 putative phytochrome A signaling protein 363 e-100
At4g15090 unknown protein 357 9e-99
At4g38180 unknown protein (At4g38180) 341 9e-94
At2g27110 Mutator-like transposase 318 5e-87
At5g18960 FAR1 - like protein 315 7e-86
At3g06250 unknown protein 314 1e-85
At4g19990 putative protein 296 2e-80
At2g32250 Mutator-like transposase 281 1e-75
At4g38170 hypothetical protein 260 2e-69
At1g10240 unknown protein 208 9e-54
At5g28530 far-red impaired response protein (FAR1) - like 178 1e-44
At3g59470 unknown protein 75 9e-14
At4g12850 putative protein 62 1e-09
At3g07500 hypothetical protein 61 2e-09
At2g05490 Mutator-like transposase 55 1e-07
At2g43280 unknown protein 54 3e-07
At2g12150 pseudogene 54 4e-07
>At1g52520 F6D8.26
Length = 703
Score = 899 bits (2323), Expect = 0.0
Identities = 430/675 (63%), Positives = 533/675 (78%), Gaps = 17/675 (2%)
Query: 2 EETSLCCED--LPHGECI-EVHKDEDAALIELDSQNGFS-EGRKEFDAPAVGMEFESYDD 57
EE +C + + G C E K E+ + D Q+G + RKEFDAPAVGMEFESYDD
Sbjct: 37 EEALVCSSEHEIDVGFCQNEERKSEEETMGGFDEQSGLLVDERKEFDAPAVGMEFESYDD 96
Query: 58 AYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKDVNHLRKETRTGCP 117
AYNYY CYA EVGF VRVKNSWFKR S+EKYGAVLCCSSQGFKR DVN +RKETRTGCP
Sbjct: 97 AYNYYNCYASEVGFRVRVKNSWFKRRSKEKYGAVLCCSSQGFKRINDVNRVRKETRTGCP 156
Query: 118 AMMRIRIAESQRWRIIEVILEHNHILGAKIHKSAKKMGNGTKRKSLPSSDSEVQTVKLYR 177
AM+R+R +S+RWR++EV L+HNH+LG K++KS K+ KRK + S S+ +T+KLYR
Sbjct: 157 AMIRMRQVDSKRWRVVEVTLDHNHLLGCKLYKSVKR-----KRKCVSSPVSDAKTIKLYR 211
Query: 178 PLVIDAGGNGNLNPSARDDRTFSEMSNK---LNVKRGDTQAIYNFLCRMQLTNPNFFYLM 234
V+D G N +NP++ ++ F + LN+KRGD+ AIYN+ CRMQLTNPNFFYLM
Sbjct: 212 ACVVDNGSN--VNPNSTLNKKFQNSTGSPDLLNLKRGDSAAIYNYFCRMQLTNPNFFYLM 269
Query: 235 DFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVGINHHGQSVLLG 294
D +DEG LRN FWADA S+ +C YFGDVI+ D++Y+S K+EIPLV F G+NHHG++ LL
Sbjct: 270 DVNDEGQLRNVFWADAFSKVSCSYFGDVIFIDSSYISGKFEIPLVTFTGVNHHGKTTLLS 329
Query: 295 CGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDRCEALQSAIIEVFPKSHHCFGLSLIM 354
CG LAGET ESY WL + W++ M SPQTI+TDRC+ L++AI +VFP+SH F L+ IM
Sbjct: 330 CGFLAGETMESYHWLLKVWLSVMKR-SPQTIVTDRCKPLEAAISQVFPRSHQRFSLTHIM 388
Query: 355 KKVPEKLGGLHNYDGIRKALIKAVYETLKISEFEAAWGIMIQRFGVSDHEWLRSLYEDRV 414
+K+PEKLGGLHNYD +RKA KAVYETLK+ EFEAAWG M+ FGV ++EWLRSLYE+R
Sbjct: 389 RKIPEKLGGLHNYDAVRKAFTKAVYETLKVVEFEAAWGFMVHNFGVIENEWLRSLYEERA 448
Query: 415 RWAPVFLKDVFFGGMSAARPGESIGPFFGRYVHKQTPLKEFLDKYELALHKKHKEESLAD 474
+WAPV+LKD FF G++AA PGE++ PFF RYVHKQTPLKEFLDKYELAL KKH+EE+L+D
Sbjct: 449 KWAPVYLKDTFFAGIAAAHPGETLKPFFERYVHKQTPLKEFLDKYELALQKKHREETLSD 508
Query: 475 TESRN-SSPVLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCFGTTQLHVDGPIVIYLV 533
ES+ ++ LKT+CS E QLSR+YTR++F KFQ EVEEM+SCF TTQ+HVDGP VI+LV
Sbjct: 509 IESQTLNTAELKTKCSFETQLSRIYTRDMFKKFQIEVEEMYSCFSTTQVHVDGPFVIFLV 568
Query: 534 KERVLIDGNRREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHALCVLNFNGVEEIPFKY 593
KERV + +RREI+DFEV+Y+R+ GE+RCICSCFNFYGYLCRHALCVLNFNGVEEIP +Y
Sbjct: 569 KERVRGESSRREIRDFEVLYNRSVGEVRCICSCFNFYGYLCRHALCVLNFNGVEEIPLRY 628
Query: 594 ILSRWKKDYKRLHVPDHSSVS-ADDTDPIQWSNQLFSSALQVVEEGTISLDHYNVALQAC 652
IL RW+KDYKRLH D+ D TD +QW +QL+ ++LQVVEEG +SLDHY VA+Q
Sbjct: 629 ILPRWRKDYKRLHFADNGLTGFVDGTDRVQWFDQLYKNSLQVVEEGAVSLDHYKVAMQVL 688
Query: 653 EESLSKVHDVDQRQD 667
+ESL KVH V+++QD
Sbjct: 689 QESLDKVHSVEEKQD 703
>At1g80010 hypothetical protein
Length = 696
Score = 577 bits (1486), Expect = e-164
Identities = 306/680 (45%), Positives = 430/680 (63%), Gaps = 29/680 (4%)
Query: 2 EETSLCCEDLPHGECIEVHKDEDAALIELDSQNGFSEGRKEFDAPAV-GMEFESYDDAYN 60
++ CC L H I + +E+ E N F+ + P GMEFESYDDAY+
Sbjct: 28 DDDDACCHGLLH---IAPNHEEETGCDE----NAFANEKCLMAPPPTPGMEFESYDDAYS 80
Query: 61 YYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKDVNHLRKETRTGCPAMM 120
+Y YA+E+GF +RVK+SW KRNS+EK GAVLCC+ QGFK KD + RKETRTGC AM+
Sbjct: 81 FYNSYARELGFAIRVKSSWTKRNSKEKRGAVLCCNCQGFKLLKDAHSRRKETRTGCQAMI 140
Query: 121 RIRIAESQRWRIIEVILEHNHIL------GAKIHK-SAKKMGNGTKRKSLPSSDSEVQTV 173
R+R+ RW++ +V L+HNH +K HK S+ TK P +V+T+
Sbjct: 141 RLRLIHFDRWKVDQVKLDHNHSFDPQRAHNSKSHKKSSSSASPATKTNPEPPPHVQVRTI 200
Query: 174 KLYRPLVIDAGGNGNLNPSARDDRTFS----EMSNKLNVKRGDTQAIYNFLCRMQLTNPN 229
KLYR L +D + S+ + S + S +L + RG +A+ +F ++QL++PN
Sbjct: 201 KLYRTLALDTPPALGTSLSSGETSDLSLDHFQSSRRLEL-RGGFRALQDFFFQIQLSSPN 259
Query: 230 FFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVGINHHGQ 289
F YLMD D+G LRN FW DAR+RAA +FGDV+ FD T LS+ YE+PLV FVGINHHG
Sbjct: 260 FLYLMDLADDGSLRNVFWIDARARAAYSHFGDVLLFDTTCLSNAYELPLVAFVGINHHGD 319
Query: 290 SVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDRCEALQSAIIEVFPKSHHCFG 349
++LLGCGLLA ++ E+Y+WLFR W+TCM G PQ IT++C+A+++A+ EVFP++HH
Sbjct: 320 TILLGCGLLADQSFETYVWLFRAWLTCMLGRPPQIFITEQCKAMRTAVSEVFPRAHHRLS 379
Query: 350 LSLIMKKVPEKLGGLHNYDGIRKALIKAVYETLKISEFEAAWGIMIQRFGVSDHEWLRSL 409
L+ ++ + + + L + D AL + VY LK+ EFE AW MI RFG++++E +R +
Sbjct: 380 LTHVLHNICQSVVQLQDSDLFPMALNRVVYGCLKVEEFETAWEEMIIRFGMTNNETIRDM 439
Query: 410 YEDRVRWAPVFLKDVFFGGMSAARPGESIGPF-FGRYVHKQTPLKEFLDKYELALHKKHK 468
++DR WAPV+LKD F G G PF F YVH+ T L+EFL+ YE L KK+
Sbjct: 440 FQDRELWAPVYLKDTFLAGALTFPLGNVAAPFIFSGYVHENTSLREFLEGYESFLDKKYT 499
Query: 469 EESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCFGTTQLHVDGPI 528
E+L D+ES P LKT E Q+++++T EIF +FQ EV M SCFG TQ+H +G
Sbjct: 500 REALCDSESLKLIPKLKTTHPYESQMAKVFTMEIFRRFQDEVSAMSSCFGVTQVHSNGSA 559
Query: 529 VIYLVKERVLIDGNRREIKDFEVVY-SRAAGELRCICSC--FNFYGYLCRHALCVLNFNG 585
Y+VKER +G++ ++DFEV+Y + AA ++RC C C F+F GY CRH L +L+ NG
Sbjct: 560 SSYVVKER---EGDK--VRDFEVIYETSAAAQVRCFCVCGGFSFNGYQCRHVLLLLSHNG 614
Query: 586 VEEIPFKYILSRWKKDYKRLHVPDHSSVSADDTDPIQWSNQLFSSALQVVEEGTISLDHY 645
++E+P +YIL RW+KD KRL+V + S D +P QW L A+QVVE+G S +H
Sbjct: 615 LQEVPPQYILQRWRKDVKRLYVAEFGSGRVDIMNPDQWYEHLHRRAMQVVEQGMRSKEHC 674
Query: 646 NVALQACEESLSKVHDVDQR 665
A +A E +KV V ++
Sbjct: 675 RAAWEAFRECANKVQFVTEK 694
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 372 bits (955), Expect = e-103
Identities = 211/629 (33%), Positives = 324/629 (50%), Gaps = 48/629 (7%)
Query: 46 PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKDV 105
P GMEFES+ +AY++Y Y++ +GF ++NS + +RE A CS G KR D
Sbjct: 70 PLNGMEFESHGEAYSFYQEYSRAMGFNTAIQNSRRSKTTREFIDAKFACSRYGTKREYDK 129
Query: 106 NHLRKETR-----------------TGCPAMMRIRIAESQRWRIIEVILEHNHILGAKIH 148
+ R R T C A M ++ +W I + EHNH L
Sbjct: 130 SFNRPRARQSKQDPENMAGRRTCAKTDCKASMHVKRRPDGKWVIHSFVREHNHEL----- 184
Query: 149 KSAKKMGNGTKRKSLPSSDSEVQTVKLYRPLVIDAGGNGNLNPSARDDRTFSEMSNKLNV 208
LP+ QT K+Y + + D ++ E L+V
Sbjct: 185 --------------LPAQAVSEQTRKIYAAMAKQFAEYKTVISLKSDSKSSFEKGRTLSV 230
Query: 209 KRGDTQAIYNFLCRMQLTNPNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNT 268
+ GD + + +FL RMQ N NFFY +D D+ ++N FW DA+SR G F DV+ D T
Sbjct: 231 ETGDFKILLDFLSRMQSLNSNFFYAVDLGDDQRVKNVFWVDAKSRHNYGSFCDVVSLDTT 290
Query: 269 YLSSKYEIPLVVFVGINHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITD 328
Y+ +KY++PL +FVG+N H Q ++LGC L++ E+ +Y WL TW+ + G +P+ +IT+
Sbjct: 291 YVRNKYKMPLAIFVGVNQHYQYMVLGCALISDESAATYSWLMETWLRAIGGQAPKVLITE 350
Query: 329 RCEALQSAIIEVFPKSHHCFGLSLIMKKVPEKLGG-LHNYDGIRKALIKAVYETLKISEF 387
+ S + E+FP + HC L ++ KV E LG + +D K +Y++ K +F
Sbjct: 351 LDVVMNSIVPEIFPNTRHCLFLWHVLMKVSENLGQVVKQHDNFMPKFEKCIYKSGKDEDF 410
Query: 388 EAAWGIMIQRFGVSDHEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFGRYVH 447
W + RFG+ D +W+ SLYEDR +WAP ++ DV GMS ++ +SI FF +Y+H
Sbjct: 411 ARKWYKNLARFGLKDDQWMISLYEDRKKWAPTYMTDVLLAGMSTSQRADSINAFFDKYMH 470
Query: 448 KQTPLKEFLDKYELALHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQ 507
K+T ++EF+ Y+ L + +EE+ AD+E N P +K+ E +S +YT +F KFQ
Sbjct: 471 KKTSVQEFVKVYDTVLQDRCEEEAKADSEMWNKQPAMKSPSPFEKSVSEVYTPAVFKKFQ 530
Query: 508 FEVEEMFSCFGTTQLHVDGPIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCICSCF 567
EV +C + + D + V++ + N +DF V +++ E+ CIC F
Sbjct: 531 IEVLGAIAC-SPREENRDATCSTFRVQD---FENN----QDFMVTWNQTKAEVSCICRLF 582
Query: 568 NFYGYLCRHALCVLNFNGVEEIPFKYILSRWKKDYKRLHVPDHSSVSADDTDPIQWSNQL 627
+ GYLCRH L VL + IP +YIL RW KD K H S + N L
Sbjct: 583 EYKGYLCRHTLNVLQCCHLSSIPSQYILKRWTKDAKSRH---FSGEPQQLQTRLLRYNDL 639
Query: 628 FSSALQVVEEGTISLDHYNVALQACEESL 656
AL++ EE ++S + YN+A A E ++
Sbjct: 640 CERALKLNEEASLSQESYNIAFLAIEGAI 668
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 363 bits (931), Expect = e-100
Identities = 212/627 (33%), Positives = 329/627 (51%), Gaps = 48/627 (7%)
Query: 50 MEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKR-TKDVNHL 108
MEFE+++DAY +Y YAK VGF +S R S+E A C G K+ + D +
Sbjct: 1 MEFETHEDAYLFYKDYAKSVGFGTAKLSSRRSRASKEFIDAKFSCIRYGSKQQSDDAINP 60
Query: 109 RKETRTGCPAMMRIRIAESQRWRIIEVILEHNHILGA------KIHKSAKKMGNGTKR-- 160
R + GC A M ++ +W + + EHNH L + H++ + + + R
Sbjct: 61 RASPKIGCKASMHVKRRPDGKWYVYSFVKEHNHDLLPEQAHYFRSHRNTELVKSNDSRLR 120
Query: 161 --KSLPSSDSEVQTVKLYRPLVIDAGGNGNLNPSARDDRTFSEMSNKLNVKRGDTQAIYN 218
K+ P +D + + Y L G N + R +L + GD + +
Sbjct: 121 RKKNTPLTDCK--HLSAYHDLDFIDGYMRNQHDKGR----------RLVLDTGDAEILLE 168
Query: 219 FLCRMQLTNPNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPL 278
FL RMQ NP FF+ +DF ++ LRN FW DA+ F DV+ F+ +Y SKY++PL
Sbjct: 169 FLMRMQEENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFVSKYKVPL 228
Query: 279 VVFVGINHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDRCEALQSAII 338
V+FVG+NHH Q VLLGCGLLA +T +Y+WL ++W+ M G P+ ++TD+ A+++AI
Sbjct: 229 VLFVGVNHHVQPVLLGCGLLADDTVYTYVWLMQSWLVAMGGQKPKVMLTDQNNAIKAAIA 288
Query: 339 EVFPKSHHCFGLSLIMKKVPEKLGGLHNY-DGIRKALIKAVYETLKISEFEAAWGIMIQR 397
V P++ HC+ L ++ ++P L + D K L K +Y + EF+ W +I +
Sbjct: 289 AVLPETRHCYCLWHVLDQLPRNLDYWSMWQDTFMKKLFKCIYRSWSEEEFDRRWLKLIDK 348
Query: 398 FGVSDHEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFGRYVHKQTPLKEFLD 457
F + D W+RSLYE+R WAP F++ + F G+S ES+ F RYVH +T LKEFL+
Sbjct: 349 FHLRDVPWMRSLYEERKFWAPTFMRGITFAGLSMRCRSESVNSLFDRYVHPETSLKEFLE 408
Query: 458 KYELALHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCF 517
Y L L +++EE+ AD ++ + +P LK+ E Q+ +Y+ EIF +FQ EV +C
Sbjct: 409 GYGLMLEDRYEEEAKADFDAWHEAPELKSPSPFEKQMLLVYSHEIFRRFQLEVLGAAAC- 467
Query: 518 GTTQLHVDGPIVIYLVKERVLIDGNRREIKDFE------VVYSRAAGELRCICSCFNFYG 571
+L KE +G +KDF+ V + ++ C C F + G
Sbjct: 468 -------------HLTKESE--EGTTYSVKDFDDEQKYLVDWDEFKSDIYCSCRSFEYKG 512
Query: 572 YLCRHALCVLNFNGVEEIPFKYILSRWKKDYKRLHVPDHSSVSADDTDPIQWSNQLFSSA 631
YLCRHA+ VL +GV IP Y+L RW + H + I+ N L A
Sbjct: 513 YLCRHAIVVLQMSGVFTIPINYVLQRWTNAARNRHQISRNLELVQSN--IRRFNDLCRRA 570
Query: 632 LQVVEEGTISLDHYNVALQACEESLSK 658
+ + EEG++S + Y++A+ A +E+ +
Sbjct: 571 IILGEEGSLSQESYDIAMFAMKEAFKQ 597
>At4g15090 unknown protein
Length = 768
Score = 357 bits (917), Expect = 9e-99
Identities = 200/610 (32%), Positives = 328/610 (52%), Gaps = 27/610 (4%)
Query: 71 FCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKDVN--HLRKET--RTGCPAMMRIRIAE 126
F +KNS + +++ A CS G + + R+ T +T C A M ++
Sbjct: 17 FTTSIKNSRRSKKTKDFIDAKFACSRYGVTPESESSGSSSRRSTVKKTDCKASMHVKRRP 76
Query: 127 SQRWRIIEVILEHNHIL--GAKIHKSAKKMGNGTKRKSLPSSDS-EVQTVKLYRPLVIDA 183
+W I E + +HNH L H ++ ++ ++ + +T K+Y + +
Sbjct: 77 DGKWIIHEFVKDHNHELLPALAYHFRIQRNVKLAEKNNIDILHAVSERTKKMYVEMSRQS 136
Query: 184 GGNGNLNPSARDDRTFS-EMSNKLNVKRGDTQAIYNFLCRMQLTNPNFFYLMDFDDEGHL 242
GG N+ + D + + L ++ GD+Q + + R++ NP FFY +D +++ L
Sbjct: 137 GGYKNIGSLLQTDVSSQVDKGRYLALEEGDSQVLLEYFKRIKKENPKFFYAIDLNEDQRL 196
Query: 243 RNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVGINHHGQSVLLGCGLLAGET 302
RN FWADA+SR F DV+ FD TY+ ++PL +F+G+NHH Q +LLGC L+A E+
Sbjct: 197 RNLFWADAKSRDDYLSFNDVVSFDTTYVKFNDKLPLALFIGVNHHSQPMLLGCALVADES 256
Query: 303 TESYIWLFRTWVTCMSGCSPQTIITDRCEALQSAIIEVFPKSHHCFGLSLIMKKVPEKLG 362
E+++WL +TW+ M G +P+ I+TD+ + L SA+ E+ P + HCF L +++K+PE
Sbjct: 257 METFVWLIKTWLRAMGGRAPKVILTDQDKFLMSAVSELLPNTRHCFALWHVLEKIPEYFS 316
Query: 363 G-LHNYDGIRKALIKAVYETLKISEFEAAWGIMIQRFGVSDHEWLRSLYEDRVRWAPVFL 421
+ ++ K ++ + EF+ W M+ +FG+ + EWL L+E R +W P F+
Sbjct: 317 HVMKRHENFLLKFNKCIFRSWTDDEFDMRWWKMVSQFGLENDEWLLWLHEHRQKWVPTFM 376
Query: 422 KDVFFGGMSAARPGESIGPFFGRYVHKQTPLKEFLDKYELALHKKHKEESLADTESRNSS 481
DVF GMS ++ ES+ FF +Y+HK+ LKEFL +Y + L +++EES+AD ++ +
Sbjct: 377 SDVFLAGMSTSQRSESVNSFFDKYIHKKITLKEFLRQYGVILQNRYEEESVADFDTCHKQ 436
Query: 482 PVLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCFGTTQLHVDGPIVIYLVKERVLIDG 541
P LK+ E Q++ YT IF KFQ EV + +C + D + + V++ D
Sbjct: 437 PALKSPSPWEKQMATTYTHTIFKKFQVEVLGVVACHPRKEKE-DENMATFRVQDCEKDD- 494
Query: 542 NRREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHALCVLNFNGVEEIPFKYILSRWKKD 601
DF V +S+ EL C C F + G+LCRHAL +L G IP +YIL RW KD
Sbjct: 495 ------DFLVTWSKTKSELCCFCRMFEYKGFLCRHALMILQMCGFASIPPQYILKRWTKD 548
Query: 602 YKRLHVPDHSSVSADDTDPIQWS----NQLFSSALQVVEEGTISLDHYNVALQACEESLS 657
K ++ + D IQ N L S A ++ EEG +S ++YN+AL+ E+L
Sbjct: 549 AK------SGVLAGEGADQIQTRVQRYNDLCSRATELSEEGCVSEENYNIALRTLVETLK 602
Query: 658 KVHDVDQRQD 667
D++ ++
Sbjct: 603 NCVDMNNARN 612
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 341 bits (874), Expect = 9e-94
Identities = 205/634 (32%), Positives = 335/634 (52%), Gaps = 33/634 (5%)
Query: 46 PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVL----CCSSQGF-- 99
P G+EFES + A +Y YA+ +GF RV +S +R+ R+ GA++ C+ +GF
Sbjct: 73 PYDGLEFESEEAAKAFYNSYARRIGFSTRVSSS--RRSRRD--GAIIQRQFVCAKEGFRN 128
Query: 100 ---KRTKD--VNHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHILGA--KIH--KS 150
KRTKD + R TR GC A + +++ +S +W + + +HNH L ++H +S
Sbjct: 129 MNEKRTKDREIKRPRTITRVGCKASLSVKMQDSGKWLVSGFVKDHNHELVPPDQVHCLRS 188
Query: 151 AKKMGNGTKRKSLPSSDSEVQTVKLYRPLVIDAGGNGNLNPSARDDRTFSEMSNKLNVKR 210
+++ K + + ++ L+ + GG + + D R + + + +++
Sbjct: 189 HRQISGPAKTLIDTLQAAGMGPRRIMSALIKEYGGISKVGFTEVDCRNYMRNNRQKSIE- 247
Query: 211 GDTQAIYNFLCRMQLTNPNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYL 270
G+ Q + ++L +M NPNFFY + ++ + N FWAD ++ +FGD + FD TY
Sbjct: 248 GEIQLLLDYLRQMNADNPNFFYSVQGSEDQSVGNVFWADPKAIMDFTHFGDTVTFDTTYR 307
Query: 271 SSKYEIPLVVFVGINHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDRC 330
S++Y +P F G+NHHGQ +L GC + ET S++WLF TW+ MS P +I TD
Sbjct: 308 SNRYRLPFAPFTGVNHHGQPILFGCAFIINETEASFVWLFNTWLAAMSAHPPVSITTDHD 367
Query: 331 EALQSAIIEVFPKSHHCFGLSLIMKKVPEKLGGLH-NYDGIRKALIKAVYETLKISEFEA 389
+++AI+ VFP + H F I+KK EKL + + K V T + +FE
Sbjct: 368 AVIRAAIMHVFPGARHRFCKWHILKKCQEKLSHVFLKHPSFESDFHKCVNLTESVEDFER 427
Query: 390 AWGIMIQRFGVSDHEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFGRYVHKQ 449
W ++ ++ + DHEWL+++Y DR +W PV+L+D FF MS +SI +F Y++
Sbjct: 428 CWFSLLDKYELRDHEWLQAIYSDRRQWVPVYLRDTFFADMSLTHRSDSINSYFDGYINAS 487
Query: 450 TPLKEFLDKYELALHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQFE 509
T L +F YE AL + ++E AD ++ NS PVLKT +E Q S +YTR++FM+FQ E
Sbjct: 488 TNLSQFFKLYEKALESRLEKEVKADYDTMNSPPVLKTPSPMEKQASELYTRKLFMRFQEE 547
Query: 510 VEEMFSCFGTTQLHVDGPIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCICSCFNF 569
+ + F ++ DG +V Y V + + ++ F V+ RA C C F F
Sbjct: 548 LVGTLT-FMASKADDDGDLVTYQVAK--YGEAHKAHFVKFNVLEMRA----NCSCQMFEF 600
Query: 570 YGYLCRHALCVLNFNGVEEIPFKYILSRWKKDYKRLHVPD----HSSVSADDTDPIQWSN 625
G +CRH L V + +P YIL RW ++ K + D H+ + ++ +++ N
Sbjct: 601 SGIICRHILAVFRVTNLLTLPPYYILKRWTRNAKSSVIFDDYNLHAYANYLESHTVRY-N 659
Query: 626 QLFSSALQVVEEGTISLDHYNVALQACEESLSKV 659
L A V+E SL +VA+ A +E+ V
Sbjct: 660 TLRHKASNFVQEAGKSLYTCDVAVVALQEAAKTV 693
>At2g27110 Mutator-like transposase
Length = 851
Score = 318 bits (816), Expect = 5e-87
Identities = 197/624 (31%), Positives = 308/624 (48%), Gaps = 35/624 (5%)
Query: 46 PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKDV 105
P VGMEF S +A ++Y Y++++GF ++ R CSS KR+K
Sbjct: 49 PCVGMEFNSEKEAKSFYDEYSRQLGFTSKL----LPRTDGSVSVREFVCSSSS-KRSK-- 101
Query: 106 NHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHILGAK--IHKSAKKMGNGTKRKSL 163
+ C AM+RI + ++W + + + EH H L + +H + KS
Sbjct: 102 ----RRLSESCDAMVRIELQGHEKWVVTKFVKEHTHGLASSNMLHCLRPRRHFANSEKSS 157
Query: 164 PSSDSEVQTVKLYRPLVIDAGGNGNLNPSARDDRTFSEMSNKLNVKRGDTQAIYNFLCRM 223
V + +Y + ++ G N + + RT D + + RM
Sbjct: 158 YQEGVNVPSGMMYVSMDANSRGARNASMATNTKRTIGR----------DAHNLLEYFKRM 207
Query: 224 QLTNPNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVG 283
Q NP FFY + D++ + N FWAD+RSR A +FGD + D Y +++ +P F G
Sbjct: 208 QAENPGFFYAVQLDEDNQMSNVFWADSRSRVAYTHFGDTVTLDTRYRCNQFRVPFAPFTG 267
Query: 284 INHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDRCEALQSAIIEVFPK 343
+NHHGQ++L GC L+ E+ S+IWLF+T++T M P +++TD+ A+Q A +VFP
Sbjct: 268 VNHHGQAILFGCALILDESDTSFIWLFKTFLTAMRDQPPVSLVTDQDRAIQIAAGQVFPG 327
Query: 344 SHHCFGLSLIMKKVPEKLGGL-HNYDGIRKALIKAVYETLKISEFEAAWGIMIQRFGVSD 402
+ HC ++++ EKL + Y + L + T I EFE++W +I ++ +
Sbjct: 328 ARHCINKWDVLREGQEKLAHVCLAYPSFQVELYNCINFTETIEEFESSWSSVIDKYDLGR 387
Query: 403 HEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFGRYVHKQTPLKEFLDKYELA 462
HEWL SLY R +W PV+ +D FF + ++ G S G FF YV++QT L F YE A
Sbjct: 388 HEWLNSLYNARAQWVPVYFRDSFFAAVFPSQ-GYS-GSFFDGYVNQQTTLPMFFRLYERA 445
Query: 463 LHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCFGTTQL 522
+ + E AD ++ N+ PVLKT +E Q + ++TR+IF KFQ E+ E F+ ++
Sbjct: 446 MESWFEMEIEADLDTVNTPPVLKTPSPMENQAANLFTRKIFGKFQEELVETFA-HTANRI 504
Query: 523 HVDGPIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHALCVLN 582
DG + V + K + V + C C F G LCRH L V
Sbjct: 505 EDDGTTSTFRVA------NFENDNKAYIVTFCYPEMRANCSCQMFEHSGILCRHVLTVFT 558
Query: 583 FNGVEEIPFKYILSRWKKDYKRL-HVPDHSSVSADDTDPIQWSNQLFSSALQVVEEGTIS 641
+ +P YIL RW ++ K + + +H S + D+ I N L A++ EEG I+
Sbjct: 559 VTNILTLPPHYILRRWTRNAKSMVELDEHVSENGHDSS-IHRYNHLCREAIKYAEEGAIT 617
Query: 642 LDHYNVALQACEESLSKVHDVDQR 665
+ YN+AL E KV V +R
Sbjct: 618 AEAYNIALGQLREGGKKVSVVRKR 641
>At5g18960 FAR1 - like protein
Length = 788
Score = 315 bits (806), Expect = 7e-86
Identities = 202/618 (32%), Positives = 318/618 (50%), Gaps = 48/618 (7%)
Query: 46 PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKDV 105
P G+EF S ++A +Y YA+ VGF VR+ + + CS +GF+
Sbjct: 211 PYAGLEFGSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGSITSRRFVCSREGFQHP--- 267
Query: 106 NHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHILGAKIHKSAKKMGNGTKRKSLPS 165
+R GC A MRI+ +S W + + +HNH L + KK G K+ +P
Sbjct: 268 ------SRMGCGAYMRIKRQDSGGWIVDRLNKDHNHDL-----EPGKKNDAGMKK--IPD 314
Query: 166 SDSE-VQTVKLYRPLVIDAGGNGNLNPSARDDRTFSEMSNKLNVKRGDTQAIYNFLCRMQ 224
+ + +V L + ++ GN ++ + R++R E L ++ Q
Sbjct: 315 DGTGGLDSVDL---IELNDFGNNHIKKT-RENRIGKEWYPLL----------LDYFQSRQ 360
Query: 225 LTNPNFFYLMDFD-DEGHLRNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVG 283
+ FFY ++ D + G + FWAD+R+R AC FGD + FD +Y Y +P +G
Sbjct: 361 TEDMGFFYAVELDVNNGSCMSIFWADSRARFACSQFGDSVVFDTSYRKGSYSVPFATIIG 420
Query: 284 INHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDRCEALQSAIIEVFPK 343
NHH Q VLLGC ++A E+ E+++WLF+TW+ MSG P++I+ D+ +Q A+++VFP
Sbjct: 421 FNHHRQPVLLGCAMVADESKEAFLWLFQTWLRAMSGRRPRSIVADQDLPIQQALVQVFPG 480
Query: 344 SHHCFGLSLIMKKVPEKLGGLHNYDGIRKALIKAVYETLKISEFEAAWGIMIQRFGVSDH 403
+HH + I +K E L + + K +Y+T I EF++ W +I ++G+ D
Sbjct: 481 AHHRYSAWQIREKERENL--IPFPSEFKYEYEKCIYQTQTIVEFDSVWSALINKYGLRDD 538
Query: 404 EWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFGRYVHKQTPLKEFLDKYELAL 463
WLR +YE R W P +L+ FF G+ +I PFFG + TPL+EF+ +YE AL
Sbjct: 539 VWLREIYEQRENWVPAYLRASFFAGIPI---NGTIEPFFGASLDALTPLREFISRYEQAL 595
Query: 464 HKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCFGTTQLH 523
++ +EE D S N P L+T+ +E Q R+YT +F FQ E+ + ++ + + +
Sbjct: 596 EQRREEERKEDFNSYNLQPFLQTKEPVEEQCRRLYTLTVFRIFQNELVQSYN-YLCLKTY 654
Query: 524 VDGPIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHALCVLNF 583
+G I +LV++ GN E + V +S + C C F G LCRH L V N
Sbjct: 655 EEGAISRFLVRK----CGN--ESEKHAVTFSASNLNSSCSCQMFEHEGLLCRHILKVFNL 708
Query: 584 NGVEEIPFKYILSRWKKDYKRLHVPD-HSSVSADDTDPIQ-WSNQLFSSALQVVEEGTIS 641
+ E+P +YIL RW K+ + V D S VSA D + WS L +A + +E GT S
Sbjct: 709 LDIRELPSRYILHRWTKNAEFGFVRDMESGVSAQDLKALMVWS--LREAASKYIEFGTSS 766
Query: 642 LDHYNVALQACEESLSKV 659
L+ Y +A + E K+
Sbjct: 767 LEKYKLAYEIMREGGKKL 784
Score = 62.0 bits (149), Expect = 1e-09
Identities = 36/119 (30%), Positives = 57/119 (47%), Gaps = 11/119 (9%)
Query: 30 LDSQNGFSEGRKEFDA--PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREK 87
L NG SE + P VG+EF++ ++A +Y YA GF VR + R
Sbjct: 25 LHHNNGISEDEEGGSGVEPYVGLEFDTAEEAREFYNAYAARTGFKVRTGQLYRSRTDGTV 84
Query: 88 YGAVLCCSSQGFKRTKDVNHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHILGAK 146
CS +GF + +RTGC A +R++ ++ +W + ++ EHNH LG +
Sbjct: 85 SSRRFVCSKEGF---------QLNSRTGCTAFIRVQRRDTGKWVLDQIQKEHNHELGGE 134
>At3g06250 unknown protein
Length = 764
Score = 314 bits (804), Expect = 1e-85
Identities = 196/622 (31%), Positives = 307/622 (48%), Gaps = 59/622 (9%)
Query: 46 PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKDV 105
P G+EF S ++A +Y YA+ VGF VR+ + + CS +GF+
Sbjct: 190 PYAGLEFNSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGSITSRRFVCSKEGFQH---- 245
Query: 106 NHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHILGAKIHKSAKKMGNGTKRKSLPS 165
+R GC A MRI+ +S W + + +HNH L + KK N +K
Sbjct: 246 -----PSRMGCGAYMRIKRQDSGGWIVDRLNKDHNHDL-----EPGKK--NAGMKKITDD 293
Query: 166 SDSEVQTVKLYRPLVIDAGGNGNLNPSARDDRTFSEMSNKLNVKRGDT------QAIYNF 219
+ +V L +++SN ++ R +T + ++
Sbjct: 294 VTGGLDSVDLIE---------------------LNDLSNHISSTRENTIGKEWYPVLLDY 332
Query: 220 LCRMQLTNPNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLV 279
Q + FFY ++ D G + FWAD+RSR AC FGD + FD +Y Y +P
Sbjct: 333 FQSKQAEDMGFFYAIELDSNGSCMSIFWADSRSRFACSQFGDAVVFDTSYRKGDYSVPFA 392
Query: 280 VFVGINHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDRCEALQSAIIE 339
F+G NHH Q VLLG L+A E+ E++ WLF+TW+ MSG P++++ D+ +Q A+ +
Sbjct: 393 TFIGFNHHRQPVLLGGALVADESKEAFSWLFQTWLRAMSGRRPRSMVADQDLPIQQAVAQ 452
Query: 340 VFPKSHHCFGLSLIMKKVPEKLGGLHNYDGIRKALIKAVYETLKISEFEAAWGIMIQRFG 399
VFP +HH F I K E L N + K +Y++ EF+ W ++ ++G
Sbjct: 453 VFPGTHHRFSAWQIRSKERENLRSFPN--EFKYEYEKCLYQSQTTVEFDTMWSSLVNKYG 510
Query: 400 VSDHEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFGRYVHKQTPLKEFLDKY 459
+ D+ WLR +YE R +W P +L+ FFGG+ + PF+G ++ T L+EF+ +Y
Sbjct: 511 LRDNMWLREIYEKREKWVPAYLRASFFGGIHV---DGTFDPFYGTSLNSLTSLREFISRY 567
Query: 460 ELALHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCFGT 519
E L ++ +EE D S N P L+T+ +E Q R+YT IF FQ E+ + ++ G
Sbjct: 568 EQGLEQRREEERKEDFNSYNLQPFLQTKEPVEEQCRRLYTLTIFRIFQSELAQSYNYLG- 626
Query: 520 TQLHVDGPIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHALC 579
+ + +G I +LV++ GN E V +S + C C F + G LCRH L
Sbjct: 627 LKTYEEGAISRFLVRK----CGNENE--KHAVTFSASNLNASCSCQMFEYEGLLCRHILK 680
Query: 580 VLNFNGVEEIPFKYILSRWKKDYKRLHVPD-HSSVSADDTDPIQ-WSNQLFSSALQVVEE 637
V N + E+P +YIL RW K+ + V D S V++ D + WS L +A + +E
Sbjct: 681 VFNLLDIRELPSRYILHRWTKNAEFGFVRDVESGVTSQDLKALMIWS--LREAASKYIEF 738
Query: 638 GTISLDHYNVALQACEESLSKV 659
GT SL+ Y +A + E K+
Sbjct: 739 GTSSLEKYKLAYEIMREGGKKL 760
Score = 68.6 bits (166), Expect = 1e-11
Identities = 42/162 (25%), Positives = 74/162 (44%), Gaps = 10/162 (6%)
Query: 32 SQNGFSEGRKEFDA-PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGA 90
+ NG +E + P VG+EF++ ++A +YY YA GF VR + R
Sbjct: 13 TNNGIAENEGDSGLEPYVGLEFDTAEEARDYYNSYATRTGFKVRTGQLYRSRTDGTVSSR 72
Query: 91 VLCCSSQGFKRTKDVNHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHILGAKIHKS 150
CS +GF+ +RTGCPA +R++ ++ +W + ++ EHNH LG I ++
Sbjct: 73 RFVCSKEGFQLN---------SRTGCPAFIRVQRRDTGKWVLDQIQKEHNHDLGGHIEEA 123
Query: 151 AKKMGNGTKRKSLPSSDSEVQTVKLYRPLVIDAGGNGNLNPS 192
++++ + + + V+D G PS
Sbjct: 124 QTTPRPSVQQRAPAPTKLGISVPHRPKMKVVDEADKGRSCPS 165
>At4g19990 putative protein
Length = 672
Score = 296 bits (758), Expect = 2e-80
Identities = 188/632 (29%), Positives = 305/632 (47%), Gaps = 94/632 (14%)
Query: 49 GMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKDVNHL 108
G EFES ++A+ +Y YA VGF +K S R + + A C+ G K+ L
Sbjct: 24 GREFESKEEAFEFYKEYANSVGFTTIIKASRRSRMTGKFIDAKFVCTRYGSKKEDIDTGL 83
Query: 109 -----------------RKETRTGCPAMMRIRIAESQRWRIIEVILEHNHILGAKIHKSA 151
R ++T C A + ++ + RW + ++ EHNH + S
Sbjct: 84 GTDGFNIPQARKRGRINRSSSKTDCKAFLHVKRRQDGRWVVRSLVKEHNHEIFTGQADSL 143
Query: 152 KKMGNGTKRKSLPSSDSEVQTVKLYRPLVIDAGGNGNLNPSARDDRTFSEMSNKLNVKRG 211
+++ K + L + + V+ VK S KL + G
Sbjct: 144 RELSGRRKLEKL--NGAIVKEVK----------------------------SRKL--EDG 171
Query: 212 DTQAIYNFLCRMQLTNPNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYLS 271
D + + NF MQ LRN FW DA+ R F DV+ D T++
Sbjct: 172 DVERLLNFFTDMQ----------------SLRNIFWVDAKGRFDYTCFSDVVSIDTTFIK 215
Query: 272 SKYEIPLVVFVGINHHGQSVLLGCGLLAGETTES-YIWLFRTWVTCMSGCSPQTIITDRC 330
++Y++PLV F G+NHHGQ +LLG GLL + ++S ++WLFR W+ M GC P+ I+T
Sbjct: 216 NEYKLPLVAFTGVNHHGQFLLLGFGLLLTDESKSGFVWLFRAWLKAMHGCRPRVILTKHD 275
Query: 331 EALQSAIIEVFPKSHHCFGLSLIMKKVPEKLGGLHNYDGIRKALIK----AVYETLKISE 386
+ L+ A++EVFP S HCF + + ++PEKLG + + K L+ A+Y + + +
Sbjct: 276 QMLKEAVLEVFPSSRHCFYMWDTLGQMPEKLGHVIR---LEKKLVDEINDAIYGSCQSED 332
Query: 387 FEAAWGIMIQRFGVSDHEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFGRYV 446
FE W ++ RF + D+ WL+SLYEDR W PV++KDV GM A+ +S+ +Y+
Sbjct: 333 FEKNWWEVVDRFHMRDNVWLQSLYEDREYWVPVYMKDVSLAGMCTAQRSDSVNSGLDKYI 392
Query: 447 HKQTPLKEFLDKYELALHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKF 506
++T K FL++Y+ + ++++EE ++ E+ P LK+ Q++ +YTRE+F KF
Sbjct: 393 QRKTTFKAFLEQYKKMIQERYEEEEKSEIETLYKQPGLKSPSPFGKQMAEVYTREMFKKF 452
Query: 507 QFEVEEMFSCFGTTQLHVDGPIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCICSC 566
Q EV +C + DG V +R + + + F VV++ + E+ C C
Sbjct: 453 QVEVLGGVACHPKKESEEDG------VNKRTFRVQDYEQNRSFVVVWNSESSEVVCSCRL 506
Query: 567 FNFYGYLCRHALCVLNFNGVEEIPFKYILSRWKKDYKRLHVPDHSSVSADDTDPIQWSNQ 626
F G L IP +Y+L RW KD K V + + T ++ +
Sbjct: 507 FELKGEL--------------SIPSQYVLKRWTKDAKSREVMESDQTDVESTKAQRYKD- 551
Query: 627 LFSSALQVVEEGTISLDHYNVALQACEESLSK 658
L +L++ EE ++S + YN + E+L K
Sbjct: 552 LCLRSLKLSEEASLSEESYNAVVNVLNEALRK 583
>At2g32250 Mutator-like transposase
Length = 684
Score = 281 bits (718), Expect = 1e-75
Identities = 168/554 (30%), Positives = 266/554 (47%), Gaps = 46/554 (8%)
Query: 49 GMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKDVN-H 107
GM+FES + AY +Y YA+ VGF + +K S + S + + CS G KR K +
Sbjct: 41 GMDFESKEAAYYFYREYARSVGFGITIKASRRSKRSGKFIDVKIACSRFGTKREKATAIN 100
Query: 108 LRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHILGAKIHKSAKKMGNGTKRKSLPSSD 167
R +TGC A + ++ E ++W I + EHNH + + K+ P+
Sbjct: 101 PRSCPKTGCKAGLHMKRKEDEKWVIYNFVKEHNHEI------CPDDFYVSVRGKNKPAG- 153
Query: 168 SEVQTVKLYRPLVIDAGGNGNLNPSARDDRTFSEMSNKLNVKRGDTQAIYNFLCRMQLTN 227
L I G +L ++ D + + MQ
Sbjct: 154 ----------ALAIKKG-------------------LQLALEEEDLKLLLEHFMEMQDKQ 184
Query: 228 PNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVGINHH 287
P FFY +DFD + +RN FW DA+++ F DV+ FD Y+ + Y IP F+G++HH
Sbjct: 185 PGFFYAVDFDSDKRVRNVFWLDAKAKHDYCSFSDVVLFDTFYVRNGYRIPFAPFIGVSHH 244
Query: 288 GQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDRCEALQSAIIEVFPKSHHC 347
Q VLLGC L+ + +Y WLFRTW+ + G +P +ITD+ + L ++EVFP H
Sbjct: 245 RQYVLLGCALIGEVSESTYSWLFRTWLKAVGGQAPGVMITDQDKLLSDIVVEVFPDVRHI 304
Query: 348 FGLSLIMKKVPEKLGG-LHNYDGIRKALIKAVYETLKISEFEAAWGIMIQRFGVSDHEWL 406
F L ++ K+ E L + DG ++ V + FE W MI +F ++++EW+
Sbjct: 305 FCLWSVLSKISEMLNPFVSQDDGFMESFGNCVASSWTDEHFERRWSNMIGKFELNENEWV 364
Query: 407 RSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFGRYVHKQTPLKEFLDKYELALHKK 466
+ L+ DR +W P + + G+S SI F +Y++ + K+F + Y L +
Sbjct: 365 QLLFRDRKKWVPHYFHGICLAGLSGPERSGSIASHFDKYMNSEATFKDFFELYMKFLQYR 424
Query: 467 HKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCFGTTQLHVDG 526
E+ D E ++ P L++ + E QLS +YT F KFQ EV + SC + DG
Sbjct: 425 CDVEAKDDLEYQSKQPTLRSSLAFEKQLSLIYTDAAFKKFQAEVPGVVSC-QLQKEREDG 483
Query: 527 PIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHALCVLNFNGV 586
I+ +++ E ++F V + + C C F + G+LC+HA+ VL V
Sbjct: 484 TTAIFRIED-------FEERQNFFVALNNELLDACCSCHLFEYQGFLCKHAILVLQSADV 536
Query: 587 EEIPFKYILSRWKK 600
+P +YIL RW K
Sbjct: 537 SRVPSQYILKRWSK 550
>At4g38170 hypothetical protein
Length = 531
Score = 260 bits (665), Expect = 2e-69
Identities = 152/453 (33%), Positives = 227/453 (49%), Gaps = 14/453 (3%)
Query: 216 IYNFLCRMQLTNPNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYLSSK-Y 274
+ N+L R QL NP F Y ++ DD G N FWAD R YFGD + FD TY K Y
Sbjct: 7 VLNYLKRRQLENPGFLYAIE-DDCG---NVFWADPTCRLNYTYFGDTLVFDTTYRRGKRY 62
Query: 275 EIPLVVFVGINHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDRCEALQ 334
++P F G NHHGQ VL GC L+ E+ S+ WLF+TW+ MS P +I + +Q
Sbjct: 63 QVPFAAFTGFNHHGQPVLFGCALILNESESSFAWLFQTWLQAMSAPPPPSITVEPDRLIQ 122
Query: 335 SAIIEVFPKSHHCFGLSLIMKKVPEKLGGLHN-YDGIRKALIKAVYETLKISEFEAAWGI 393
A+ VF ++ F LI ++ EKL + + I V ET +EFEA+W
Sbjct: 123 VAVSRVFSQTRLRFSQPLIFEETEEKLAHVFQAHPTFESEFINCVTETETAAEFEASWDS 182
Query: 394 MIQRFGVSDHEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFGRYVHKQTPLK 453
+++R+ + D++WL+S+Y R +W VF++D F+G +S + FF +V T ++
Sbjct: 183 IVRRYYMEDNDWLQSIYNARQQWVRVFIRDTFYGELSTNEGSSILNSFFQGFVDASTTMQ 242
Query: 454 EFLDKYELALHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQFEVEEM 513
+ +YE A+ ++E AD E+ NS+PV+KT +E Q + +YTR F+KFQ E E
Sbjct: 243 MLIKQYEKAIDSWREKELKADYEATNSTPVMKTPSPMEKQAASLYTRAAFIKFQEEFVET 302
Query: 514 FSCFGTTQLHVDGPIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCICSCFNFYGYL 573
+ + G Y V + + K V + + C C F + G +
Sbjct: 303 LA-IPANIISDSGTHTTYRVAKFGEVH------KGHTVSFDSLEVKANCSCQMFEYSGII 355
Query: 574 CRHALCVLNFNGVEEIPFKYILSRWKKDYKRLHVPDHSSVSADDTDPIQWS-NQLFSSAL 632
CRH L V + V +P +Y+L RW K+ K + S + + N L A
Sbjct: 356 CRHILAVFSAKNVLALPSRYLLRRWTKEAKIRGTEEQPEFSNGCQESLNLCFNSLRQEAT 415
Query: 633 QVVEEGTISLDHYNVALQACEESLSKVHDVDQR 665
+ VEEG S+ Y VA+ A +E+ KV R
Sbjct: 416 KYVEEGAKSIQIYKVAMDALDEAAKKVAAASNR 448
>At1g10240 unknown protein
Length = 680
Score = 208 bits (529), Expect = 9e-54
Identities = 167/659 (25%), Positives = 288/659 (43%), Gaps = 40/659 (6%)
Query: 22 DEDAALIELDSQNGFSEGRKEFDAPAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFK 81
D+ ++ E N S P +G F ++D AY +Y +AK GF +R + K
Sbjct: 24 DDASSTEESPDDNNLSLEAVHNAIPYLGQIFLTHDTAYEFYSTFAKRCGFSIRRHRTEGK 83
Query: 82 ----RNSREKYGAVLCCSSQGFKRTKDVNH-----LRKETRTGCPAMMRI-RIAE--SQR 129
+ +Y C G K ++ R+ +R GC A +RI ++ E S
Sbjct: 84 DGVGKGLTRRY---FVCHRAGNTPIKTLSEGKPQRNRRSSRCGCQAYLRISKLTELGSTE 140
Query: 130 WRIIEVILEHNHIL----GAKIHKSAKKMGNGTKRKSLPSSDSEVQTVKLYRPLVIDAGG 185
WR+ HNH L + + + + + K + L S + + ++ R L ++
Sbjct: 141 WRVTGFANHHNHELLEPNQVRFLPAYRSISDADKSRILMFSKTGISVQQMMRLLELEKCV 200
Query: 186 NGNLNP-SARDDRTFSEMSNKLNVKRGDTQAIYNFLCR-MQLTNPNFFYLMDFDDEGHLR 243
P + +D R + KL+ + D + +C+ ++ +PNF + D L
Sbjct: 201 EPGFLPFTEKDVRNLLQSFKKLDPE--DENIDFLRMCQSIKEKDPNFKFEFTLDANDKLE 258
Query: 244 NAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVGINHHGQSVLLGCGLLAGETT 303
N W+ A S + FGD + FD T+ S E+PL ++VG+N++G GC LL E
Sbjct: 259 NIAWSYASSIQSYELFGDAVVFDTTHRLSAVEMPLGIWVGVNNYGVPCFFGCVLLRDENL 318
Query: 304 ESYIWLFRTWVTCMSGCSPQTIITDRCEALQSAIIEVFPKSHHCFGLSLIMKKVPEKL-G 362
S+ W + + M+G +PQTI+TD L+ AI P + H + +++ K P
Sbjct: 319 RSWSWALQAFTGFMNGKAPQTILTDHNMCLKEAIAGEMPATKHALCIWMVVGKFPSWFNA 378
Query: 363 GLHNYDGIRKALIKAVYETLKISEFEAAWGIMIQRFGVSDHEWLRSLYEDRVRWAPVFLK 422
GL KA +Y + EFE W M+ FG+ + + +LY R W+ +L+
Sbjct: 379 GLGERYNDWKAEFYRLYHLESVEEFELGWRDMVNSFGLHTNRHINNLYASRSLWSLPYLR 438
Query: 423 DVFFGGMSAARPGESIGPFFGRYVHKQTPLKEFLDKYELALHKKHKEESLADTESRNSSP 482
F GM+ ++I F R++ QT L F+++ + + K + + +
Sbjct: 439 SHFLAGMTLTGRSKAINAFIQRFLSAQTRLAHFVEQVAVVVDFKDQATEQQTMQQNLQNI 498
Query: 483 VLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCFGTTQLHVDGPIVIYLVKERVLIDGN 542
LKT +E + + T F K Q E++ +D YLV+ +DG
Sbjct: 499 SLKTGAPMESHAASVLTPFAFSKLQ---EQLVLAAHYASFQMDEG---YLVRHHTKLDGG 552
Query: 543 RREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHALCVLNFNGVEEIPFKYILSRWKKDY 602
R+ V + G + C C F F G+LCRHAL VL+ ++P +Y+ RW++
Sbjct: 553 RK------VYWVPQEGIISCSCQLFEFSGFLCRHALRVLSTGNCFQVPDRYLPLRWRRIS 606
Query: 603 KRLHVPDHSSVSADDTDPIQWSNQLFSSALQVVEEGTISLDHYNVALQACEESLSKVHD 661
S+ + D + +Q L S+ +V E S + ++A + LS++ +
Sbjct: 607 TSFSKTFRSN-AEDHGERVQLLQNLVST---LVSESAKSKERLDIATEQTSILLSRIRE 661
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 178 bits (451), Expect = 1e-44
Identities = 171/681 (25%), Positives = 283/681 (41%), Gaps = 68/681 (9%)
Query: 19 VHKDEDAALIELDSQNGFSEGRKEFDA---PAVGMEFESYDDAYNYYICYAKEVGFCVRV 75
+ +ED + I ++D P VG F + D+A+ YY +A++ GF +R
Sbjct: 40 IRLEEDESTITKSEIESTPTPTSQYDTVFTPYVGQIFTTDDEAFEYYSTFARKSGFSIRK 99
Query: 76 KNSWFKRNSREKYGAVLCCSSQGFKRTK---DVNHLR--KETRTGCPAMMRIR---IAES 127
S +N Y C GF + + +V H R K R GC + + +
Sbjct: 100 ARSTESQNLGV-YRRDFVCYRSGFNQPRKKANVEHPRERKSVRCGCDGKLYLTKEVVDGV 158
Query: 128 QRWRIIEVILEHNHIL----GAKIHKSAKKMGNGTKRKSLPSSDSEVQTVKLYRPLVIDA 183
W + + HNH L ++ + +K+ + + L S + ++ + L ++
Sbjct: 159 SHWYVSQFSNVHNHELLEDDQVRLLPAYRKIQQSDQERILLLSKAGFPVNRIVKLLELEK 218
Query: 184 GG-NGNLNPSARDDRTFSEMSNK---------LNVKRGDTQAIYNFLCRMQLTNPNFFYL 233
G +G L +D R F K + DT + + + +F Y
Sbjct: 219 GVVSGQLPFIEKDVRNFVRACKKSVQENDAFMTEKRESDTLELLECCKGLAERDMDFVYD 278
Query: 234 MDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVGINHHGQSVLL 293
D+ + N WA S FGDV+ FD +Y S Y + L VF GI+++G+++LL
Sbjct: 279 CTSDENQKVENIAWAYGDSVRGYSLFGDVVVFDTSYRSVPYGLLLGVFFGIDNNGKAMLL 338
Query: 294 GCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDRCEALQSAIIEVFPKSHHCFGLSLI 353
GC LL E+ S+ W +T+V M G PQTI+TD L+ AI P ++H +S I
Sbjct: 339 GCVLLQDESCRSFTWALQTFVRFMRGRHPQTILTDIDTGLKDAIGREMPNTNHVVFMSHI 398
Query: 354 MKKV----PEKLGGLHNYDGIRKALIKAVYETLKISEFEAAWGIMIQRFGVSDHEWLRSL 409
+ K+ + LG +Y+ R A + + EFE W +++ RFG+ L
Sbjct: 399 VSKLASWFSQTLGS--HYEEFR-AGFDMLCRAGNVDEFEQQWDLLVTRFGLVPDRHAALL 455
Query: 410 YEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFGRYVHKQTPLKEFLDKYELALHKKHKE 469
Y R W P +++ F + SI F R V T ++ L+ E AL
Sbjct: 456 YSCRASWLPCCIREHFVAQTMTSEFNLSIDSFLKRVVDGATCMQLLLE--ESALQVSAAA 513
Query: 470 ESLADTESRNSSPVLKTRCSLELQLSRMYT--------REIFMKFQFEVEEMFSCFGTTQ 521
R + P LKT +E + T E+ + Q+ V EM
Sbjct: 514 SLAKQILPRFTYPSLKTCMPMEDHARGILTPYAFSVLQNEMVLSVQYAVAEM-------- 565
Query: 522 LHVDGPIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHALCVL 581
+GP +++ K+ ++G + V+++ E++C C F G LCRH L VL
Sbjct: 566 --ANGPFIVHHYKK---MEG------ECCVIWNPENEEIQCSCKEFEHSGILCRHTLRVL 614
Query: 582 NFNGVEEIPFKYILSRWKKDYKRLHVPDHSSVSADDTDPIQWSNQLFSSALQ-VVEEGTI 640
IP +Y L RW+++ + + + D S Q F S + ++ E I
Sbjct: 615 TVKNCFHIPEQYFLLRWRQESPHVATENQNGQGIGDD-----SAQTFHSLTETLLTESMI 669
Query: 641 SLDHYNVALQACEESLSKVHD 661
S D + A Q + +V +
Sbjct: 670 SKDRLDYANQELSLLIDRVRN 690
>At3g59470 unknown protein
Length = 251
Score = 75.5 bits (184), Expect = 9e-14
Identities = 43/104 (41%), Positives = 56/104 (53%), Gaps = 5/104 (4%)
Query: 44 DAPAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGF---- 99
D P VG EFES A+ +Y YA +VGF +RV R+ G L C+ +G+
Sbjct: 68 DEPYVGQEFESEAAAHGFYNAYATKVGFVIRVSKLSRSRHDGSPIGRQLVCNKEGYRLPS 127
Query: 100 KRTKDVNHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHIL 143
KR K + R ETR GC AM+ IR S +W I + + EHNH L
Sbjct: 128 KRDKVIRQ-RAETRVGCKAMILIRKENSGKWVITKFVKEHNHSL 170
>At4g12850 putative protein
Length = 183
Score = 62.0 bits (149), Expect = 1e-09
Identities = 33/106 (31%), Positives = 55/106 (51%), Gaps = 1/106 (0%)
Query: 39 GRKEFDAPAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQG 98
G + D P VG++FES ++A ++Y+ Y+K +GF VR+ L C+ QG
Sbjct: 3 GDLKLDEPYVGLKFESEEEAKDFYVEYSKRLGFVVRMMQRRRSGIDGRTLARRLGCNKQG 62
Query: 99 F-KRTKDVNHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHIL 143
F + + +R GC A + +++ +S +W + I EHNH L
Sbjct: 63 FGPNNQRSSSSSSSSREGCKATILVKMEKSGKWVVTRFIKEHNHSL 108
>At3g07500 hypothetical protein
Length = 214
Score = 60.8 bits (146), Expect = 2e-09
Identities = 45/147 (30%), Positives = 74/147 (49%), Gaps = 16/147 (10%)
Query: 6 LCCEDLPHGECIEVHKDEDAALIE-LDSQNGFSEGRKEFDAPAVGMEFESYDDAYNYYIC 64
+CC G + +ED ++E D ++ P +GMEFES + A ++Y
Sbjct: 3 VCCVVAVEGNSLP---EEDVEMMENSDVMKTVTDEASPMVEPFIGMEFESEEAAKSFYDN 59
Query: 65 YAKEVGFCVRVKNSWFKRNSREKYGAV----LCCSSQGFKRTK----DVNHLRKETRTGC 116
YA +GF +RV F+R+ R+ G V L C+ +GF+R++ + R TR GC
Sbjct: 60 YATCMGFVMRV--DAFRRSMRD--GTVVWRRLVCNKEGFRRSRPRRSESRKPRAITREGC 115
Query: 117 PAMMRIRIAESQRWRIIEVILEHNHIL 143
A++ ++ +S W + + EHNH L
Sbjct: 116 KALIVVKREKSGTWLVTKFEKEHNHPL 142
>At2g05490 Mutator-like transposase
Length = 942
Score = 55.5 bits (132), Expect = 1e-07
Identities = 41/156 (26%), Positives = 65/156 (41%), Gaps = 22/156 (14%)
Query: 206 LNVKRGDTQAIYNFLCRMQLTNPNFFYLMDFDDEG---HLRNAFWADARSR--------- 253
+N RGD A Y FL P + YL+ + G HL + A R R
Sbjct: 510 VNSIRGDDMASYRFL-------PTYLYLLQLANPGTICHLHSTPEAKGRQRFKYVFVSLS 562
Query: 254 ---AACGYFGDVIYFDNTYLSSKYEIPLVVFVGINHHGQSVLLGCGLLAGETTESYIWLF 310
Y V+ D T L +Y+ L++ + + Q L G++ GET S+IW F
Sbjct: 563 ASIKGWKYMRKVVVVDGTQLVGRYKGCLLIACAQDRNFQIFPLAFGVVDGETDASWIWFF 622
Query: 311 RTWVTCMSGCSPQTIITDRCEALQSAIIEVFPKSHH 346
+ I++D+ ++ + V+PK+HH
Sbjct: 623 EKLSEIVPDTDNLMIVSDKHSSIYKGVSVVYPKAHH 658
>At2g43280 unknown protein
Length = 206
Score = 53.9 bits (128), Expect = 3e-07
Identities = 37/124 (29%), Positives = 57/124 (45%), Gaps = 13/124 (10%)
Query: 46 PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLC----CSSQGF-- 99
P G+ FES D A +Y Y++ +GF +RV + EK G +L C+ +G
Sbjct: 22 PREGIIFESEDAAKMFYDDYSRRLGFVMRVMSC----RRSEKDGRILARRFGCNKEGHCV 77
Query: 100 ---KRTKDVNHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHILGAKIHKSAKKMGN 156
+ V R TR GC AM+ ++ S +W I + + EHNH L ++ +
Sbjct: 78 SIRGKFGSVRKPRPSTREGCKAMIHVKYDRSGKWVITKFVKEHNHPLVVSPREARHTLDE 137
Query: 157 GTKR 160
KR
Sbjct: 138 KDKR 141
>At2g12150 pseudogene
Length = 942
Score = 53.5 bits (127), Expect = 4e-07
Identities = 42/149 (28%), Positives = 68/149 (45%), Gaps = 8/149 (5%)
Query: 206 LNVKRGDTQAIYNFLCR----MQLTNPNFFYLMDF--DDEGHLRNAFWADARSRAACG-- 257
+N RGD A Y FL +QL NP + +D+G R + + S + G
Sbjct: 510 VNSIRGDDMASYRFLPTYLYLLQLANPGTICHLHSTPEDKGRQRFKYVFVSLSASIKGWK 569
Query: 258 YFGDVIYFDNTYLSSKYEIPLVVFVGINHHGQSVLLGCGLLAGETTESYIWLFRTWVTCM 317
Y VI D T L +Y+ L++ + + Q L G++ GET S+IW F +
Sbjct: 570 YMRKVIVVDGTQLVGRYKGCLLIACAQDGNFQIFPLAFGVVDGETDASWIWFFEKLSEIV 629
Query: 318 SGCSPQTIITDRCEALQSAIIEVFPKSHH 346
I++DR ++ + V+PK++H
Sbjct: 630 PDTDDLMIVSDRHSSIYKGVSVVYPKANH 658
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,860,676
Number of Sequences: 26719
Number of extensions: 705572
Number of successful extensions: 1596
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 1430
Number of HSP's gapped (non-prelim): 116
length of query: 667
length of database: 11,318,596
effective HSP length: 106
effective length of query: 561
effective length of database: 8,486,382
effective search space: 4760860302
effective search space used: 4760860302
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0066.5


