

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0066.1
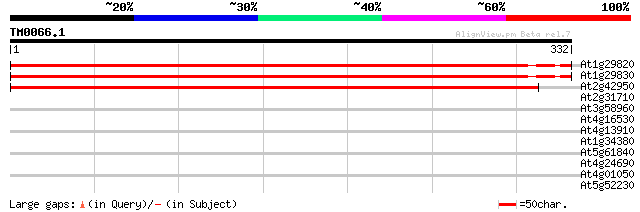
(332 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g29820 hypothetical protein 537 e-153
At1g29830 unknown protein 491 e-139
At2g42950 unknown protein 471 e-133
At2g31710 unknown protein 34 0.13
At3g58960 putative protein 32 0.48
At4g16530 hypothetical protein 30 2.4
At4g13910 putative disease resistance protein 28 5.3
At1g34380 DNA polymerase type I like protein 28 5.3
At5g61840 unknown protein 28 6.9
At4g24690 unknown protein 28 6.9
At4g01050 unknown protein 28 6.9
At5g52230 unknown protein 28 9.1
>At1g29820 hypothetical protein
Length = 540
Score = 537 bits (1383), Expect = e-153
Identities = 268/332 (80%), Positives = 301/332 (89%), Gaps = 6/332 (1%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRVAGGLLFELLGQS GDP++ EDD+P+V RSWQA+NFLV+VMHIKG+V+ NVLGI
Sbjct: 214 VPVRVAGGLLFELLGQSVGDPVISEDDVPVVFRSWQAKNFLVSVMHIKGNVTNTNVLGIT 273
Query: 61 EVQELLSGGGYNVPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMNRRNYEDL 120
EV+ELL GGYNVPRTVHEVIA LACRLSRWDDRLFRKSIFG ADEIELKFMNRRNYEDL
Sbjct: 274 EVEELLYAGGYNVPRTVHEVIAHLACRLSRWDDRLFRKSIFGAADEIELKFMNRRNYEDL 333
Query: 121 NLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMIE 180
NLF IILNQEIRKLS QVIRVKWSLHAREEI+FELLQHL+G AR LL+G+RK+TREM+E
Sbjct: 334 NLFSIILNQEIRKLSRQVIRVKWSLHAREEIIFELLQHLRGNIARHLLDGLRKNTREMLE 393
Query: 181 EQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIP 240
EQEAVRGRLFTIQDV QS+VRAWLQD+SLRV+HNLAVF G G+VLTII GLFGINVDGIP
Sbjct: 394 EQEAVRGRLFTIQDVMQSSVRAWLQDKSLRVSHNLAVFGGCGLVLTIIVGLFGINVDGIP 453
Query: 241 GAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKKPIAEEKVEVRKLELQELVKMFQHEA 300
GA++TPYAFGLFT ++V +GA+L+VVGLVYLGLKKPI EE+VEVRKLELQ++VK+FQHEA
Sbjct: 454 GAQNTPYAFGLFTFLMVLLGAILIVVGLVYLGLKKPITEEQVEVRKLELQDVVKIFQHEA 513
Query: 301 ETHAQVRKNVSRNNLPPTAGDAFRRDVDYLLL 332
ETHAQ+R RNNL PTAGD F D DY+L+
Sbjct: 514 ETHAQLR----RNNLSPTAGDVF--DADYILI 539
>At1g29830 unknown protein
Length = 533
Score = 491 bits (1264), Expect = e-139
Identities = 246/332 (74%), Positives = 279/332 (83%), Gaps = 6/332 (1%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRV GGLLFELLGQS GDP++ EDD+P+V RSWQA+NFLV+VMHIKG+VS+ NVLGI
Sbjct: 207 VPVRVDGGLLFELLGQSMGDPVIGEDDVPVVFRSWQAKNFLVSVMHIKGNVSKSNVLGIT 266
Query: 61 EVQELLSGGGYNVPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMNRRNYEDL 120
EV+ELL G YNVPRT+HEVIA LACRLSRWDDRLFRKSIFG ADEIELKFMNRRN+EDL
Sbjct: 267 EVEELLYAGSYNVPRTIHEVIAHLACRLSRWDDRLFRKSIFGAADEIELKFMNRRNHEDL 326
Query: 121 NLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMIE 180
NLF IILNQEIRKL+ Q IRVKWSLHAREEI+ ELLQHL+G R LLEG+R +TREM+E
Sbjct: 327 NLFSIILNQEIRKLARQTIRVKWSLHAREEIILELLQHLRGNIPRHLLEGLRNNTREMLE 386
Query: 181 EQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIP 240
EQEAVRGRLFTIQD QS +R+WLQD+SL +HNLA+F G G+VLTII GLF +N+DG+P
Sbjct: 387 EQEAVRGRLFTIQDNIQSNIRSWLQDQSLNGSHNLAIFGGCGLVLTIILGLFSVNLDGVP 446
Query: 241 GAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKKPIAEEKVEVRKLELQELVKMFQHEA 300
G +HTPYAF LF+ LV IG VL+ GL YLG KKPI EE VE RKLELQ +VK+FQHEA
Sbjct: 447 GVKHTPYAFVLFSVFLVLIGIVLIAFGLRYLGPKKPITEEHVEARKLELQNVVKIFQHEA 506
Query: 301 ETHAQVRKNVSRNNLPPTAGDAFRRDVDYLLL 332
ETHAQ+R RNNL PTAGD F D DY L+
Sbjct: 507 ETHAQLR----RNNLSPTAGDVF--DADYFLI 532
>At2g42950 unknown protein
Length = 501
Score = 471 bits (1211), Expect = e-133
Identities = 235/313 (75%), Positives = 266/313 (84%)
Query: 1 VPVRVAGGLLFELLGQSAGDPLVEEDDIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIA 60
VPVRVAGGLLFELLGQSAGDP ++EDDIPIVLRSWQAQNFLVT +H+KG I+VLGI
Sbjct: 167 VPVRVAGGLLFELLGQSAGDPFIQEDDIPIVLRSWQAQNFLVTALHVKGFALNISVLGIT 226
Query: 61 EVQELLSGGGYNVPRTVHEVIAQLACRLSRWDDRLFRKSIFGVADEIELKFMNRRNYEDL 120
EVQE+L GG +PRTVHE+IA LACRL+RWDDRLFRK IFG ADE+EL FMN+R YED
Sbjct: 227 EVQEMLIAGGACIPRTVHELIAHLACRLARWDDRLFRKYIFGAADEVELMFMNKRLYEDP 286
Query: 121 NLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMIE 180
NLF ILNQEIR+LSTQVIRVKWSLHAREEIVFELLQ LKG + LLEGI+KSTR+MI
Sbjct: 287 NLFTTILNQEIRRLSTQVIRVKWSLHAREEIVFELLQQLKGNRTKDLLEGIKKSTRDMIN 346
Query: 181 EQEAVRGRLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIP 240
EQEAVRGRLFTIQDV Q+TVRAWLQD+SL VTHNL +F GVG+++TI+TGLFGINVDGIP
Sbjct: 347 EQEAVRGRLFTIQDVMQNTVRAWLQDQSLTVTHNLGIFGGVGLLITIVTGLFGINVDGIP 406
Query: 241 GAEHTPYAFGLFTAILVFIGAVLVVVGLVYLGLKKPIAEEKVEVRKLELQELVKMFQHEA 300
GA+ P AF LF+ +L G VLVV GL+YLGLK+P+AEE VE RKLEL E+VK FQ EA
Sbjct: 407 GAKDFPQAFALFSVVLFVSGLVLVVAGLIYLGLKEPVAEENVETRKLELDEMVKKFQQEA 466
Query: 301 ETHAQVRKNVSRN 313
E+HAQV K V +N
Sbjct: 467 ESHAQVCKKVPQN 479
>At2g31710 unknown protein
Length = 105
Score = 33.9 bits (76), Expect = 0.13
Identities = 24/82 (29%), Positives = 41/82 (49%), Gaps = 11/82 (13%)
Query: 217 VFAGVGMVLTIITGLFGINVDGIPGAEH-TPYAFGLFTAILVFIGAVLVVVGLVYLGLKK 275
+ A + M + I L+G N D +PG+ +P++ L + L + +V+V + L LK+
Sbjct: 9 LIASMFMWILPIAILYGFNNDLLPGSTTLSPHSLTLLSGFLAVVSVNVVIVFYICLALKE 68
Query: 276 P----------IAEEKVEVRKL 287
P +AE K V+KL
Sbjct: 69 PADKHKPDASFVAEAKDSVKKL 90
>At3g58960 putative protein
Length = 475
Score = 32.0 bits (71), Expect = 0.48
Identities = 21/49 (42%), Positives = 26/49 (52%), Gaps = 4/49 (8%)
Query: 270 YLGLKKPIAEEKVEVRKLELQELVKMFQHE--AETHAQVRKNVSRNNLP 316
Y G K + K + KL ELVK+F HE E H Q+R N+ NLP
Sbjct: 411 YSGTKIELKHMKHFLEKLSCLELVKVFSHERDEEEHVQLRTNLL--NLP 457
>At4g16530 hypothetical protein
Length = 773
Score = 29.6 bits (65), Expect = 2.4
Identities = 23/78 (29%), Positives = 36/78 (45%), Gaps = 1/78 (1%)
Query: 110 KFMNRRNYEDLNLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLE 169
KF+++ E N+F+ E L+ Q W + + ELL L I S++
Sbjct: 181 KFLDKILEEASNVFLYPYRVEDWSLALQTFVKMWIQLEKTGVTVELLMALMEIWISSVVN 240
Query: 170 GIRKSTREMIEEQEAVRG 187
++K EM +EQ VRG
Sbjct: 241 SVKKLV-EMKKEQFLVRG 257
>At4g13910 putative disease resistance protein
Length = 509
Score = 28.5 bits (62), Expect = 5.3
Identities = 25/119 (21%), Positives = 54/119 (45%), Gaps = 8/119 (6%)
Query: 46 HIKGSVSRINVLGIAEVQELLSGGGY---NVPRTVHEVIAQLACRLSRWDDR--LFRKSI 100
H G + N+ + +Q L G +PR++ +++ LS W+ R + SI
Sbjct: 40 HFSGPLEIGNISSQSNLQILYIGENNFDGPIPRSISKLVGLSELSLSFWNTRRSIVDFSI 99
Query: 101 F---GVADEIELKFMNRRNYEDLNLFVIILNQEIRKLSTQVIRVKWSLHAREEIVFELL 156
F + ++L ++N R+ + +LF +++ LS ++ +LH ++ L+
Sbjct: 100 FLHLKSLESLDLSYLNTRSMVEFSLFSPLMSLGYLDLSGISLKFSSTLHLPSSLIEYLI 158
>At1g34380 DNA polymerase type I like protein
Length = 343
Score = 28.5 bits (62), Expect = 5.3
Identities = 23/110 (20%), Positives = 48/110 (42%), Gaps = 11/110 (10%)
Query: 146 HAREEIVFELLQHLKGIGARSLLEGIRKSTREMIEEQEAVRGRLFTIQDVTQSTVRAWLQ 205
H +++V L++ G R+++ K +++I E + L ++ + T++ +
Sbjct: 165 HEADDVVATLMEQAVQRGYRAVIASPDKDFKQLISENVQIVIPLADLRRWSFYTLKHYHA 224
Query: 206 DRSLRVTHNLAVFAGVGMVLTIITGLFGINVDGIPGAEHTPYAFGLFTAI 255
+ +L+ + G VDG+PG +H AFG TA+
Sbjct: 225 QYNCDPQSDLSFRC-----------IMGDEVDGVPGIQHMVPAFGRKTAM 263
>At5g61840 unknown protein
Length = 415
Score = 28.1 bits (61), Expect = 6.9
Identities = 15/39 (38%), Positives = 21/39 (53%), Gaps = 5/39 (12%)
Query: 91 WDDRLFRKSIFG-----VADEIELKFMNRRNYEDLNLFV 124
W RL IFG +AD+I L F + +ED+ +FV
Sbjct: 290 WSPRLVEAVIFGCIPVIIADDIVLPFADAIPWEDIGVFV 328
>At4g24690 unknown protein
Length = 704
Score = 28.1 bits (61), Expect = 6.9
Identities = 21/93 (22%), Positives = 42/93 (44%), Gaps = 13/93 (13%)
Query: 27 DIPIVLRSWQAQNFLVTVMHIKGSVSRINVLGIAEV---QELLSGGGYNVPRTVHEVIAQ 83
D+P+ L+ +N + M + + +G E+ +E+L YN+ ++V +
Sbjct: 604 DVPVPLQEDIEKNDVEITM-----LKELEEMGFKEIDLNKEILRDNEYNLEQSVDAL--- 655
Query: 84 LACRLSRWDDRLFRKSIFGVADEIELKFMNRRN 116
C +S WD L G D++ K + ++N
Sbjct: 656 --CGVSEWDPILEELQEMGFCDDVTNKRLLKKN 686
>At4g01050 unknown protein
Length = 466
Score = 28.1 bits (61), Expect = 6.9
Identities = 30/116 (25%), Positives = 51/116 (43%), Gaps = 7/116 (6%)
Query: 177 EMIEEQEAVRG--RLFTIQDVTQSTVRAWLQDRSLRVTHNLAVFAGVGMVLTIITGLFGI 234
E++ E A+ G + I+D + R WL + + + + I+G+FG
Sbjct: 214 ELVAELVALNGFKSAYAIKDGAEGP-RGWLNSSLPWIEPKKTLSLDLSSLTDSISGVFGE 272
Query: 235 NVDGIPGAEHTPYAFGLFTAILVFIGAVLVVVG---LVYLGLKKPI-AEEKVEVRK 286
+ DG+ A A GL I +L ++G LV L KK + AE++ + K
Sbjct: 273 SSDGVSVALGVAAAAGLSVFAFTEIETILQLLGSAALVQLAGKKLLFAEDRKQTLK 328
>At5g52230 unknown protein
Length = 746
Score = 27.7 bits (60), Expect = 9.1
Identities = 15/56 (26%), Positives = 29/56 (51%)
Query: 130 EIRKLSTQVIRVKWSLHAREEIVFELLQHLKGIGARSLLEGIRKSTREMIEEQEAV 185
E + ++ +V R + EE+V +L ++L A+S + + S R ++EAV
Sbjct: 180 EKQLIAKRVTRSQTKASTTEEVVVDLKRNLSSSNAKSEKDSVNSSVRSQKPKKEAV 235
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.141 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,760,652
Number of Sequences: 26719
Number of extensions: 259077
Number of successful extensions: 737
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 729
Number of HSP's gapped (non-prelim): 12
length of query: 332
length of database: 11,318,596
effective HSP length: 100
effective length of query: 232
effective length of database: 8,646,696
effective search space: 2006033472
effective search space used: 2006033472
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0066.1


