

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
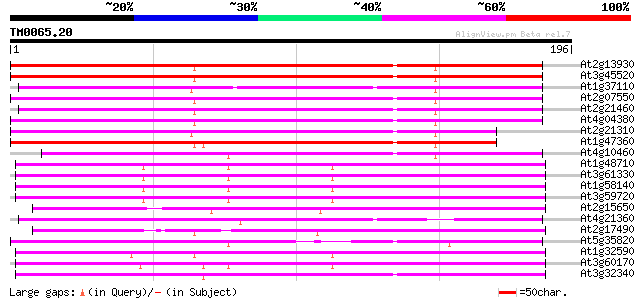
Query= TM0065.20
(196 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g13930 putative retroelement pol polyprotein 149 1e-36
At3g45520 copia-like polyprotein 146 8e-36
At1g37110 143 5e-35
At2g07550 putative retroelement pol polyprotein 143 7e-35
At2g21460 putative retroelement pol polyprotein 136 6e-33
At4g04380 putative polyprotein 132 2e-31
At2g21310 putative retroelement pol polyprotein 132 2e-31
At1g47360 polyprotein, putative 129 1e-30
At4g10460 putative retrotransposon 125 1e-29
At1g48710 hypothetical protein 122 1e-28
At3g61330 copia-type polyprotein 122 2e-28
At1g58140 hypothetical protein 122 2e-28
At3g59720 copia-type reverse transcriptase-like protein 120 6e-28
At2g15650 putative retroelement pol polyprotein 119 1e-27
At4g21360 putative transposable element 118 2e-27
At2g17490 putative retroelement pol polyprotein 116 9e-27
At5g35820 copia-like retrotransposable element 114 3e-26
At1g32590 hypothetical protein, 5' partial 113 6e-26
At3g60170 putative protein 110 4e-25
At3g32340 hypothetical protein 109 1e-24
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 149 bits (375), Expect = 1e-36
Identities = 78/191 (40%), Positives = 120/191 (61%), Gaps = 6/191 (3%)
Query: 1 SSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSL 60
+S+G+ L +VR++P + RNLIS+G L+D G G K+ KG + +G+KR +L
Sbjct: 333 NSDGSQVILTDVRYMPNMTRNLISLGTLEDRGCWFKSQDGILKIVKGCSTILKGQKRDTL 392
Query: 61 YMV----AEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHC 116
Y++ E + + E + +++WH RLGHMS+KGM+I+ KG + +L CE C
Sbjct: 393 YILDGVTEEGESHSSAEVKDETALWHSRLGHMSQKGMEILVKKGCLRREVIKELEFCEDC 452
Query: 117 ILGKQRKVSFSKAGRKSKLEKLKLVHTDVWG-PAPVKSLGGSRYYVTFIDDSTRKV*VYF 175
+ GKQ +VSF+ A +K EKL VH+D+WG P SLG S+Y+++F+DD +RKV +YF
Sbjct: 453 VYGKQHRVSFAPAQHVTK-EKLAYVHSDLWGSPHNPASLGNSQYFISFVDDYSRKVWIYF 511
Query: 176 LKNKSDVFSVF 186
L+ K + F F
Sbjct: 512 LRKKDEAFEKF 522
>At3g45520 copia-like polyprotein
Length = 1363
Score = 146 bits (368), Expect = 8e-36
Identities = 75/191 (39%), Positives = 116/191 (60%), Gaps = 6/191 (3%)
Query: 1 SSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSL 60
+ NG TL NVR++P + RNL+S+G + G+ G ++ GN V+ G++ +L
Sbjct: 364 NDNGLTVTLQNVRYIPDMDRNLLSLGTFEKAGHKFESENGMLRIKSGNQVLLEGRRYDTL 423
Query: 61 YMV----AEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHC 116
Y++ A ++ +AV A + + +WH+RL HMS+K M ++ KG + K L CE C
Sbjct: 424 YILHGKPATDESLAVARANDDTVLWHRRLCHMSQKNMSLLIKKGFLDKKKVSMLDTCEDC 483
Query: 117 ILGKQRKVSFSKAGRKSKLEKLKLVHTDVWG-PAPVKSLGGSRYYVTFIDDSTRKV*VYF 175
I G+ +K+ F+ A +K +KL+ VH+D+WG P SLG +Y+++FIDD TRKV VYF
Sbjct: 484 IYGRAKKIGFNLAQHDTK-KKLEYVHSDLWGAPTVPMSLGNCQYFISFIDDYTRKVWVYF 542
Query: 176 LKNKSDVFSVF 186
LK K + F F
Sbjct: 543 LKTKDEAFEKF 553
>At1g37110
Length = 1356
Score = 143 bits (361), Expect = 5e-35
Identities = 78/189 (41%), Positives = 111/189 (58%), Gaps = 8/189 (4%)
Query: 4 GTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYM- 62
GT+ L NV++VP ++RNLIS G LD GY G G + K N RG LY+
Sbjct: 359 GTIKILENVKYVPHLRRNLISTGTLDKLGYRHEGGEGKVRYFKNNKTALRGSLSNGLYVL 418
Query: 63 ----VAEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHCIL 118
V E A T+ V ++ +WH RLGHMS +K++A KG + + +L CEHC++
Sbjct: 419 DGSTVMSELCNAETDKVKTA-LWHSRLGHMSMNNLKVLAGKGLIDRKEINELEFCEHCVM 477
Query: 119 GKQRKVSFSKAGRKSKLEKLKLVHTDVWG-PAPVKSLGGSRYYVTFIDDSTRKV*VYFLK 177
GK +KVSF+ G+ + + L VH D+WG P S+ G +Y+++ IDD TRKV +YFLK
Sbjct: 478 GKSKKVSFN-VGKHTSEDALSYVHADLWGSPNVTPSISGKQYFLSIIDDKTRKVWLYFLK 536
Query: 178 NKSDVFSVF 186
+K + F F
Sbjct: 537 SKDETFDKF 545
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 143 bits (360), Expect = 7e-35
Identities = 76/191 (39%), Positives = 115/191 (59%), Gaps = 6/191 (3%)
Query: 1 SSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSL 60
+S+G L NVR++P + RNL+S+G + GY G ++ GN V+ G++ +L
Sbjct: 357 NSDGLTIVLTNVRYIPDMDRNLLSLGTFEKAGYKFESEDGILRIKAGNQVLLTGRRYDTL 416
Query: 61 YMV----AEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHC 116
Y++ + +AV + + + +WHQRL HMS+K M+I+ KG + K L VCE C
Sbjct: 417 YLLNWKPVASESLAVVKRADDTVLWHQRLCHMSQKNMEILVRKGFLDKKKVSSLDVCEDC 476
Query: 117 ILGKQRKVSFSKAGRKSKLEKLKLVHTDVWG-PAPVKSLGGSRYYVTFIDDSTRKV*VYF 175
I GK ++ SFS A +K EKL+ +H+D+WG P SLG +Y+++ IDD TRKV VYF
Sbjct: 477 IYGKAKRKSFSLAHHDTK-EKLEYIHSDLWGAPFVPLSLGKCQYFMSIIDDFTRKVWVYF 535
Query: 176 LKNKSDVFSVF 186
+K K + F F
Sbjct: 536 MKTKDEAFEKF 546
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 136 bits (343), Expect = 6e-33
Identities = 70/188 (37%), Positives = 113/188 (59%), Gaps = 6/188 (3%)
Query: 4 GTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMV 63
G + L NVR++P + RNL+S+G + GY G + G+ V+ ++ +LY++
Sbjct: 338 GMVVRLTNVRYIPEMDRNLLSLGTFEKSGYSFKLENGTLSIIAGDSVLLTVRRCYTLYLL 397
Query: 64 ----AEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHCILG 119
E+ ++V + + + +WH+RLGHMS+K M ++ KG + K L CE CI G
Sbjct: 398 QWRPVTEESLSVVKRQDDTILWHRRLGHMSQKNMDLLLKKGLLDKKKVSKLETCEDCIYG 457
Query: 120 KQRKVSFSKAGRKSKLEKLKLVHTDVWG-PAPVKSLGGSRYYVTFIDDSTRKV*VYFLKN 178
K +++ F+ A ++ EKL+ VH+D+WG P+ SLG +Y+++FIDD TRKV +YFLK
Sbjct: 458 KAKRIGFNLAQHDTR-EKLEYVHSDLWGAPSVPFSLGKCQYFISFIDDYTRKVRIYFLKT 516
Query: 179 KSDVFSVF 186
K + F F
Sbjct: 517 KDEAFDKF 524
>At4g04380 putative polyprotein
Length = 778
Score = 132 bits (331), Expect = 2e-31
Identities = 72/191 (37%), Positives = 109/191 (56%), Gaps = 6/191 (3%)
Query: 1 SSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSL 60
+ NG TL NVR++ + R +S+G + G +V G V+ G++ +L
Sbjct: 247 NDNGLTVTLQNVRYISDMDRIFLSLGTFEKAGRKFESENDMLRVNAGEQVLLEGRRYDTL 306
Query: 61 YMV----AEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHC 116
Y++ A ++ +AV +A + +WH+RL HMS+K M ++ KG + K L CE C
Sbjct: 307 YILHGKPATDESLAVAKANDDIVLWHRRLCHMSQKNMSLLVKKGFLDKKKVSMLDTCEDC 366
Query: 117 ILGKQRKVSFSKAGRKSKLEKLKLVHTDVWG-PAPVKSLGGSRYYVTFIDDSTRKV*VYF 175
I GK +K+ F+ A +K EKL+ VH D+WG P +LG +Y+++FIDD TRKV VYF
Sbjct: 367 IYGKAKKIGFNFAQHDTK-EKLEYVHYDLWGAPTMPMALGNCQYFISFIDDHTRKVCVYF 425
Query: 176 LKNKSDVFSVF 186
LK K + F F
Sbjct: 426 LKTKDEAFEKF 436
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 132 bits (331), Expect = 2e-31
Identities = 71/175 (40%), Positives = 105/175 (59%), Gaps = 6/175 (3%)
Query: 1 SSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSL 60
+ + T L NVR+VP + +NLIS+G L+D+G G KV KG + + +GKK G+L
Sbjct: 353 NDDNTTVLLKNVRYVPSMSKNLISMGTLEDQGCWFQSKAGTLKVVKGCMTLLKGKKVGTL 412
Query: 61 YM----VAEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHC 116
Y+ V + AVT + + S IWH RL HMS++ + ++ KG + K L CE C
Sbjct: 413 YLLQGVVVTGNANAVTSSKDESKIWHSRLCHMSQRNIDVLIKKGCLQAEKINGLEFCEDC 472
Query: 117 ILGKQRKVSFSKAGRKSKLEKLKLVHTDVWG-PAPVKSLGGSRYYVTFIDDSTRK 170
+ GK +V F A ++ EKL+ +H+D+WG P+ SLG +Y++TFIDD TRK
Sbjct: 473 VYGKTHRVGFGSAKHVTR-EKLEYIHSDLWGAPSVPNSLGNCQYFITFIDDLTRK 526
>At1g47360 polyprotein, putative
Length = 1182
Score = 129 bits (323), Expect = 1e-30
Identities = 70/175 (40%), Positives = 109/175 (62%), Gaps = 6/175 (3%)
Query: 1 SSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSL 60
+++GT L +VR+VP + RNLIS+G L+D+G G K+ KG + +G+KR +L
Sbjct: 323 NNDGTQVILTDVRYVPNMARNLISLGTLEDKGCWFKSQDGVLKIVKGCSTILKGQKRETL 382
Query: 61 YMV---AEE-DMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHC 116
Y++ AE + + E + +S+WH RLGHMS+KGM+I+ K + +L CE C
Sbjct: 383 YILEGLAENGESHSSAELKDETSLWHSRLGHMSQKGMEILVKKDCLQRETIKELKFCEDC 442
Query: 117 ILGKQRKVSFSKAGRKSKLEKLKLVHTDVWG-PAPVKSLGGSRYYVTFIDDSTRK 170
+ GK +VSF+ A +K EKL +H+D+WG P SLG +Y+++F+DD +RK
Sbjct: 443 VYGKNHRVSFAPAQHVTK-EKLAYIHSDLWGSPHNPASLGNCQYFISFVDDYSRK 496
>At4g10460 putative retrotransposon
Length = 1230
Score = 125 bits (314), Expect = 1e-29
Identities = 66/180 (36%), Positives = 107/180 (58%), Gaps = 6/180 (3%)
Query: 12 VRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMIAV 71
V+++P + RNL+S+G L++ GY G V +G + G + LY++ + ++
Sbjct: 352 VKYIPDMDRNLLSMGTLEEHGYSFESKNGVLVVKEGTRTLLIGSRHEKLYLLQGKPEVSH 411
Query: 72 TEAV----NSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHCILGKQRKVSFS 127
+ V + + +WH+RLGH+S+K M I+ KG + K L +CE CI GK R++SF
Sbjct: 412 SMTVERRNDDTVLWHRRLGHISQKNMDILVKKGYLDGKKVSKLELCEDCIYGKARRLSFV 471
Query: 128 KAGRKSKLEKLKLVHTDVWG-PAPVKSLGGSRYYVTFIDDSTRKV*VYFLKNKSDVFSVF 186
A ++ +KL VH+D+WG P+ SLG +Y+++FID +RK VYFLK+K + F F
Sbjct: 472 VATHNTE-DKLNYVHSDLWGAPSVPLSLGKCQYFISFIDVYSRKTWVYFLKHKDEAFGTF 530
>At1g48710 hypothetical protein
Length = 1352
Score = 122 bits (306), Expect = 1e-28
Identities = 69/192 (35%), Positives = 109/192 (55%), Gaps = 7/192 (3%)
Query: 3 NGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVT--KGNLVVARGKKRGSL 60
NG + NV ++P +K N++S+GQL ++GY + + NL+ + +
Sbjct: 385 NGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRM 444
Query: 61 YMVAEEDMIAVTEAV---NSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG--VCEH 115
+++ + IA + S +WH R GH++ G+++++ K + L ++ VCE
Sbjct: 445 FVLNIRNDIAQCLKMCYKEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQVCEG 504
Query: 116 CILGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV*VYF 175
C+LGKQ K+SF K + L+L+HTDV GP KSLG S Y++ FIDD +RK VYF
Sbjct: 505 CLLGKQFKMSFPKESSSRAQKSLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYF 564
Query: 176 LKNKSDVFSVFK 187
LK KS+VF +FK
Sbjct: 565 LKEKSEVFEIFK 576
>At3g61330 copia-type polyprotein
Length = 1352
Score = 122 bits (305), Expect = 2e-28
Identities = 69/192 (35%), Positives = 109/192 (55%), Gaps = 7/192 (3%)
Query: 3 NGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVT--KGNLVVARGKKRGSL 60
NG + NV ++P +K N++S+GQL ++GY + + NL+ + +
Sbjct: 385 NGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRM 444
Query: 61 YMVAEEDMIAVTEAV---NSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG--VCEH 115
+++ + IA + S +WH R GH++ G+++++ K + L ++ VCE
Sbjct: 445 FVLNIRNDIAQCLKMCYKEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQVCEG 504
Query: 116 CILGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV*VYF 175
C+LGKQ K+SF K + L+L+HTDV GP KSLG S Y++ FIDD +RK VYF
Sbjct: 505 CLLGKQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYF 564
Query: 176 LKNKSDVFSVFK 187
LK KS+VF +FK
Sbjct: 565 LKEKSEVFEIFK 576
>At1g58140 hypothetical protein
Length = 1320
Score = 122 bits (305), Expect = 2e-28
Identities = 69/192 (35%), Positives = 109/192 (55%), Gaps = 7/192 (3%)
Query: 3 NGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVT--KGNLVVARGKKRGSL 60
NG + NV ++P +K N++S+GQL ++GY + + NL+ + +
Sbjct: 385 NGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRM 444
Query: 61 YMVAEEDMIAVTEAV---NSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG--VCEH 115
+++ + IA + S +WH R GH++ G+++++ K + L ++ VCE
Sbjct: 445 FVLNIRNDIAQCLKMCYKEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQVCEG 504
Query: 116 CILGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV*VYF 175
C+LGKQ K+SF K + L+L+HTDV GP KSLG S Y++ FIDD +RK VYF
Sbjct: 505 CLLGKQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYF 564
Query: 176 LKNKSDVFSVFK 187
LK KS+VF +FK
Sbjct: 565 LKEKSEVFEIFK 576
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 120 bits (300), Expect = 6e-28
Identities = 68/192 (35%), Positives = 108/192 (55%), Gaps = 7/192 (3%)
Query: 3 NGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVT--KGNLVVARGKKRGSL 60
NG + NV ++P +K N++S+GQL ++GY + + NL+ + +
Sbjct: 385 NGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSIRDKESNLITKVPMSKNRM 444
Query: 61 YMVAEEDMIAVTEAV---NSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG--VCEH 115
+++ + IA + S +WH R GH++ G+++++ K + L ++ VCE
Sbjct: 445 FVLNIRNDIAQCLKMCYKEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQVCEG 504
Query: 116 CILGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV*VYF 175
C+LG Q K+SF K + L+L+HTDV GP KSLG S Y++ FIDD +RK VYF
Sbjct: 505 CLLGNQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYF 564
Query: 176 LKNKSDVFSVFK 187
LK KS+VF +FK
Sbjct: 565 LKEKSEVFEIFK 576
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 119 bits (297), Expect = 1e-27
Identities = 72/189 (38%), Positives = 102/189 (53%), Gaps = 15/189 (7%)
Query: 9 LHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDM 68
+ NV VPG+++NL+S+ Q+ GY F + N GK+ ++ M +
Sbjct: 381 IKNVFLVPGLEKNLLSVPQIISSGYWVRFQDKRCIIQDAN-----GKEIMNIEMTDKSFK 435
Query: 69 I--------AVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKH--VDLGVCEHCIL 118
I A+T V + WH+RLGH+S K ++ M K ++ L V C+ C L
Sbjct: 436 IKLSSVEEEAMTANVQTEETWHKRLGHVSNKRLQQMQDKELVNGLPRFKVTKETCKACNL 495
Query: 119 GKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV*VYFLKN 178
GKQ + SF K + EKL++VHTDV GP +S+ GSRYYV F+DD T VYFLK
Sbjct: 496 GKQSRKSFPKESQTKTREKLEIVHTDVCGPMQHQSIDGSRYYVLFLDDYTHMCWVYFLKQ 555
Query: 179 KSDVFSVFK 187
KS+ F+ FK
Sbjct: 556 KSETFATFK 564
>At4g21360 putative transposable element
Length = 1308
Score = 118 bits (295), Expect = 2e-27
Identities = 69/187 (36%), Positives = 99/187 (52%), Gaps = 14/187 (7%)
Query: 4 GTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMV 63
G++ L NVR VP ++RNLIS G LD GY G G + K N G LY++
Sbjct: 360 GSIRVLKNVRFVPNLRRNLISTGTLDKLGYKHEGGDGKVRFYKENKTALCGNLVNGLYVL 419
Query: 64 AEEDMIAVTEAVNSSS----IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHCILG 119
++ V S+ +WH RLGHMS MKI+A KG + +L CE+C++G
Sbjct: 420 DGHTVVNENCNVEGSNEKTELWHCRLGHMSLNNMKILAEKGLLEKKDIKELSFCENCVMG 479
Query: 120 KQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV*VYFLKNK 179
K +K+SF+ G+ E L +H D+W G +Y+++ IDD +RKV + FLK K
Sbjct: 480 KSKKLSFN-VGKHITDEVLGYIHADLW---------GKQYFLSIIDDKSRKVWLMFLKTK 529
Query: 180 SDVFSVF 186
+ F F
Sbjct: 530 DETFERF 536
>At2g17490 putative retroelement pol polyprotein
Length = 822
Score = 116 bits (290), Expect = 9e-27
Identities = 75/192 (39%), Positives = 102/192 (53%), Gaps = 21/192 (10%)
Query: 9 LHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMV----- 63
+ +V VP + +NL+S+ Q+ GY T + AR KK G + MV
Sbjct: 85 IRDVLLVPKLGKNLLSVPQMIINGYQVTLKNNYCTIHDS----AR-KKIGEVEMVNKSFH 139
Query: 64 ------AEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLK--HVDLGVCEH 115
E M+A EA + +WH+RLGH +KI+ SK ++ L +V+ G CE
Sbjct: 140 LRWLSNEETAMVAKDEA---TELWHKRLGHTGHSNLKILQSKEMVTGLPKFNVEEGKCES 196
Query: 116 CILGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV*VYF 175
CIL K + F K KL+L+H+DV GP S+ GSRY +TFIDD+TR V VYF
Sbjct: 197 CILSKHSRDPFPKESETRAKHKLELIHSDVCGPMQNSSINGSRYILTFIDDATRMVWVYF 256
Query: 176 LKNKSDVFSVFK 187
LK KS+VF FK
Sbjct: 257 LKAKSEVFQTFK 268
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 114 bits (285), Expect = 3e-26
Identities = 65/191 (34%), Positives = 105/191 (54%), Gaps = 22/191 (11%)
Query: 1 SSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSL 60
+ +G+ L +VR++P + +NLIS+G L+D+G G + K +L V GKK +L
Sbjct: 360 NEDGSTILLTDVRYIPEMSKNLISLGTLEDKGCWFESKKGILTIFKNDLTVLTGKKESTL 419
Query: 61 YMVAEEDMIAVTEAV----NSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHC 116
Y + + + + +S+WH RLGH+ KG++++ SKG H+D
Sbjct: 420 YFLQGTTLAGEANVIDKEKDETSLWHSRLGHIGAKGLQVLVSKG------HLD------- 466
Query: 117 ILGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVK-SLGGSRYYVTFIDDSTRKV*VYF 175
K +SF A +K +KL VH+D+WG V S+G +Y++TFIDD TR+ +YF
Sbjct: 467 ---KNIMISFGAAKHVTK-DKLDYVHSDLWGSTNVPFSIGKCQYFITFIDDFTRRTWIYF 522
Query: 176 LKNKSDVFSVF 186
++ K + FS F
Sbjct: 523 IRTKDEAFSKF 533
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 113 bits (283), Expect = 6e-26
Identities = 64/200 (32%), Positives = 101/200 (50%), Gaps = 15/200 (7%)
Query: 3 NGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGA---WKVTKGNLVVARGKKRGS 59
+G + + +V VPG+K NL S+GQL +G G W T+ +V+ +
Sbjct: 307 DGRIQVISDVYFVPGLKNNLFSVGQLQQKGLRFIIEGDVCEVWHKTEKRMVMHSTMTKNR 366
Query: 60 LYMV--------AEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG 111
+++V E+ + ++++WH+R GH++ +G++ +A K + L DLG
Sbjct: 367 MFVVFAAVKKSKETEETRCLQVIGKANNMWHKRFGHLNHQGLRSLAEKEMVKGLPKFDLG 426
Query: 112 ----VCEHCILGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDS 167
VC+ C+ GKQ + S K + L+LVHTD+ GP S G RY + FIDD
Sbjct: 427 EEEAVCDICLKGKQIRESIPKESAWKSTQVLQLVHTDICGPINPASTSGKRYILNFIDDF 486
Query: 168 TRKV*VYFLKNKSDVFSVFK 187
+RK Y L KS+ F FK
Sbjct: 487 SRKCWTYLLSEKSETFQFFK 506
>At3g60170 putative protein
Length = 1339
Score = 110 bits (276), Expect = 4e-25
Identities = 66/196 (33%), Positives = 103/196 (51%), Gaps = 11/196 (5%)
Query: 3 NGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKV---TKGNLVVARGKKRGS 59
NG + V +VP ++ NL+S+GQL + G G KV +KG ++
Sbjct: 349 NGVTQVIPEVYYVPELRNNLLSLGQLQERGLAILIRDGTCKVYHPSKGAIMETNMSGNRM 408
Query: 60 LYMVAEE----DMIAVTEAV--NSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG-- 111
+++A + + TE V + +WH R GH++++G+K++A K + L +
Sbjct: 409 FFLLASKPQKNSLCLQTEEVMDKENHLWHCRFGHLNQEGLKLLAHKKMVIGLPILKATKE 468
Query: 112 VCEHCILGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV 171
+C C+ GKQ + S SK +L+LVH+D+ GP S G RY ++FIDD TRK
Sbjct: 469 ICAICLTGKQHRESMSKKTSWKSSTQLQLVHSDICGPITPISHSGKRYILSFIDDFTRKT 528
Query: 172 *VYFLKNKSDVFSVFK 187
VYFL KS+ F+ FK
Sbjct: 529 WVYFLHEKSEAFATFK 544
>At3g32340 hypothetical protein
Length = 871
Score = 109 bits (272), Expect = 1e-24
Identities = 59/192 (30%), Positives = 100/192 (51%), Gaps = 8/192 (4%)
Query: 3 NGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYM 62
+G + L N +VP I +N++S+ LD EG+H + + ++ LY+
Sbjct: 203 SGMVLELKNCYYVPAINKNIVSVSCLDMEGFHFSIKNKCCSFDRDDMFYGSAPLENGLYV 262
Query: 63 VAEE-------DMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEH 115
+ + I + +N + +WH RLGH++EK ++ + S G +++ + CE
Sbjct: 263 LNQSMPVYNIRTKIFKSNDLNPTFLWHCRLGHINEKRIQKLHSDGLLNSFDYESYETCES 322
Query: 116 CILGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV*VYF 175
C+LGK K FS ++ + L L+HTDV GP + G +Y++TF DD +R VY
Sbjct: 323 CLLGKMTKAPFSGHSERAG-DLLGLIHTDVCGPMSTSARGNYQYFITFTDDFSRYSYVYL 381
Query: 176 LKNKSDVFSVFK 187
+K+KS+ F FK
Sbjct: 382 MKHKSESFEKFK 393
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,407,314
Number of Sequences: 26719
Number of extensions: 176709
Number of successful extensions: 595
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 86
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 372
Number of HSP's gapped (non-prelim): 102
length of query: 196
length of database: 11,318,596
effective HSP length: 94
effective length of query: 102
effective length of database: 8,807,010
effective search space: 898315020
effective search space used: 898315020
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0065.20


