

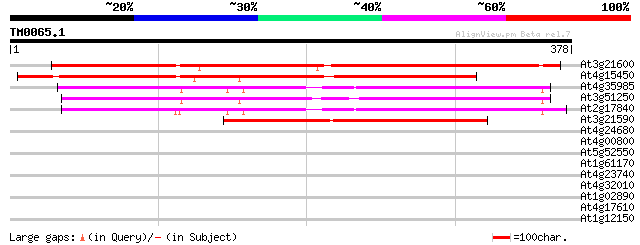
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0065.1
(378 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g21600 unknown protein 377 e-105
At4g15450 unknown protein 313 1e-85
At4g35985 unknown protein 208 3e-54
At3g51250 unknown protein 204 5e-53
At2g17840 putative senescence-related protein 196 1e-50
At3g21590 hypothetical protein 147 7e-36
At4g24680 unknown protein 32 0.57
At4g00800 31 0.98
At5g52550 unknown protein 30 1.7
At1g61170 unknown protein 30 1.7
At4g23740 putative receptor kinase 30 2.8
At4g32010 predicted protein 29 4.8
At1g02890 hypothetical protein 29 4.8
At4g17610 putative protein 28 8.3
At1g12150 hypothetical protein 28 8.3
>At3g21600 unknown protein
Length = 374
Score = 377 bits (968), Expect = e-105
Identities = 192/352 (54%), Positives = 262/352 (73%), Gaps = 17/352 (4%)
Query: 29 KPKNLIRQEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWP 88
+P+ + +EVL+QIP CRV+L+ E +AVELA G F ++KV D+ V+LA I+++G+ +QWP
Sbjct: 28 QPQTMRAEEVLLQIPRCRVHLIGESEAVELASGDFKLVKVSDNGVTLAMIVRIGHDLQWP 87
Query: 89 LTKDEPVVKVDTLHYLFTLPVKDGGSEPLSYGVTFPEQ-----CYGSMGLLESFLKDHSC 143
+ +DEPVVK+D YLFTLPVKDG +PLSYGVTF S+ LL+ FL ++SC
Sbjct: 88 VIRDEPVVKLDARDYLFTLPVKDG--DPLSYGVTFSGDDRDVALVNSLKLLDQFLSENSC 145
Query: 144 FSDLKTGK-KGGLDWEQFAPRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQ 202
FS + K G+DW++FAPR+EDYN+ +AKAIAGGTG I++GIF SNAY+NQV GG
Sbjct: 146 FSSTASSKVNNGIDWQEFAPRIEDYNNVVAKAIAGGTGHIIRGIFSLSNAYSNQVHKGGD 205
Query: 203 TILT---DNKNNGLVRQSVSNNKTADATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGI 259
++T +++ NG S +N ++ KKN +N +L+RVR+L+ TE LSK++L+G G+
Sbjct: 206 IMITKAEESQRNG----SYNNGNSSGNEKKNGINTHLQRVRKLSKATENLSKTMLNGAGV 261
Query: 260 MSGSVMAPVLKSQPGQAFLKMLPGEVLLASLDAVNKVFEATEAAQKQSLSATSQAATRMV 319
+SGSVM P++KS+PG AF M+PGEVLLASLDA+NK+ +ATEAA++Q+LSATS+AATRMV
Sbjct: 262 VSGSVMVPMMKSKPGMAFFSMVPGEVLLASLDALNKILDATEAAERQTLSATSRAATRMV 321
Query: 320 SNRYGEEAGEATEHVFATAGHAANTAWNVSKIRKAINPVTPASTAGGALRNS 371
S R+G+ AGEAT V ATAGHAA TAWNV KIRK P +S G ++N+
Sbjct: 322 SERFGDNAGEATGDVLATAGHAAGTAWNVLKIRKTFYP--SSSLTSGIVKNA 371
>At4g15450 unknown protein
Length = 331
Score = 313 bits (801), Expect = 1e-85
Identities = 163/327 (49%), Positives = 230/327 (69%), Gaps = 30/327 (9%)
Query: 6 SKPEEEEPTMMRTSTLVPIAEYSKPKNLIRQEVLIQIPGCRVYLMDEGQAVELAQGQFMI 65
SK E ++P+ + Y + EVL+QI GCR +L++ +AVELA G F +
Sbjct: 9 SKTETQQPSSYGQQQDAMYSSYGT----VSDEVLLQIHGCRAHLINGSEAVELAAGDFEL 64
Query: 66 IKVMDDNVSLATIIKVGNGVQWPLTKDEPVVKVDTLHYLFTLPVKDGGSEPLSYGVTF-- 123
++V+D NV+LA ++++GN +QWP+ KDEPVVK+D+ YLFTLPVKDG EPLSYG+TF
Sbjct: 65 VQVLDSNVALAMVVRIGNDLQWPVIKDEPVVKLDSRDYLFTLPVKDG--EPLSYGITFFP 122
Query: 124 ----PEQCYGSMGLLESFLKDHSCFSDLKTGKKG-----------GLDWEQFAPRVEDYN 168
S+ LL+ FL+++SCFS + G+DW++FAP++EDYN
Sbjct: 123 IDENDIVFVNSIELLDDFLRENSCFSSSPSSSSSSSSSSSSSVNNGIDWKEFAPKIEDYN 182
Query: 169 HFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTD-NKNNGLVRQSVSNNKTADAT 227
+ +AKAIAGGTG I++G+F CSNAYTNQV GG+ ++T K NG +++K T
Sbjct: 183 NVVAKAIAGGTGHIIRGMFKCSNAYTNQVHKGGEIMITKAEKKNG------ASSKRNATT 236
Query: 228 KKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEVLL 287
KN +N+NL+RVR+L+ TEKLSK++L+GVG++SGSVM PV+KS+PG+AF M+PGEVLL
Sbjct: 237 NKNQINKNLQRVRKLSRATEKLSKTMLNGVGVVSGSVMGPVVKSKPGKAFFSMVPGEVLL 296
Query: 288 ASLDAVNKVFEATEAAQKQSLSATSQA 314
ASLDA+NK+ +A EAA++Q+LSATS+A
Sbjct: 297 ASLDALNKLLDAAEAAERQTLSATSKA 323
>At4g35985 unknown protein
Length = 431
Score = 208 bits (530), Expect = 3e-54
Identities = 130/375 (34%), Positives = 202/375 (53%), Gaps = 54/375 (14%)
Query: 33 LIRQEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKD 92
L +EV++ I G V+L+D+ +VELA G I++++ ++++A +VG+ +QWPLTKD
Sbjct: 66 LATEEVILTIHGAMVHLIDKSYSVELACGDLEILRLVQGDITVAVFARVGDEIQWPLTKD 125
Query: 93 EPVVKVDTLHYLFTL-PVKDGGS----------EPLSYGVTFPEQCYGSM-GLLESFLKD 140
EP VKVD HY F+L PVK+ S E L+YG+T + M L+ L D
Sbjct: 126 EPAVKVDESHYFFSLRPVKESESSDHSVNETENEMLNYGLTMASKGQEPMLEKLDKILAD 185
Query: 141 HSCFS----------------------DLKTGKKGGLD------WEQFAPRVEDYNHFLA 172
+S F+ +LK +K ++ W AP VEDY+ A
Sbjct: 186 YSSFTAEEKQKEENVLDLTAAKETSPEELKGKRKKMVEKQCTAYWTTLAPNVEDYSGVAA 245
Query: 173 KAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTDNKNNGLVRQSVSNNKTADATKKNAM 232
K IA G+GQ++KGI C + +++ G N +++ +S + +
Sbjct: 246 KLIAAGSGQLIKGILWCGDLTMDRLMWG----------NDFMKKKLSKAEKERQVSPGTL 295
Query: 233 NENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEVLLASLDA 292
+ LKRV+++T MTEK++ +L GV +SG + V+ S+ GQ +LPGE++LA+LD
Sbjct: 296 -KRLKRVKKMTKMTEKVANGVLSGVVKVSGFFSSSVINSKAGQKLFGLLPGEMVLATLDG 354
Query: 293 VNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTAWNVSKIR 352
NKV +A E A + + TS T +V ++YG + +AT + AGHA TAW V KIR
Sbjct: 355 FNKVCDAVEVAGRHVMKTTSDVTTEIVDHKYGAKTAQATNEGLSAAGHAFGTAWTVFKIR 414
Query: 353 KAINP---VTPASTA 364
+A+NP + P+S A
Sbjct: 415 QALNPKSAMKPSSLA 429
>At3g51250 unknown protein
Length = 463
Score = 204 bits (520), Expect = 5e-53
Identities = 126/387 (32%), Positives = 198/387 (50%), Gaps = 69/387 (17%)
Query: 36 QEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKDEPV 95
+EVLI++PG + L+D+ +VELA G F I++++ +A + VGN +QWPLTK+E
Sbjct: 75 EEVLIRVPGAILNLIDKSYSVELACGDFTIVRIIQGQNIVAVLANVGNEIQWPLTKNEVA 134
Query: 96 VKVDTLHYLFTL-PVKDGGS--------------------EPLSYGVTFPEQCYGSMGL- 133
KVD HY F++ P K+ G E L+YG+T + ++ L
Sbjct: 135 AKVDGSHYFFSIHPPKEKGQGSGSDSDDEQGQKSKSKSDDEILNYGLTIASKGQENVLLV 194
Query: 134 LESFLKDHSCFSDLKTGKKG---------------------------------GLDWEQF 160
L+ L+D+SCF++ + +K W
Sbjct: 195 LDQVLRDYSCFTEQRMSEKAKETGEEVLGNSVVADTSPEELKGERKDVVEGQCAAYWTTL 254
Query: 161 APRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTDNKNNGLVRQSVSN 220
AP VEDY H AK IA G+G++++GI C + +++ G + + N L R
Sbjct: 255 APNVEDYTHSTAKMIASGSGKLIRGILWCGDVTVERLKKGNEVM-----KNRLSRAEKEK 309
Query: 221 NKTADATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKM 280
+ + + ++ +KRV+++T MTEK++ +L GV +SG + + S+ G+ +
Sbjct: 310 DVSPETLRR------IKRVKRVTQMTEKVATGVLSGVVKVSGFITGSMANSKAGKKLFGL 363
Query: 281 LPGEVLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGH 340
LPGE++LASLD +K+ +A E A K +S +S T +V++RYG +A EAT AGH
Sbjct: 364 LPGEIVLASLDGFSKICDAVEVAGKNVMSTSSTVTTELVNHRYGTKAAEATNEGLDAAGH 423
Query: 341 AANTAWNVSKIRKAINP---VTPASTA 364
A TAW KIRKA NP + P+S A
Sbjct: 424 AFGTAWVAFKIRKAFNPKNVIKPSSLA 450
>At2g17840 putative senescence-related protein
Length = 452
Score = 196 bits (499), Expect = 1e-50
Identities = 132/393 (33%), Positives = 205/393 (51%), Gaps = 64/393 (16%)
Query: 36 QEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKDEPV 95
+EV+++I G ++L+D+ +VELA G II+++ +A + V + +QWPLTKDE
Sbjct: 69 EEVILKISGAILHLIDKSYSVELACGDLEIIRIVQGENVVAVLASVSDEIQWPLTKDENS 128
Query: 96 VKVDTLHYLFTL-PVK------------DGG---SEPLSYGVTFPEQCYGSMGL-LESFL 138
VKVD HY FTL P K DGG +E L+YG+T + + + LE L
Sbjct: 129 VKVDESHYFFTLRPTKEISHDSSDEEDGDGGKNTNEMLNYGLTIASKGQEHLLVELEKIL 188
Query: 139 KDHSCFS------DLKTGKKGGLD---------------------------WEQFAPRVE 165
+D+S FS + K + LD W AP VE
Sbjct: 189 EDYSSFSVQEVSEEAKEAGEKVLDVTVARETSPVELTGERKEIVERQCSAYWTTLAPNVE 248
Query: 166 DYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTDNKNNGLVRQSVSNNKTAD 225
DY+ AK IA G+G ++KGI C + +++ G NG +++ +S +
Sbjct: 249 DYSGKAAKLIATGSGHLIKGILWCGDVTMDRLIWG----------NGFMKRRLSKAEKES 298
Query: 226 ATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEV 285
+ + + ++RV+++T MTE ++ S+L GV +SG + V ++ G+ F +LPGEV
Sbjct: 299 EVHPDTL-KRIRRVKRMTKMTESVANSILSGVLKVSGFFTSSVANTKVGKKFFSLLPGEV 357
Query: 286 LLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTA 345
+LASLD NKV +A E A + +S +S T +V ++YG +A EAT AG+A TA
Sbjct: 358 ILASLDGFNKVCDAVEVAGRNVMSTSSTVTTELVDHKYGGKAAEATNEGLDAAGYALGTA 417
Query: 346 WNVSKIRKAINP---VTPASTAGGALRNSIKNR 375
W KIRKAINP + P++ A A+R++ +
Sbjct: 418 WVAFKIRKAINPKSVLKPSTLAKTAIRSAASQK 450
>At3g21590 hypothetical protein
Length = 241
Score = 147 bits (372), Expect = 7e-36
Identities = 77/179 (43%), Positives = 117/179 (65%), Gaps = 2/179 (1%)
Query: 145 SDLKTGKKGG-LDWEQFAPRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQT 203
S +GK +DW++FAP+ EDY + +AKAIA GTG I+KGIF CSN+Y+ ++ G
Sbjct: 64 SSSTSGKNNNEIDWKKFAPKAEDYKNGVAKAIAVGTGHIIKGIFTCSNSYSKKIHEEGTI 123
Query: 204 ILTDNKNNGLVRQSVSNNKTADATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGS 263
D + +G + Q + + KKN +N+NL+R +L ++E + + L+G ++S S
Sbjct: 124 AEEDEERSGDISQ-IDGGGNNETNKKNKLNKNLQRAEKLWKVSEAIGMAALEGGDLVSSS 182
Query: 264 VMAPVLKSQPGQAFLKMLPGEVLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNR 322
++APV+KS+ G+A L PGEV+LASLD+ + + A EAA+ Q+ ATS AATR+VS R
Sbjct: 183 MIAPVVKSKLGKALLSTAPGEVILASLDSFHNIIGAAEAAEIQTHYATSMAATRLVSKR 241
>At4g24680 unknown protein
Length = 1480
Score = 32.0 bits (71), Expect = 0.57
Identities = 20/75 (26%), Positives = 32/75 (42%)
Query: 287 LASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTAW 346
LA L+ +N+ + E +++ A S A+ + G + AT V T G NT
Sbjct: 769 LAKLEELNRRSQIYEEGSVKNMEAASNASPADMPTDPGSHSSNATNSVEPTGGSGKNTTQ 828
Query: 347 NVSKIRKAINPVTPA 361
N + N V P+
Sbjct: 829 NTRTSTEYANNVGPS 843
>At4g00800
Length = 979
Score = 31.2 bits (69), Expect = 0.98
Identities = 33/117 (28%), Positives = 52/117 (44%), Gaps = 5/117 (4%)
Query: 230 NAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEVL-LA 288
+A +E L +V ++ S D VG+ +GSV A +S G++ L+ V +A
Sbjct: 144 SASSELLLQVSNQEEDDHEVLSSNGDSVGVAAGSVSADDFRSFGGESLLEDEDNGVSGVA 203
Query: 289 SLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTA 345
SL+ KV E + +SL+ VS+ + E +TE T A N A
Sbjct: 204 SLEDEAKVMEVQASDITESLNPDLVT----VSSGFDSEGNVSTEKEAETTMEAGNAA 256
>At5g52550 unknown protein
Length = 360
Score = 30.4 bits (67), Expect = 1.7
Identities = 13/46 (28%), Positives = 23/46 (49%)
Query: 216 QSVSNNKTADATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMS 261
++ +ADA KK + L+R++Q +L KS+ IM+
Sbjct: 67 EAADEEDSADAAKKKQERDELERIKQAENKKNRLEKSIATSAAIMA 112
>At1g61170 unknown protein
Length = 251
Score = 30.4 bits (67), Expect = 1.7
Identities = 32/115 (27%), Positives = 52/115 (44%), Gaps = 17/115 (14%)
Query: 223 TADATKKNAMNENLKRVRQLTTMTEKLSKSLL--DGVGIMSGSVMAPVL-------KSQP 273
T D +N ++N V + T + E LS++ L + + PV + +P
Sbjct: 10 TTDRENENVSDKNTTIVEEETVVKEVLSETTLFTPSSSFRTSTFKDPVKTKIREDEEKKP 69
Query: 274 GQAFLKMLPGEVLL--ASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEE 326
G FLKM VL+ S+D E +E ++ SLS + ++ +V N Y EE
Sbjct: 70 G--FLKMASDPVLIRPGSVDPE----EGSEVSEICSLSLSESVSSTVVMNGYDEE 118
>At4g23740 putative receptor kinase
Length = 638
Score = 29.6 bits (65), Expect = 2.8
Identities = 14/28 (50%), Positives = 19/28 (67%)
Query: 145 SDLKTGKKGGLDWEQFAPRVEDYNHFLA 172
SD K KKGG+ E+F R+ED N+ L+
Sbjct: 294 SDNKLQKKGGMSPEKFVSRMEDVNNRLS 321
>At4g32010 predicted protein
Length = 675
Score = 28.9 bits (63), Expect = 4.8
Identities = 28/114 (24%), Positives = 49/114 (42%), Gaps = 18/114 (15%)
Query: 197 VQSGGQTILTDNKNNGLVRQSVSNNKTADATKKNAMNENLKRVRQLTTMTEKLSKSLLDG 256
+++GG T ++ K +GL+ +VS+ A E++ V + T + L +D
Sbjct: 32 LENGGVTCISCAKKSGLISMNVSHESNGKDFPSFASAEHVGSVLERTNLKHLLHFQRID- 90
Query: 257 VGIMSGSVMAPVLKSQPGQAFLKMLPGEVLL-ASLDAVNKVFEATEAAQKQSLS 309
P + L+M E LL +SLDA+ E E + + +LS
Sbjct: 91 ----------------PTHSSLQMKQEESLLPSSLDALRHKTERKELSAQPNLS 128
>At1g02890 hypothetical protein
Length = 1240
Score = 28.9 bits (63), Expect = 4.8
Identities = 46/190 (24%), Positives = 68/190 (35%), Gaps = 34/190 (17%)
Query: 86 QWPLTKDEPVVKVDTLHYLFTLPVKDGGSEPLSYGVTFPEQCYGSMGLLESFLKDHSCFS 145
+ P + P V T ++ G P V+F Y G + L S ++
Sbjct: 412 EMPKEYERPSASVLTRRQAHKDSLRGGILNPQDIEVSFENFPYFLSGTTKDVLMI-STYA 470
Query: 146 DLKTGKKGG---LDWEQFAPRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQ 202
+K GK+ D PR E Y LAKA+A Q G +
Sbjct: 471 HIKYGKEYAEYASDLPTACPRSEIYQEMLAKALA--------------------KQCGAK 510
Query: 203 TILTDNKNNGLVRQSVSNNKTADATKKNAMNENL----KRVRQLTTMTEKLSKSLLDGV- 257
++ D+ L+ S K AD TK+++ E L KR Q K + V
Sbjct: 511 LMIVDS----LLLPGGSTPKEADTTKESSRRERLSVLAKRAVQAAQAAVLQHKKPISSVE 566
Query: 258 -GIMSGSVMA 266
GI GS ++
Sbjct: 567 AGITGGSTLS 576
>At4g17610 putative protein
Length = 1850
Score = 28.1 bits (61), Expect = 8.3
Identities = 22/61 (36%), Positives = 32/61 (52%), Gaps = 5/61 (8%)
Query: 218 VSNNKTADATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAF 277
VS + + N++N ++ R T EK +KSL GVG + GSV + + Q QAF
Sbjct: 372 VSETISEKKPEGNSVNRSMTRKE---TWAEKEAKSL--GVGELYGSVDSGLTSQQGWQAF 426
Query: 278 L 278
L
Sbjct: 427 L 427
>At1g12150 hypothetical protein
Length = 548
Score = 28.1 bits (61), Expect = 8.3
Identities = 17/36 (47%), Positives = 25/36 (69%), Gaps = 2/36 (5%)
Query: 286 LLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSN 321
L A+L A+ ++ +ATE AQK + SA +AA RMV +
Sbjct: 501 LEANLKAIEEMKQATELAQKSAESA--EAAKRMVES 534
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.130 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,994,525
Number of Sequences: 26719
Number of extensions: 327422
Number of successful extensions: 633
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 607
Number of HSP's gapped (non-prelim): 19
length of query: 378
length of database: 11,318,596
effective HSP length: 101
effective length of query: 277
effective length of database: 8,619,977
effective search space: 2387733629
effective search space used: 2387733629
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0065.1


