

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0063.5
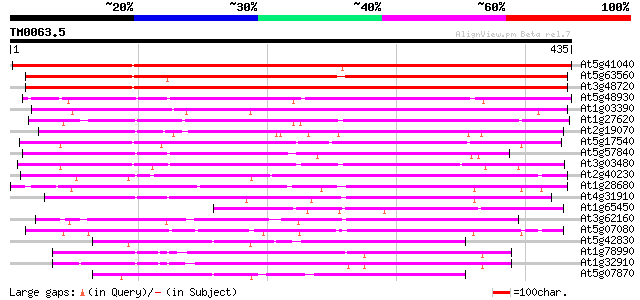
(435 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g41040 N-hydroxycinnamoyl/benzoyltransferase-like protein 580 e-166
At5g63560 acyltransferase-like protein 503 e-143
At3g48720 unknown protein 496 e-140
At5g48930 anthranilate N-benzoyltransferase 263 1e-70
At1g03390 hypothetical protein 243 2e-64
At1g27620 putative hypersensitivity-related protein 218 7e-57
At2g19070 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 196 2e-50
At5g17540 unknown protein 196 3e-50
At5g57840 N-hydroxycinnamoyl/benzoyltransferase 190 1e-48
At3g03480 putative hypersensitivity-related gene 189 3e-48
At2g40230 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 174 1e-43
At1g28680 anthranilate N-hydroxycinnamoyl/benzoyltransferase lik... 164 1e-40
At4g31910 unknown protein 161 7e-40
At1g65450 unknown protein 145 6e-35
At3g62160 unknown protein 141 8e-34
At5g07080 unknown protein 139 4e-33
At5g42830 N-hydroxycinnamoyl/benzoyltransferase-like protein 134 1e-31
At1g78990 123 2e-28
At1g32910 hypothetical protein 116 2e-26
At5g07870 N-hydroxycinnamoyl/benzoyltransferase - like protein 115 6e-26
>At5g41040 N-hydroxycinnamoyl/benzoyltransferase-like protein
Length = 457
Score = 580 bits (1495), Expect = e-166
Identities = 284/436 (65%), Positives = 348/436 (79%), Gaps = 4/436 (0%)
Query: 3 NADMSNIGINVRQGEPTRVHPAEETEKGLYFLSNLDQNIAVPVRTVYCFKSSSRGNEDAA 62
N +NI + V Q EP V P ET KGLYFLSNLDQNIAV VRT+YCFKS RGNE+A
Sbjct: 23 NIKGTNIHLEVHQKEPALVKPESETRKGLYFLSNLDQNIAVIVRTIYCFKSEERGNEEAV 82
Query: 63 HVIRVALSKILVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPD 122
VI+ ALS++LV YYP+AG+L IS EGKL VDC T EG VFVEAEA+C +++IGD+TKPD
Sbjct: 83 QVIKKALSQVLVHYYPLAGRLTISPEGKLTVDC-TEEGVVFVEAEANCKMDEIGDITKPD 141
Query: 123 PDSLGKLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSE 182
P++LGKL+Y+ A++ILE+P +T QVTKFKCGGF +GL + HCM DG+ AMEFVN+W +
Sbjct: 142 PETLGKLVYDVVDAKNILEIPPVTAQVTKFKCGGFVLGLCMNHCMFDGIGAMEFVNSWGQ 201
Query: 183 TARDLDLKTPPFLDRTILKARDPPIIEFKHHEYDEIVDLSDTKKLYEEEKMLYRSFCFDP 242
AR L L TPPF DRTIL AR+PP IE H E++EI D S+ LY +E LYRSFCFDP
Sbjct: 202 VARGLPLTTPPFSDRTILNARNPPKIENLHQEFEEIEDKSNINSLYTKEPTLYRSFCFDP 261
Query: 243 KKLELLKKKATEDG---VVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRF 299
+K++ LK +ATE+ + C++FEALS FVWRART++LKM DQKTKLLFAVDGR +F
Sbjct: 262 EKIKKLKLQATENSESLLGNSCTSFEALSAFVWRARTKSLKMLSDQKTKLLFAVDGRAKF 321
Query: 300 VPPIPDRYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTR 359
P +P YFGN IVLT+S+C+AGE++E PLSF+VGLV AI +VTD YMRS+IDYFEVTR
Sbjct: 322 EPQLPKGYFGNGIVLTNSICEAGELIEKPLSFAVGLVREAIKMVTDGYMRSAIDYFEVTR 381
Query: 360 ARPSLTATLLITTWTRLSFHTTDFGWGEPLCSGPVTLPEKEVILFLSNGQDRKSINVLLG 419
ARPSL++TLLITTW+RL FHTTDFGWGEP+ SGPV LPEKEV LFLS+G+ R+SINVLLG
Sbjct: 382 ARPSLSSTLLITTWSRLGFHTTDFGWGEPILSGPVALPEKEVTLFLSHGEQRRSINVLLG 441
Query: 420 LPASAMETFEALMMQV 435
LPA+AM+ F+ +Q+
Sbjct: 442 LPATAMDVFQEQFLQI 457
>At5g63560 acyltransferase-like protein
Length = 426
Score = 503 bits (1295), Expect = e-143
Identities = 254/422 (60%), Positives = 320/422 (75%), Gaps = 8/422 (1%)
Query: 13 VRQGEPTRVHPAEETEKGLYFLSNLDQNIAVPVRTVYCFKSSSRGNEDAAHVIRVALSKI 72
V + EP V PA ET KGL++LSNLDQNIA+ V+T Y FKS+SR NE++ VI+ +LS++
Sbjct: 9 VTRKEPVLVSPASETPKGLHYLSNLDQNIAIIVKTFYYFKSNSRSNEESYEVIKKSLSEV 68
Query: 73 LVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKP--DPDSLGKLI 130
LV YYP AG+L IS EGK+ VDC T EG V VEAEA+C IE I P++L KL+
Sbjct: 69 LVHYYPAAGRLTISPEGKIAVDC-TGEGVVVVEAEANCGIEKIKKAISEIDQPETLEKLV 127
Query: 131 YNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETARDLDLK 190
Y+ PGAR+ILE+P + VQVT FKCGGF +GL + H M DG++AMEF+N+W+ETAR L L
Sbjct: 128 YDVPGARNILEIPPVVVQVTNFKCGGFVLGLGMNHNMFDGIAAMEFLNSWAETARGLPLS 187
Query: 191 TPPFLDRTILKARDPPIIEFKHHEYDEIVDLSDTKKLYEEEKMLYRSFCFDPKKLELLKK 250
PPFLDRT+L+ R PP IEF H+E++++ D+S T KLY +EK++Y+SF F P+KLE LK
Sbjct: 188 VPPFLDRTLLRPRTPPKIEFPHNEFEDLEDISGTGKLYSDEKLVYKSFLFGPEKLERLKI 247
Query: 251 KATEDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRFVPPIPDRYFGN 310
A + +TF+ L+GF+WRAR +AL ++PDQ+ KLLFA DGR+RFVP +P Y GN
Sbjct: 248 MAET-----RSTTFQTLTGFLWRARCQALGLKPDQRIKLLFAADGRSRFVPELPKGYSGN 302
Query: 311 AIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTRARPSLTATLLI 370
IV T+ V AGE+ NPLS SV LV A+++V D +MRS+IDYFEVTRARPSLTATLLI
Sbjct: 303 GIVFTYCVTTAGEVTLNPLSHSVCLVKRAVEMVNDGFMRSAIDYFEVTRARPSLTATLLI 362
Query: 371 TTWTRLSFHTTDFGWGEPLCSGPVTLPEKEVILFLSNGQDRKSINVLLGLPASAMETFEA 430
T+W +LSFHT DFGWGEP+ SGPV LPEKEVILFL G D KSINVLLGLP SAM+ F+
Sbjct: 363 TSWAKLSFHTKDFGWGEPVVSGPVGLPEKEVILFLPCGSDTKSINVLLGLPGSAMKVFQG 422
Query: 431 LM 432
+M
Sbjct: 423 IM 424
>At3g48720 unknown protein
Length = 430
Score = 496 bits (1276), Expect = e-140
Identities = 243/421 (57%), Positives = 323/421 (76%), Gaps = 2/421 (0%)
Query: 13 VRQGEPTRVHPAEETEKGLYFLSNLDQNIAVPVRTVYCFKSSSRGNEDAAHVIRVALSKI 72
V + P + P ET G Y+LSNLDQNIA+ V+T+Y +KS SR N+++ +VI+ +LS++
Sbjct: 9 VTRKNPELIPPVSETPNGHYYLSNLDQNIAIIVKTLYYYKSESRTNQESYNVIKKSLSEV 68
Query: 73 LVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGD-LTKPDPDSLGKLIY 131
LV YYP+AG+L IS EGK+ V+C T EG V VEAEA+C I+ I + +++ ++L KL+Y
Sbjct: 69 LVHYYPVAGRLTISPEGKIAVNC-TGEGVVVVEAEANCGIDTIKEAISENRMETLEKLVY 127
Query: 132 NTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETARDLDLKT 191
+ PGAR+ILE+P + VQVT FKCGGF +GL + H M DG++A EF+N+W E A+ L L
Sbjct: 128 DVPGARNILEIPPVVVQVTNFKCGGFVLGLGMSHNMFDGVAAAEFLNSWCEMAKGLPLSV 187
Query: 192 PPFLDRTILKARDPPIIEFKHHEYDEIVDLSDTKKLYEEEKMLYRSFCFDPKKLELLKKK 251
PPFLDRTIL++R+PP IEF H+E+DEI D+SDT K+Y+EEK++Y+SF F+P+KLE LK
Sbjct: 188 PPFLDRTILRSRNPPKIEFPHNEFDEIEDISDTGKIYDEEKLIYKSFLFEPEKLEKLKIM 247
Query: 252 ATEDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRFVPPIPDRYFGNA 311
A E+ K STF+AL+GF+W++R EAL+ +PDQ+ KLLFA DGR+RF+P +P Y GN
Sbjct: 248 AIEENNNNKVSTFQALTGFLWKSRCEALRFKPDQRVKLLFAADGRSRFIPRLPQGYCGNG 307
Query: 312 IVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTRARPSLTATLLIT 371
IVLT V +GE+V NPLS SVGLV +++VTD +MRS++DYFEV R RPS+ ATLLIT
Sbjct: 308 IVLTGLVTSSGELVGNPLSHSVGLVKRLVELVTDGFMRSAMDYFEVNRTRPSMNATLLIT 367
Query: 372 TWTRLSFHTTDFGWGEPLCSGPVTLPEKEVILFLSNGQDRKSINVLLGLPASAMETFEAL 431
+W++L+ H DFGWGEP+ SGPV LP +EVILFL +G D KSINV LGLP SAME FE L
Sbjct: 368 SWSKLTLHKLDFGWGEPVFSGPVGLPGREVILFLPSGDDMKSINVFLGLPTSAMEVFEEL 427
Query: 432 M 432
M
Sbjct: 428 M 428
>At5g48930 anthranilate N-benzoyltransferase
Length = 433
Score = 263 bits (672), Expect = 1e-70
Identities = 159/443 (35%), Positives = 240/443 (53%), Gaps = 30/443 (6%)
Query: 11 INVRQGEPTRVHPAEETEKGLYFLSNLDQNIAVP---VRTVYCFKSSSRGNEDAAHVIRV 67
IN+R + T V PA ET + SN+D + +P +VY ++ + N V++
Sbjct: 3 INIR--DSTMVRPATETPITNLWNSNVD--LVIPRFHTPSVYFYRPTGASNFFDPQVMKE 58
Query: 68 ALSKILVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDPDSLG 127
ALSK LVP+YPMAG+L +G++ +DCN G +FV A+ I+D GD +L
Sbjct: 59 ALSKALVPFYPMAGRLKRDDDGRIEIDCNGA-GVLFVVADTPSVIDDFGDFAPTL--NLR 115
Query: 128 KLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETARDL 187
+LI + I PL+ +QVT FKCGG ++G+ + H DG S + F+N WS+ AR L
Sbjct: 116 QLIPEVDHSAGIHSFPLLVLQVTFFKCGGASLGVGMQHHAADGFSGLHFINTWSDMARGL 175
Query: 188 DLKTPPFLDRTILKARDPPIIEFKHHEYDEI----VDLSDTKKLYEEEKMLYRSFCFDPK 243
DL PPF+DRT+L+ARDPP F H EY + L +K E F
Sbjct: 176 DLTIPPFIDRTLLRARDPPQPAFHHVEYQPAPSMKIPLDPSKS--GPENTTVSIFKLTRD 233
Query: 244 KLELLKKKATEDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRFVPPI 303
+L LK K+ EDG S++E L+G VWR+ +A + DQ+TKL A DGR+R P +
Sbjct: 234 QLVALKAKSKEDGNTVSYSSYEMLAGHVWRSVGKARGLPNDQETKLYIATDGRSRLRPQL 293
Query: 304 PDRYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTRARPS 363
P YFGN I + AG+++ P ++ G +H + + D+Y+RS++DY E+ +P
Sbjct: 294 PPGYFGNVIFTATPLAVAGDLLSKPTWYAAGQIHDFLVRMDDNYLRSALDYLEM---QPD 350
Query: 364 LTA-----------TLLITTWTRLSFHTTDFGWGEPLCSGPVTLPEKEVILFLSNGQDRK 412
L+A L IT+W RL + DFGWG P+ GP +P + + L + +
Sbjct: 351 LSALVRGAHTYKCPNLGITSWVRLPIYDADFGWGRPIFMGPGGIPYEGLSFVLPSPTNDG 410
Query: 413 SINVLLGLPASAMETFEALMMQV 435
S++V + L + M+ FE + ++
Sbjct: 411 SLSVAIALQSEHMKLFEKFLFEI 433
>At1g03390 hypothetical protein
Length = 461
Score = 243 bits (619), Expect = 2e-64
Identities = 153/430 (35%), Positives = 236/430 (54%), Gaps = 16/430 (3%)
Query: 18 PTRVHPAEETEKGLYFLSNLDQNIAVPVRT--VYCFKSSSRGNEDAAHVIRVALSKILVP 75
P+ PA ++ +LSNLD I V T VY + S++ ++ ALS++LVP
Sbjct: 27 PSSPTPANQSPHHSLYLSNLDDIIGARVFTPSVYFYPSTNNRESFVLKRLQDALSEVLVP 86
Query: 76 YYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDPDSLGKLIYNTPG 135
YYP++G+L GKL V +G + V A + D+ D+GDLT P+P L LI+ PG
Sbjct: 87 YYPLSGRLREVENGKLEVFFGEEQGVLMVSANSSMDLADLGDLTVPNPAWL-PLIFRNPG 145
Query: 136 --ARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETAR--DLDLKT 191
A ILEMPL+ QVT F CGGF++G+ L HC+ DG AM+F+ +W+ TA+ L
Sbjct: 146 EEAYKILEMPLLIAQVTFFTCGGFSLGIRLCHCICDGFGAMQFLGSWAATAKTGKLIADP 205
Query: 192 PPFLDRTILKARDPPIIEFKHHEYDEIVDLSDTKKLYEEEKMLYRSFCFDPKKLELLKKK 251
P DR K R+PP++++ HHEY I + S+ + K L + + + +K
Sbjct: 206 EPVWDRETFKPRNPPMVKYPHHEYLPIEERSNLTNSLWDTKPLQKCYRISKEFQCRVKSI 265
Query: 252 ATEDGVVKKCSTFEALSGFVWRARTEALKMEP-DQKTKLLFAVDGRTRF-VPPIPDRYFG 309
A + CSTF+A++ +WR+ +AL ++P D +L F+V+ RTR + ++G
Sbjct: 266 AQGEDPTLVCSTFDAMAAHIWRSWVKALDVKPLDYNLRLTFSVNVRTRLETLKLRKGFYG 325
Query: 310 NAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTR-ARPSLTATL 368
N + L ++ ++ + LS + LV A V++ Y+RS +DY +V R R L
Sbjct: 326 NVVCLACAMSSVESLINDSLSKTTRLVQDARLRVSEDYLRSMVDYVDVKRPKRLEFGGKL 385
Query: 369 LITTWTRLS-FHTTDFGWGEPLCSGPVTL-PEKEVILFLSNG----QDRKSINVLLGLPA 422
IT WTR + T DFGWG+P+ +GP+ L P +V + L G + +S+ V L LP
Sbjct: 386 TITQWTRFEMYETADFGWGKPVYAGPIDLRPTPQVCVLLPQGGVESGNDQSMVVCLCLPP 445
Query: 423 SAMETFEALM 432
+A+ TF L+
Sbjct: 446 TAVHTFTRLL 455
>At1g27620 putative hypersensitivity-related protein
Length = 442
Score = 218 bits (554), Expect = 7e-57
Identities = 138/429 (32%), Positives = 219/429 (50%), Gaps = 21/429 (4%)
Query: 15 QGEPTRVHPAEETEKGLYFLSNLDQN--IAVPVRTVYCFKSSSRGNEDAAHVIRVALSKI 72
+ +PT + P T +LSNLD + + ++ +Y F+ S + ++ +LS++
Sbjct: 11 ENQPTLITPLSPTPNHSLYLSNLDDHHFLRFSIKYLYLFQKSI-----SPLTLKDSLSRV 65
Query: 73 LVPYYPMAGKLVISTEG-KLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDPDSLGKLIY 131
LV YYP AG++ +S EG KL VDCN EGAVF EA D +D L+ S KL++
Sbjct: 66 LVDYYPFAGRIRVSDEGSKLEVDCNG-EGAVFAEAFMDITCQDFVQLSPKPNKSWRKLLF 124
Query: 132 NTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSE-TARDLDLK 190
A+S L++P + +QVT +CGG + + HC+ DG+ +F++AW+ T L
Sbjct: 125 KVQ-AQSFLDIPPLVIQVTYLRCGGMILCTAINHCLCDGIGTSQFLHAWAHATTSQAHLP 183
Query: 191 TPPFLDRTILKARDPPIIEFKHHEYDEI--VDLSDT---KKLYEEEKMLYRSFCFDPKKL 245
T PF R +L R+PP + H + VD S T K + + + + F+ L
Sbjct: 184 TRPFHSRHVLDPRNPPRVTHSHPGFTRTTTVDKSSTFDISKYLQSQPLAPATLTFNQSHL 243
Query: 246 ELLKKKATEDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRFVPPIPD 305
LKK KC+TFEAL+ WR+ ++L + KLLF+V+ R R P +P
Sbjct: 244 LRLKKTCAPS---LKCTTFEALAANTWRSWAQSLDLPMTMLVKLLFSVNMRKRLTPELPQ 300
Query: 306 RYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTRARPSLT 365
Y+GN VL + K ++V + +V + A +TD Y+RS+ID E + ++
Sbjct: 301 GYYGNGFVLACAESKVQDLVNGNIYHAVKSIQEAKSRITDEYVRSTIDLLEDKTVKTDVS 360
Query: 366 ATLLITTWTRLSFHTTDFGWGEPLCSGPVTLPEKEVILFLSNGQDRKSINVLLGLPASAM 425
+L+I+ W +L D G G+P+ GP+T LFL D +I V + LP +
Sbjct: 361 CSLVISQWAKLGLEELDLGGGKPMYMGPLT--SDIYCLFLPVASDNDAIRVQMSLPEEVV 418
Query: 426 ETFEALMMQ 434
+ E M++
Sbjct: 419 KRLEYCMVK 427
>At2g19070 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 451
Score = 196 bits (499), Expect = 2e-50
Identities = 138/437 (31%), Positives = 215/437 (48%), Gaps = 36/437 (8%)
Query: 23 PAEETEKGLYFLSNLDQ-NIAVPVRTVYCF-KSSSRGNEDAAHVIRVALSKILVPYYPMA 80
PAE T G + L+ DQ + T+Y + K S + +++ +LS++LV +YPMA
Sbjct: 14 PAEPTWSGRFPLAEWDQVGTITHIPTLYFYDKPSESFQGNVVEILKTSLSRVLVHFYPMA 73
Query: 81 GKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDPDS---LGKLIYNTPGAR 137
G+L G+ ++CN EG F+EAE++ + D D + P P+ + ++ Y P
Sbjct: 74 GRLRWLPRGRFELNCNA-EGVEFIEAESEGKLSDFKDFS-PTPEFENLMPQVNYKNP--- 128
Query: 138 SILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETARDLDLKTPPFLDR 197
I +PL QVTKFKCGG ++ +N+ H + DG SA+ ++ W AR L+T PFLDR
Sbjct: 129 -IETIPLFLAQVTKFKCGGISLSVNVSHAIVDGQSALHLISEWGRLARGEPLETVPFLDR 187
Query: 198 TILKARDP--PII---EFKHHEYDEIVDLSDTKKLYEE--EKMLYRSFCFDPKKLELLKK 250
IL A +P P + +F H E+D+ L EE +K + +L+ L+
Sbjct: 188 KILWAGEPLPPFVSPPKFDHKEFDQPPFLIGETDNVEERKKKTIVVMLPLSTSQLQKLRS 247
Query: 251 KAT---EDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRFVPPIPDRY 307
KA K + +E ++G VWR +A P+Q T L +D R+R PP+P Y
Sbjct: 248 KANGSKHSDPAKGFTRYETVTGHVWRCACKARGHSPEQPTALGICIDTRSRMEPPLPRGY 307
Query: 308 FGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDY---------FEVT 358
FGNA + + +GE++ N L F+ L+ AI VT+ Y+ I+Y F+
Sbjct: 308 FGNATLDVVAASTSGELISNELGFAASLISKAIKNVTNEYVMIGIEYLKNQKDLKKFQDL 367
Query: 359 RARPSL------TATLLITTWTRLSFHTTDFGWGEPLCSGPVTLPEKEVILFLSNGQDRK 412
A S L + +W L + DFGWG+ +GP T L L + +
Sbjct: 368 HALGSTEGPFYGNPNLGVVSWLTLPMYGLDFGWGKEFYTGPGTHDFDGDSLILPDQNEDG 427
Query: 413 SINVLLGLPASAMETFE 429
S+ + L + ME F+
Sbjct: 428 SVILATCLQVAHMEAFK 444
>At5g17540 unknown protein
Length = 461
Score = 196 bits (497), Expect = 3e-50
Identities = 135/433 (31%), Positives = 215/433 (49%), Gaps = 16/433 (3%)
Query: 8 NIGINVRQGEPTRVHPAEETEKGLYFLSNLD--QNIAVPVRTVYCFKSSSRGNEDAAHVI 65
++ + + +P V PA+ T + L LS++D + + + T++ ++ + N D VI
Sbjct: 4 SLTFKIYRQKPELVSPAKPTPRELKPLSDIDDQEGLRFHIPTIFFYRHNPTTNSDPVAVI 63
Query: 66 RVALSKILVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIG--DLTKPDP 123
R AL++ LV YYP AG+L KL VDC T EG +F+EA+AD + + D KP
Sbjct: 64 RRALAETLVYYYPFAGRLREGPNRKLAVDC-TGEGVLFIEADADVTLVEFEEKDALKPPF 122
Query: 124 DSLGKLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSET 183
+L++N G+ +L PLM +QVT+ KCGGF + + H M D F+ E
Sbjct: 123 PCFEELLFNVEGSCEMLNTPLMLMQVTRLKCGGFIFAVRINHAMSDAGGLTLFLKTMCEF 182
Query: 184 ARDLDLKT-PPFLDRTILKARDPPIIEFKHHEYDEIVDLSDTKKLYEEEKMLYRSFCFDP 242
R T P +R +L AR + H EYDE+ + T+ + ++ RS F P
Sbjct: 183 VRGYHAPTVAPVWERHLLSARVLLRVTHAHREYDEMPAIG-TELGSRRDNLVGRSLFFGP 241
Query: 243 KKLELLKKKATEDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRFV-P 301
++ + ++ +V + E L+ F+WR RT AL+ + D++ +L+ V+ R++ P
Sbjct: 242 CEMSAI-RRLLPPNLVNSSTNMEMLTSFLWRYRTIALRPDQDKEMRLILIVNARSKLKNP 300
Query: 302 PIPDRYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTRAR 361
P+P Y+GNA ++ A E+ + PL F++ L+ A VT+ YMRS D V + R
Sbjct: 301 PLPRGYYGNAFAFPVAIATANELTKKPLEFALRLIKEAKSSVTEEYMRSLADLM-VIKGR 359
Query: 362 PSLTATLLITTWTRLSFHTTDFG-WGEPLCSGPVT-----LPEKEVILFLSNGQDRKSIN 415
PS ++ F DFG WG+P+ G T LP + I
Sbjct: 360 PSFSSDGAYLVSDVRIFADIDFGIWGKPVYGGIGTAGVEDLPGASFYVSFEKRNGEIGIV 419
Query: 416 VLLGLPASAMETF 428
V + LP AM+ F
Sbjct: 420 VPVCLPEKAMQRF 432
>At5g57840 N-hydroxycinnamoyl/benzoyltransferase
Length = 443
Score = 190 bits (483), Expect = 1e-48
Identities = 136/398 (34%), Positives = 195/398 (48%), Gaps = 29/398 (7%)
Query: 11 INVRQGEPTRVHPAEETEKGLYFLSNLDQ-NIAVPVRTVYCFKSSSRGNEDAAHVIRVAL 69
+ +R + T V PAEET +LSNLD + + + T+Y +K S + + AL
Sbjct: 1 MKIRVKQATIVKPAEETPTHNLWLSNLDLIQVRLHMGTLYFYKPCSSSDRPNTQSLIEAL 60
Query: 70 SKILVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDPDSLGKL 129
SK+LV +YP AG+L +T G+L V CN EG +FVEAE+D ++DIG LT+ L +L
Sbjct: 61 SKVLVFFYPAAGRLQKNTNGRLEVQCNG-EGVLFVEAESDSTVQDIGLLTQSL--DLSQL 117
Query: 130 IYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETARDLDL 189
+ A I PL+ QVT FKCG +G ++ H + S + AWS TAR L +
Sbjct: 118 VPTVDYAGDISSYPLLLFQVTYFKCGTICVGSSIHHTFGEATSLGYIMEAWSLTARGLLV 177
Query: 190 KTPPFLDRTILKARDPPIIEFKHHEYDEIVDLSDTKKLYEEEKMLYRS--------FCFD 241
K PFLDRT+L AR+PP F H EY S + + + YRS
Sbjct: 178 KLTPFLDRTVLHARNPPSSVFPHTEYQ-----SPPFHNHPMKSLAYRSNPESDSAIATLK 232
Query: 242 PKKLELLKKKATEDGVVKKCSTFEALSGFVWRARTEALK-MEPDQKTKLLFAVDGRTRFV 300
+L+L KA + K ST+E L WR + A + + + T+L +DGR R
Sbjct: 233 LTRLQLKALKARAEIADSKFSTYEVLVAHTWRCASFANEDLSEEHSTRLHIIIDGRPRLQ 292
Query: 301 PPIPDRYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFE---- 356
P +P Y GN + V G S +V VH I + + Y+RS+IDY E
Sbjct: 293 PKLPQGYIGNTLFHARPVSPLGAFHRESFSETVERVHREIRKMDNEYLRSAIDYLERHPD 352
Query: 357 VTRARP-------SLTATLLITTWTRLSFHTTDFGWGE 387
+ + P S A I + + + + DFGWG+
Sbjct: 353 LDQLVPGEGNTIFSCAANFCIVSLIKQTAYEMDFGWGK 390
>At3g03480 putative hypersensitivity-related gene
Length = 454
Score = 189 bits (479), Expect = 3e-48
Identities = 135/440 (30%), Positives = 217/440 (48%), Gaps = 25/440 (5%)
Query: 7 SNIGINVRQGEPTRVHPAEETEKGLYFLSNLD--QNIAVPVRTVYCFKSSSRGNEDAAHV 64
+ + V + + V PA+ T + L LS++D Q + + ++ ++ + + D V
Sbjct: 13 TGLSFKVHRQQRELVTPAKPTPRELKPLSDIDDQQGLRFQIPVIFFYRPNLSSDLDPVQV 72
Query: 65 IRVALSKILVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEAD---CDIEDIGDLTKP 121
I+ AL+ LV YYP AG+L + KL VDC T EG +F+EAEAD ++E+ L P
Sbjct: 73 IKKALADALVYYYPFAGRLRELSNRKLAVDC-TGEGVLFIEAEADVALAELEEADALLPP 131
Query: 122 DPDSLGKLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWS 181
P L +L+++ G+ +L PL+ VQVT+ KC GF L H M DG F+ +
Sbjct: 132 FP-FLEELLFDVEGSSDVLNTPLLLVQVTRLKCCGFIFALRFNHTMTDGAGLSLFLKSLC 190
Query: 182 ETARDLDL-KTPPFLDRTILK-ARDPPIIEFKHHEYDEIVDLSDTKKLYEEEKMLYRSFC 239
E A L PP +R +L + + H EYD+ V + + ++ RSF
Sbjct: 191 ELACGLHAPSVPPVWNRHLLTVSASEARVTHTHREYDDQVGID---VVATGHPLVSRSFF 247
Query: 240 FDPKKLELLKKKATEDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRF 299
F +++ ++K D ++FEALS F+WR RT AL +P+ + +L ++ R++
Sbjct: 248 FRAEEISAIRKLLPPD---LHNTSFEALSSFLWRCRTIALNPDPNTEMRLTCIINSRSKL 304
Query: 300 -VPPIPDRYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVT 358
PP+ Y+GN V+ ++ A +++E PL F++ L+ VT+ Y+R S+ T
Sbjct: 305 RNPPLEPGYYGNVFVIPAAIATARDLIEKPLEFALRLIQETKSSVTEDYVR-SVTALMAT 363
Query: 359 RARPSLTAT--LLITTWTRLSFHTTDFG-WGEPLCSGP-----VTLPEKEVILFLSNGQD 410
R RP A+ +I+ DFG WG+P+ G P + N +
Sbjct: 364 RGRPMFVASGNYIISDLRHFDLGKIDFGPWGKPVYGGTAKAGIALFPGVSFYVPFKNKKG 423
Query: 411 RKSINVLLGLPASAMETFEA 430
V + LP AMETF A
Sbjct: 424 ETGTVVAISLPVRAMETFVA 443
>At2g40230 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 433
Score = 174 bits (440), Expect = 1e-43
Identities = 125/434 (28%), Positives = 219/434 (49%), Gaps = 16/434 (3%)
Query: 9 IGINVRQGEPTRVHPAEETEKGLYFLSNLDQNIAVPVRTVYC--FKSSSRGNEDAAHVIR 66
+G V E T + P+++T + LS LD + + Y + S + ++
Sbjct: 1 MGSLVHVKEATVITPSDQTPSSVLSLSALDSQLFLRFTIEYLLVYPPVSDPEYSLSGRLK 60
Query: 67 VALSKILVPYYPMAGKLVISTEGK--LIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDPD 124
ALS+ LVPY+P +G++ +G L V+C +GA+F+EA +D I D KP
Sbjct: 61 SALSRALVPYFPFSGRVREKPDGGGGLEVNCRG-QGALFLEAVSD--ILTCLDFQKPPRH 117
Query: 125 SLG-KLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSET 183
+ + + + P + VQ+T + GG + + + HC+ DG+ + EF+ ++E
Sbjct: 118 VTSWRKLLSLHVIDVLAGAPPLVVQLTWLRDGGAALAVGVNHCVSDGIGSAEFLTLFAEL 177
Query: 184 ARD----LDLKTPPFLDRTILKARDPPIIEFKHHEYDEIVDLSDTKKLYEEEKMLYRSFC 239
++D +LK DR +L P H E++ + DL + E+++ S
Sbjct: 178 SKDSLSQTELKRKHLWDRQLLMP-SPIRDSLSHPEFNRVPDLCGFVNRFNAERLVPTSVV 236
Query: 240 FDPKKLELLKKKATEDGVVK-KCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTR 298
F+ +KL LKK A+ G K ++FE LS VWR+ +L + +Q KLLF+V+ R R
Sbjct: 237 FERQKLNELKKLASRLGEFNSKHTSFEVLSAHVWRSWARSLNLPSNQVLKLLFSVNIRDR 296
Query: 299 FVPPIPDRYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVT 358
P +P ++GNA V+ + ++ E LS++ LV A + V D Y+RS ++
Sbjct: 297 VKPSLPSGFYGNAFVVGCAQTTVKDLTEKGLSYATMLVKQAKERVGDEYVRSVVEAVSKE 356
Query: 359 RARPSLTATLLITTWTRLSFHTTDFGWGEPLCSGPVTLPEKEVILFLSNGQDRKSINVLL 418
RA P L+++ W+RL DFG G+P+ G V ++L + D ++ V++
Sbjct: 357 RASPDSVGVLILSQWSRLGLEKLDFGLGKPVHVGSVCCDRYCLLLPIPEQND--AVKVMV 414
Query: 419 GLPASAMETFEALM 432
+P+S+++T+E L+
Sbjct: 415 AVPSSSVDTYENLV 428
>At1g28680 anthranilate N-hydroxycinnamoyl/benzoyltransferase like
protein
Length = 451
Score = 164 bits (414), Expect = 1e-40
Identities = 131/456 (28%), Positives = 212/456 (45%), Gaps = 40/456 (8%)
Query: 1 MANADMSNIGINVRQGEPTRVHPAEETEKGLYFLSNLDQNIAVPVR--TVYCFKSSSRGN 58
MA ++++I + +P+ + + L L N D N+ V R VY SS+
Sbjct: 1 MATLEITDIALV----QPSHQPLSNDQTLSLSHLDN-DNNLHVSFRYLRVYSSSSSTVAG 55
Query: 59 EDAAHVIRVALSKILVPYYPMAGKLVIS-TEGKLIVDCNTMEGAVFVEAEADCDIEDIGD 117
E + V+ +L+ LV YYP+AG L S ++ + + C+ + V A +C +E +G
Sbjct: 56 ESPSAVVSASLATALVHYYPLAGSLRRSASDNRFELLCSAGQSVPLVNATVNCTLESVGY 115
Query: 118 LTKPDPDSLGKLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFV 177
L PDP + +L+ + ++ ++ QVT F+CGG+ +G ++ H + DGL A F
Sbjct: 116 LDGPDPGFVERLVPDPTREEGMVNPCIL--QVTMFQCGGWVLGASIHHAICDGLGASLFF 173
Query: 178 NAWSETARDLD-LKTPPFLDRT-ILKARDPPIIEFKHHEYDEIVDLSDTKKLYEEEKMLY 235
NA +E AR + P DR +L R+ P + ++ + L Y +
Sbjct: 174 NAMAELARGATKISIEPVWDRERLLGPREKPWVGAPVRDF---LSLDKDFDPYGQAIGDV 230
Query: 236 RSFCF-------DPKKLELLKKKATEDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTK 288
+ CF D K +LL+K +TFEAL ++WRA+ A K E + K
Sbjct: 231 KRDCFFVTDDSLDQLKAQLLEKSGLN------FTTFEALGAYIWRAKVRAAKTEEKENVK 284
Query: 289 LLFAVDGRTRFVPPIPDRYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYM 348
+++++ R PP+P Y+GN V ++ KAGE++E P+ + L+ + +D Y+
Sbjct: 285 FVYSINIRRLMNPPLPKGYWGNGCVPMYAQIKAGELIEQPIWKTAELIKQSKSNTSDEYV 344
Query: 349 RSSIDYFEVTR--ARPSLTATLLITTWTRLSFHTTDFGWGEPLCSGPVT---LPEKEVIL 403
RS ID+ E+ + T T W L T DFGWG P+ P++ L E
Sbjct: 345 RSFIDFQELHHKDGINAGTGVTGFTDWRYLGHSTIDFGWGGPVTVLPLSNKLLGSMEPCF 404
Query: 404 FLSNGQDR-------KSINVLLGLPASAMETFEALM 432
FL D VL+ L SAM F+ M
Sbjct: 405 FLPYSTDAAAGSKKDSGFKVLVNLRESAMPEFKEAM 440
>At4g31910 unknown protein
Length = 458
Score = 161 bits (407), Expect = 7e-40
Identities = 127/423 (30%), Positives = 205/423 (48%), Gaps = 35/423 (8%)
Query: 28 EKGLYFLSNLDQNIAVPVRTVYCFKSSSRGNEDAAHV-IRVALSKILVPYYPMAGKLVIS 86
+K L LSNLD+ + + +V+ +K+++ + D+ +++ L + + +YP AG+L +
Sbjct: 26 KKKLITLSNLDRQCPLLMYSVFFYKNTTTRDFDSVFSNLKLGLEETMSVWYPAAGRLGLD 85
Query: 87 TEG-KLIVDCNTMEGAVFVEAEAD-CDIEDIGDLTKPDPDSLGKLIYNTPGARSILEMPL 144
G KL + CN GAV VEA A + ++GDLT+ + + L+Y MPL
Sbjct: 86 GGGCKLNIRCND-GGAVMVEAVATGVKLSELGDLTQYN-EFYENLVYKPSLDGDFSVMPL 143
Query: 145 MTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSE------------TARDLDLKTP 192
+ QVT+F CGG+++G+ H + DG+SA EF++AW+ T + D+
Sbjct: 144 VVAQVTRFACGGYSIGIGTSHSLFDGISAYEFIHAWASNSHIHNKSNSKITNKKEDVVIK 203
Query: 193 PFLDRTILKARDPPIIEFKHHEYDEIVDL-SDTKKLYEEEK---------MLYRSFCFDP 242
P DR L + E + L Y+E+ + ++F +
Sbjct: 204 PVHDRRNLLVNRDAVRETNAAAICHLYQLIKQAMMTYQEQNRNLELPDSGFVIKTFELNG 263
Query: 243 KKLELLKKKATEDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRFVPP 302
+E +KKK+ E + CS+FE L+ +W+ART AL + D L FAVD R R P
Sbjct: 264 DAIESMKKKSLEGFM---CSSFEFLAAHLWKARTRALGLRRDAMVCLQFAVDIRKRTETP 320
Query: 303 IPDRYFGNAIVLTHSVCKAGEIVEN-PLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTR-- 359
+P+ + GNA VL A E++E L V + A + Y+ S ++ +
Sbjct: 321 LPEGFSGNAYVLASVASTARELLEELTLESIVNKIREAKKSIDQGYINSYMEALGGSNDG 380
Query: 360 ARPSLTATLLITTWTRLSFHTTDFG-WGEPL-CSGPVTLPEKEVILFLSNGQDRKSINVL 417
P L LI+ WT++ FH FG GEP P+ P +V F+ N +D K + V
Sbjct: 381 NLPPLKELTLISDWTKMPFHNVGFGNGGEPADYMAPLCPPVPQVAYFMKNPKDAKGVLVR 440
Query: 418 LGL 420
+GL
Sbjct: 441 IGL 443
>At1g65450 unknown protein
Length = 286
Score = 145 bits (365), Expect = 6e-35
Identities = 92/282 (32%), Positives = 150/282 (52%), Gaps = 15/282 (5%)
Query: 159 MGLNLIHCMKDGLSAMEFVNAWSETARDLDLKTPPFLDRTILKARDPPIIEFKHHEYDEI 218
MG++ H DGLS F+ + + L TPP DRT+LKARDPP + F HHE +
Sbjct: 1 MGISTNHTTFDGLSFKTFLENLASLLHEKPLSTPPCNDRTLLKARDPPSVAFPHHELVKF 60
Query: 219 VDLSDTKKLYE--EEKMLYRSFCFDPKKLELLKKKATE--DGVVKKCSTFEALSGFVWRA 274
D +T ++E E + ++ F ++++ LK++A+E +G V + + F ++ VWR
Sbjct: 61 QD-CETTTVFEATSEHLDFKIFKLSSEQIKKLKERASETSNGNV-RVTGFNVVTALVWRC 118
Query: 275 RTEALKMEPDQKTKL------LFAVDGRTRFVPPIPDRYFGNAIVLTHSVCKAGEIVENP 328
+ ++ E ++T L L+AVD R R P +P Y GNA++ ++ K ++E P
Sbjct: 119 KALSVAAEEGEETNLERESTILYAVDIRGRLNPELPPSYTGNAVLTAYAKEKCKALLEEP 178
Query: 329 LSFSVGLVHSAIDIVTDSYMRSSIDYFEVTRARPSLTATLLITTWTRLSFHTTDFGWGEP 388
V +V +TD Y RS+ID+ E+ + P +L+++W +L F ++ WG+P
Sbjct: 179 FGRIVEMVGEGSKRITDEYARSAIDWGELYKGFPH--GEVLVSSWWKLGFAEVEYPWGKP 236
Query: 389 LCSGPVTLPEKE-VILFLSNGQDRKSINVLLGLPASAMETFE 429
S PV K+ V+LF D K + VL LP+ M F+
Sbjct: 237 KYSCPVVYHRKDIVLLFPDIDGDSKGVYVLAALPSKEMSKFQ 278
>At3g62160 unknown protein
Length = 428
Score = 141 bits (355), Expect = 8e-34
Identities = 112/389 (28%), Positives = 175/389 (44%), Gaps = 38/389 (9%)
Query: 21 VHPAEETEKGLYFLSNLDQNIAVPV-----RTVYCFKSSSRGNEDAAHVIRVALSKILVP 75
+ E T + LS +D +PV RT++ F DAA I+ AL+K LV
Sbjct: 13 IQACETTPSVVLDLSLIDN---IPVLRCFARTIHVFTHGP----DAARAIQEALAKALVS 65
Query: 76 YYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTK----PDPDSLGKLIY 131
YYP++G+L +GKL +DC G FV+A D +E +G P D L I
Sbjct: 66 YYPLSGRLKELNQGKLQIDCTGKTGIWFVDAVTDVKLESVGYFDNLMEIPYDDLLPDQIP 125
Query: 132 NTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETA-RDLDLK 190
A PL+ +QVT+F CGGF MGL H + DGL A +F+ A E A +L
Sbjct: 126 KDDDAE-----PLVQMQVTQFSCGGFVMGLRFCHAICDGLGAAQFLTAVGEIACGQTNLG 180
Query: 191 TPPFLDRTILKARDPPIIEFKHHEYDEIVDLS---DTKKLYEEEKMLYRSFCFDPKKLEL 247
P R + + H ++++ + + E K+ + SF +E
Sbjct: 181 VSPVWHRDFIP---------QQHRANDVISPAIPPPFPPAFPEYKLEHLSFDIPSDLIER 231
Query: 248 LKKK-ATEDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRFVPPIPDR 306
K++ G + CS FE ++ W+ RTEA+ + D + KL+F + R PP+P
Sbjct: 232 FKREFRASTGEI--CSAFEVIAACFWKLRTEAINLREDTEVKLVFFANCRHLVNPPLPIG 289
Query: 307 YFGNAIVLTHSVCKAGEIVENPLSFS-VGLVHSAIDIVTDSYMRSSIDYFEVTRARPSLT 365
++GN ++ EI + F V L+ A + + + + + A
Sbjct: 290 FYGNCFFPVKISAESHEISKEKSVFDVVKLIKQAKTKLPEEFEKFVSGDGDDPFAPAVGY 349
Query: 366 ATLLITTWTRLSFHTTDFGWGEPLCSGPV 394
TL ++ W +L F+ D+GWG PL P+
Sbjct: 350 NTLFLSEWGKLGFNQVDYGWGPPLHVAPI 378
>At5g07080 unknown protein
Length = 450
Score = 139 bits (349), Expect = 4e-33
Identities = 135/443 (30%), Positives = 203/443 (45%), Gaps = 40/443 (9%)
Query: 13 VRQGEPTRVHPAEETEKGLYFLSNLDQN--IAVPVRTVYCFKSSSRGNE--DAAHVIRVA 68
V++ + V P++ T LS LD + + +T+Y + S + D A + + A
Sbjct: 14 VKKSQVVIVKPSKATPDVSLSLSTLDNDPYLETLAKTIYVYAPPSPNDHVHDPASLFQQA 73
Query: 69 LSKILVPYYPMAGKLVI-STEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDPDSLG 127
LS LV YYP+AGKL S + +L + C+ EG FV A ADC + + L D D L
Sbjct: 74 LSDALVYYYPLAGKLHRGSHDHRLELRCSPAEGVPFVRATADCTLSSLNYLKDMDTD-LY 132
Query: 128 KLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETARDL 187
+L+ A S PL +Q+T F CGG T+ L H + DG A +F +E A
Sbjct: 133 QLVPCDVAAPSGDYNPL-ALQITLFACGGLTLATALSHSLCDGFGASQFFKTLTELAAG- 190
Query: 188 DLKTPPFL----DRTILKARDPPIIE-FKHHEYDEIVDLSD------TKKLYEEEKMLYR 236
KT P + DR L + + + + + + ++VD + T M+
Sbjct: 191 --KTQPSIIPVWDRHRLTSNNFSLNDQVEEGQAPKLVDFGEACSSAATSPYTPSNDMVCE 248
Query: 237 SFCFDPKKLELLKKKATEDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGR 296
+ + LK+K GVV +T E L+ VWRAR ALK+ PD + AV R
Sbjct: 249 ILNVTSEDITQLKEKVA--GVV---TTLEILAAHVWRARCRALKLSPDGTSLFGMAVGIR 303
Query: 297 TRFVPPIPDRYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDS-YMRSSIDYF 355
PP+P+ Y+GNA V + KAGE+ +PLS V L+ A + Y+ +
Sbjct: 304 RIVEPPLPEGYYGNAFVKANVAMKAGELSNSPLSHVVQLIKEAKKAAQEKRYVLEQLRET 363
Query: 356 EVT-----RARPSLTATLLITTWTRLS-FHTTDFGWGEPLCSGPVT---LPEKEVILFLS 406
E T A +L+T W +L DFG+G + P+ LP ++ +FL
Sbjct: 364 EKTLKMNVACEGGKGAFMLLTDWRQLGLLDEIDFGYGGSVNIIPLVPKYLP--DICIFLP 421
Query: 407 NGQDRKSINVLLGLPASAMETFE 429
Q + VL+ LP S M F+
Sbjct: 422 RKQG--GVRVLVTLPKSVMVNFK 442
>At5g42830 N-hydroxycinnamoyl/benzoyltransferase-like protein
Length = 450
Score = 134 bits (336), Expect = 1e-31
Identities = 94/302 (31%), Positives = 144/302 (47%), Gaps = 21/302 (6%)
Query: 65 IRVALSKILVPYYPMAGKLVISTEGK-----LIVDCNTMEGAVFVEAEADCDIEDIGDLT 119
++ +L+ LV +YP+AG+L T K + VDCN GA F+ A +D I+DI
Sbjct: 64 LKDSLAVTLVHFYPLAGRLSSLTTEKPKSYSVFVDCNDSPGAGFIYATSDLCIKDIVGAK 123
Query: 120 KPDPDSLGKLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNA 179
++ M L++VQVT+ G F +GL++ H M DG + +F A
Sbjct: 124 YVPSIVQSFFDHHKAVNHDGHTMSLLSVQVTELVDGIF-IGLSMNHAMGDGTAFWKFFTA 182
Query: 180 WSETAR--------DLDLKTPPFLDRTILKARDPPIIEFKHHEYDEIVDLSDTKKLYEEE 231
WSE + DL LK PP L R I + P + + DE + + YE
Sbjct: 183 WSEIFQGQESNQNDDLCLKNPPVLKRYIPEGYGP-LFSLPYSHPDEFI------RTYESP 235
Query: 232 KMLYRSFCFDPKKLELLKKKATEDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLF 291
+ R FCF + + +LK + + S+F++L+ +WR T A ++ D++T
Sbjct: 236 ILKERMFCFSSETIRMLKTRVNQICGTTSISSFQSLTAVIWRCITRARRLPLDRETSCRV 295
Query: 292 AVDGRTRFVPPIPDRYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSS 351
A D R R PP+ YFGN + + KAGE++EN L F+ VH A+ T +
Sbjct: 296 AADNRGRMYPPLHKDYFGNCLSALRTAAKAGELLENDLGFAALKVHQAVAEHTSEKVSQM 355
Query: 352 ID 353
ID
Sbjct: 356 ID 357
>At1g78990
Length = 455
Score = 123 bits (309), Expect = 2e-28
Identities = 101/374 (27%), Positives = 178/374 (47%), Gaps = 29/374 (7%)
Query: 34 LSNLDQNIA-VPVRTVYCFKSSSRGNEDAAH-VIRVALSKILVPYYPMAGKLVISTEGKL 91
LSNLD + + V +C+K A+ ++ AL++ LV YY AG+LV + G+
Sbjct: 34 LSNLDLLLPPLNVHVCFCYKKPLHFTNTVAYETLKTALAETLVSYYAFAGELVTNPTGEP 93
Query: 92 IVDCNTMEGAVFVEAEADCDIEDIGDLTKPDPDSLGKLIYNTPGARSILEMPLMTVQVTK 151
+ CN G FVEA AD ++ ++ +L PD +S+ KL+ I + ++ +QVT+
Sbjct: 94 EILCNN-RGVDFVEAGADVELREL-NLYDPD-ESIAKLV-------PIKKHGVIAIQVTQ 143
Query: 152 FKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETAR-DLDLKTPPFLDRTILKARDPPIIEF 210
KCG +G H + D S F+ +W+E +R D+ + P R++L R P +++
Sbjct: 144 LKCGSIVVGCTFDHRVADAYSMNMFLLSWAEISRSDVPISCVPSFRRSLLNPRRPLVMDP 203
Query: 211 KHHE-YDEIVDLSDTKKLYEEEKML-YRSFCFDPKKLELLKKKATEDGVVKKCSTFEALS 268
+ Y + L ++ E +L R + L+ L+ A+ K+ + E+ S
Sbjct: 204 SIDQIYMPVTSLPPPQETTNPENLLASRIYYIKANALQELQTLASSSKNGKR-TKLESFS 262
Query: 269 GFVWR--ARTEALKMEPDQKTKLLFAVDGRTRFVPPIPDRYFGNAIVLTHSVCKAGEIVE 326
F+W+ A A P + +KL VDGR R + + YFGN + + + +++
Sbjct: 263 AFLWKLVAEHAAKDPVPIKTSKLGIVVDGRRRLMEKENNTYFGNVLSVPFGGQRIDDLIS 322
Query: 327 NPLSFSVGLVHSAI-DIVTDSYMRSSIDYFEVTRARPSLT----------ATLLITTWTR 375
PLS+ VH + VT + + ID+ E R P+++ ++++
Sbjct: 323 KPLSWVTEEVHRFLKKSVTKEHFLNLIDWVETCRPTPAVSRIYSVGSDDGPAFVVSSGRS 382
Query: 376 LSFHTTDFGWGEPL 389
+ DFGWG P+
Sbjct: 383 FPVNQVDFGWGSPV 396
>At1g32910 hypothetical protein
Length = 464
Score = 116 bits (291), Expect = 2e-26
Identities = 96/377 (25%), Positives = 177/377 (46%), Gaps = 32/377 (8%)
Query: 34 LSNLDQNIA-VPVRTVYCFKSSSRGNEDAAHVIRVALSKILVPYYPMAGKLVISTEGKLI 92
LSNLD + V + +C+K R A ++ +L++ LV YY AG+LV ++ G+
Sbjct: 34 LSNLDLLLPPVQISVCFCYKKP-RHFLSVAETLKASLAEALVSYYAFAGELVKNSSGEPE 92
Query: 93 VDCNTMEGAVFVEAEADCDIEDIGDLTKPDPDSLGKLIYNTPGARSILEMPLMTVQVTKF 152
+ CN G F+EA AD ++ ++ +L PD +S+ KL+ ++ +QVT+
Sbjct: 93 ILCNN-RGVDFLEAVADVELREL-NLHDPD-ESIAKLVPKKKHG-------VIAIQVTQL 142
Query: 153 KCGGFTMGLNLIHCMKDGLSAMEFVNAWSETAR-DLDLKTPPFLDRTILKARDPPIIEFK 211
KCG +G H + D S F+ +W+E +R + + + P R+IL R P II+
Sbjct: 143 KCGSIVVGCTFDHRIADAFSMNMFLVSWAEISRFNAPISSVPSFRRSILNPRRPLIIDSS 202
Query: 212 HHE-YDEIVDLSDTKKLYEEEKMLYRSFCFDPKKLELLKKKATEDGVVKKC-----STFE 265
+ Y + L ++ + + S + K+ L + G K + E
Sbjct: 203 IDKMYMPVTSLPLPQETTNDLDNILTSRIYYIKENALEDLQTLASGSSPKTGYGQRTKLE 262
Query: 266 ALSGFVWR--ARTEALKMEPDQKTKLLFAVDGRTRFVPPIPDRYFGNAIVLTHSVCKAGE 323
+ S F+W+ A+ + ++ +K+ VDGR R + + YFGN + + +
Sbjct: 263 SFSAFLWKLVAKHTGRDLVSNKNSKMGIVVDGRRRLMEKEDNTYFGNVLSIPFGGQSIDD 322
Query: 324 IVENPLSFSVGLVHSAI-DIVTDSYMRSSIDYFEVTRARPSLT----------ATLLITT 372
+++ PLS+ VH + + VT + + ID+ E+ R P+++ ++++
Sbjct: 323 LIDKPLSWVTNEVHRFLEEAVTKEHFLNLIDWVEIHRPIPAVSRIYSTGTDDGPAFVVSS 382
Query: 373 WTRLSFHTTDFGWGEPL 389
+ DFGWG P+
Sbjct: 383 GRSFPVNKVDFGWGLPV 399
>At5g07870 N-hydroxycinnamoyl/benzoyltransferase - like protein
Length = 464
Score = 115 bits (287), Expect = 6e-26
Identities = 90/310 (29%), Positives = 140/310 (45%), Gaps = 33/310 (10%)
Query: 65 IRVALSKILVPYYPMAGKLVI-----STEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLT 119
++ +L+ LV +YP+AG+L S + VDCN GA F+ AE+D + DI T
Sbjct: 72 LKDSLAIALVHFYPLAGRLSTLKTDNSRSHSVFVDCNNSPGARFIHAESDLSVSDILGST 131
Query: 120 KPDPDSLGKLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNA 179
++ R M L++++VT+ G F +GL++ H + DG S +F N+
Sbjct: 132 YVPLVVQSLFDHHKALNRDGYTMSLLSIKVTELVDGVF-IGLSMNHSLGDGSSFWQFFNS 190
Query: 180 WSETARD----------------LDLKTPPFLDRTILKARDPPIIEFKHHEYDEIVDLSD 223
SE L LK PP I++ P+ E +E + S+
Sbjct: 191 LSEIFNSQEETIGNNNNNNNNALLCLKNPP-----IIREATGPMYSLPFSEPNESLSQSE 245
Query: 224 TKKLYEEEKMLYRSFCFDPKKLELLKKKATEDGVVKKCSTFEALSGFVWRARTEALKMEP 283
L E R F F + + LK KA ++ S+F+AL+ +WR+ T A K+
Sbjct: 246 PPVLKE------RMFHFSSETVRSLKSKANQECGTTMISSFQALTALIWRSITRARKLPN 299
Query: 284 DQKTKLLFAVDGRTRFVPPIPDRYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIV 343
DQ+T FA R+R PP+P FG I L + K G ++EN + +H A+
Sbjct: 300 DQETTCRFAAGNRSRMNPPLPTNQFGVYISLVKTTTKIGNLLENEFGWIALKLHQAVTEH 359
Query: 344 TDSYMRSSID 353
T + ID
Sbjct: 360 TGEKISYEID 369
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,873,461
Number of Sequences: 26719
Number of extensions: 425867
Number of successful extensions: 1162
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 921
Number of HSP's gapped (non-prelim): 86
length of query: 435
length of database: 11,318,596
effective HSP length: 102
effective length of query: 333
effective length of database: 8,593,258
effective search space: 2861554914
effective search space used: 2861554914
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0063.5


