

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0063.10
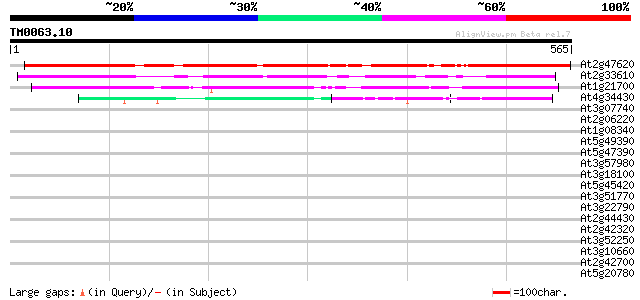
(565 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g47620 SWI/SNF family like transcription activator 491 e-139
At2g33610 putative SWI/SNF complex subunit SW13 199 3e-51
At1g21700 putative transcriptional regulatory protein 170 2e-42
At4g34430 putative protein 91 1e-18
At3g07740 unknown protein 39 0.008
At2g06220 hypothetical protein 35 0.085
At1g08340 unknown protein 35 0.085
At5g49390 putative protein 34 0.19
At5g47390 Myb-related transcription activator-like 34 0.19
At3g57980 unknown protein 34 0.25
At3g18100 putative transcription factor (MYB4R1) 34 0.25
At5g45420 unknown protein 33 0.32
At3g51770 putative protein 33 0.32
At3g22790 unknown protein 33 0.32
At2g44430 unknown protein 33 0.32
At2g42320 unknown protein 33 0.32
At3g52250 putative protein 33 0.42
At3g10660 calmodulin-domain protein kinase CDPK isoform 2 33 0.55
At2g42700 unknown protein 33 0.55
At5g20780 putative protein 32 0.72
>At2g47620 SWI/SNF family like transcription activator
Length = 512
Score = 491 bits (1263), Expect = e-139
Identities = 273/550 (49%), Positives = 363/550 (65%), Gaps = 43/550 (7%)
Query: 16 DSESELELYTIPSSSKWFAWDEIHETERTALKEFFDGTSISRTPKVYKEYRDFVINKYRE 75
D +E+ELYTIP+ S WF WD+IHE ER EFF +SI+RTPKVYKEYRDF+INK+RE
Sbjct: 5 DPSAEIELYTIPAQSSWFLWDDIHEIERREFAEFFTESSITRTPKVYKEYRDFIINKFRE 64
Query: 76 DPSRKLTFTEVRKSLVGDVTLLRKVFLFLESCGLINYGVGEAEKEEEREDEEEGEGERCK 135
D R+LTFT VRK LVGDV LL+KVFLFLE GLIN+ + + +
Sbjct: 65 DTCRRLTFTSVRKFLVGDVNLLQKVFLFLEKWGLINFSSSLKKNDHLLSVD--------N 116
Query: 136 VRVEEGAPNGIRVVATPNSLKPLSAPRNAKSGGGGGGSVSGGAGVKLPPLASFADVYGDL 195
++E+G P GIRV ATPNSL+P++AP + G+K+PPL S++DV+ DL
Sbjct: 117 AKIEQGTPAGIRVTATPNSLRPITAPPLVEE--------RVETGIKVPPLTSYSDVFSDL 168
Query: 196 MSAKE-LNCGICGDKCGFEHYRSTKDNFTICMKCFKNGNYGEKRSVEDFILNESNENSVK 254
L C CG++C Y+ K IC KCFKNGNYGE + +DF L ++
Sbjct: 169 KKPDHVLVCAHCGERCDSPFYQHNKGIVNICEKCFKNGNYGENNTADDFKLIGNS----- 223
Query: 255 QSTVWTEGETLLLLESVLKHGDDWELVAQSVQTKTKLDCISKLIELPFGELMLGSAHRND 314
+ VWTE E LLLLESVLKHGDDWEL++QSV TK++LDCISKLIELPFGE ++GSA
Sbjct: 224 AAAVWTEEEILLLLESVLKHGDDWELISQSVSTKSRLDCISKLIELPFGEFLMGSASGRL 283
Query: 315 NINIANGIVNNAIQVQSSSSDHQETSKTQNQTPELTSEKEPNGDAVNESPSKRQRVAALS 374
N +I N QVQ+ +H+ET +T+ + + +E EP P+KR+RVA +S
Sbjct: 284 NPSILTE-DENTEQVQTDGQEHEET-ETREEKEDRVNEDEP--------PAKRKRVALIS 333
Query: 375 ESSSSLMKQVGLISTSVDPHITAAAADAAITALCDENLCPREIFDVEEDYASSANSLISD 434
E SSLMKQV +++ V P + AAA AA+ ALCDE CP+EIFD +DY++
Sbjct: 334 EGDSSLMKQVAAMASKVGPSVATAAAKAALAALCDEASCPKEIFDT-DDYSNFT------ 386
Query: 435 SERAHEVEGLEMDRSTQAEKDDRGPKDDIPLPLRLRAGIATALGAAAARAKLMADQEDRE 494
+RA+ + +M+ Q EKD GP+ +P+ LR+RA +ATALGAAAA+AK++ADQE+RE
Sbjct: 387 VDRANGEKDTDMEEQ-QEEKD--GPQ-GLPVALRIRASVATALGAAAAQAKILADQEERE 442
Query: 495 IEHLVATIIEAQIGKLQHKVKHFDELELLMKKEHAEIEELKDSILTERIDVLQKTFRSGI 554
+E L AT+IE Q+ KLQ K+K D+LE +M +E IE +K++I+ ER+ VLQ FRSGI
Sbjct: 443 MEQLAATVIEQQLKKLQSKLKFLDDLESIMDEEEKVIEGVKETIIQERVSVLQCAFRSGI 502
Query: 555 TRWKDYSYVK 564
T+ D++YVK
Sbjct: 503 TKRWDHTYVK 512
>At2g33610 putative SWI/SNF complex subunit SW13
Length = 469
Score = 199 bits (507), Expect = 3e-51
Identities = 141/541 (26%), Positives = 245/541 (45%), Gaps = 116/541 (21%)
Query: 9 SNPGRTDDSESELELYTIPSSSKWFAWDEIHETERTALKEFFDGTSISRTPKVYKEYRDF 68
S + S S+++ +PS S WF+W +I++ E +L EFFD S S+ PK Y R+
Sbjct: 33 SKSAQLPSSSSDIDNIHVPSYSSWFSWTDINDCEVRSLPEFFDSRSSSKNPKFYLYLRNS 92
Query: 69 VINKYREDPSRKLTFTEVRKSLVGDVTLLRKVFLFLESCGLINYGVGEAEKEEEREDEEE 128
+I +YR+D RK++FT+VR++LV DV +R+VF FL+S GLINY + K + E++E
Sbjct: 93 IIKQYRDDHPRKISFTDVRRTLVSDVVSIRRVFDFLDSWGLINYNSSASAKPLKWEEKE- 151
Query: 129 GEGERCKVRVEEGAPNGIRVVATPNSLKPLSAPRNAKSGGGGGGSVSGGAGVKLPPLASF 188
+G G + S A
Sbjct: 152 -------------------------------------AGKSAGDAASEPATT-------- 166
Query: 189 ADVYGDLMSAKELNCGICGDKCGFEHYRSTKDNFTICMKCFKNGNYGEKRSVEDFILNES 248
+ + NC C C + K + T+C +C+ NY + +F E
Sbjct: 167 ------VKETAKRNCNGCKAICSIACFACDKYDLTLCARCYVRSNYRVGINSSEFKRVEI 220
Query: 249 NENSVKQSTVWTEGETLLLLESVLKHGDDWELVAQSVQTKTKLDCISKLIELPFGELMLG 308
+E S + W++ E LLLLE+V+ +GDDW+ VA V +T+ DC+S+ ++LPFGE +
Sbjct: 221 SEESKPE---WSDKEILLLLEAVMHYGDDWKKVASHVIGRTEKDCVSQFVKLPFGEQFVK 277
Query: 309 SAHRNDNINIANGIVNNAIQVQSSSSDHQETSKTQNQTPELTSEKEPNGDAVNESPSKRQ 368
+ D + + + Q SD E+ D SP+KR
Sbjct: 278 ESDSEDGLEMFD---------QIKDSDIPESEGI---------------DKDGSSPNKRI 313
Query: 369 RVAALSESSSSLMKQVGLISTSVDPHITAAAADAAITALCDENLCPREIFDVEEDYASSA 428
++ L+++S+ +M Q +S ++ AAA AA+ AL D D E D +S
Sbjct: 314 KLTPLADASNPIMAQAAFLSALAGTNVAEAAARAAVRALSD--------VDYEADKNASR 365
Query: 429 NSLISDSERAHEVEGLEMDRSTQAEKDDRGPKDDIPLPLRLRAGIATALGAAAARAKLMA 488
+ D+ A E T + +R A A AK +
Sbjct: 366 DPNRQDANAASSGE-------TTRNESER----------------------AWADAKSLI 396
Query: 489 DQEDREIEHLVATIIEAQIGKLQHKVKHFDELELLMKKEHAEIEELKDSILTERIDVLQK 548
++E+ E+E + +E ++ K++ ++ HF++L+L M++ ++EE+++ + +++++
Sbjct: 397 EKEEHEVEGAIKETVEVEMKKIRDRIVHFEKLDLEMERSRKQLEEVRNLLFVDQLNIFFH 456
Query: 549 T 549
T
Sbjct: 457 T 457
>At1g21700 putative transcriptional regulatory protein
Length = 807
Score = 170 bits (431), Expect = 2e-42
Identities = 142/540 (26%), Positives = 247/540 (45%), Gaps = 62/540 (11%)
Query: 23 LYTIPSSSKWFAWDEIHETERTALKEFFDGTSISRTPKVYKEYRDFVINKYREDPSRKLT 82
++ +P S WFA + + ER + +FF G S + TP+ Y E+R+ +++KY E+P + LT
Sbjct: 175 VHVLPMHSDWFAPNTVDRLERQVVPQFFSGKSPNHTPESYMEFRNAIVSKYVENPEKTLT 234
Query: 83 FTEVRKSLVG-DVTLLRKVFLFLESCGLINY-GVGEAEKEEEREDEEEGEGERCKVRVEE 140
++ + + G D+ +VF FL+ G+INY ++ R+ + E +V V
Sbjct: 235 ISDCQGLVDGVDIEDFARVFRFLDHWGIINYCATAQSHPGPLRDVSDVREDTNGEVNVPS 294
Query: 141 GAPNGIRVVATPNSLKPLSAPRNAKSGGGGGGSVSGGAGVKLPPLASFADVYGDLMSAKE 200
A I +SL P GG S+ G P L D+ +
Sbjct: 295 AALTSI------DSLIKFDKPNCRHKGGEVYSSLPSLDGDS-PDL--------DIRIREH 339
Query: 201 L---NCGICGDKCGFEHYRSTKD-NFTICMKCFKNGNYGEKRSVEDFI-LNESNENSVKQ 255
L +C C +++S K + +C CF +G + S DF+ ++ +
Sbjct: 340 LCDSHCNHCSRPLPTVYFQSQKKGDILLCCDCFHHGRFVVGHSCLDFVRVDPMKFYGDQD 399
Query: 256 STVWTEGETLLLLESVLKHGDDWELVAQSVQTKTKLDCISKLIELPFGELMLGSAHRNDN 315
WT+ ETLLLLE+V + ++W +A V +K+K CI + LP + +L DN
Sbjct: 400 GDNWTDQETLLLLEAVELYNENWVQIADHVGSKSKAQCILHFLRLPVEDGLL------DN 453
Query: 316 INIANGIVNNAIQVQSSSSDHQETSKTQNQTPELTSEKEPNGD--AVNESPSKRQRVAAL 373
+ ++ G+ N + ++ DH+ T + NGD +E S +
Sbjct: 454 VEVS-GVTNT--ENPTNGYDHKGT--------------DSNGDLPGYSEQGSDTEIKLPF 496
Query: 374 SESSSSLMKQVGLISTSVDPHITAAAADAAITALCDENLCPREIFDVEEDYASSANSLIS 433
+S + +M V ++++V P + A+ A +++ L +++ E +E AS +
Sbjct: 497 VKSPNPVMALVAFLASAVGPRVAASCAHESLSVLSEDDRMKSEGMQGKE--ASLLDGENQ 554
Query: 434 DSERAHEVEGLEMDRSTQAEKDDRGPKDDIPLPL-RLRAGIATALGAAAARAKLMADQED 492
+ AH+ G + PLP ++ A L AAA +AKL AD E+
Sbjct: 555 QQDGAHKTSS------------QNGAEAQTPLPQDKVMAAFRAGLSAAATKAKLFADHEE 602
Query: 493 REIEHLVATIIEAQIGKLQHKVKHFDELELLMKKEHAEIEELKDSILTERIDVLQKTFRS 552
REI+ L A I+ Q+ +++ K+K F E+E L+ KE ++E+ + ER +L F S
Sbjct: 603 REIQRLSANIVNHQLKRMELKLKQFAEIETLLMKECEQVEKTRQRFSAERARMLSARFGS 662
>At4g34430 putative protein
Length = 827
Score = 91.3 bits (225), Expect = 1e-18
Identities = 85/394 (21%), Positives = 156/394 (39%), Gaps = 53/394 (13%)
Query: 70 INKYREDPSRKLTFTEVRKSLVGDVTLLRKVFLFLESCGLINYGV-------GEAEKEEE 122
+ K+ +P+ ++ ++ + VGD ++V FL+ GLIN+ A ++
Sbjct: 1 MGKFHSNPNIQIELKDLTELEVGDSEAKQEVMEFLDYWGLINFHPFPPTDTGSTASDHDD 60
Query: 123 REDEEEGEGERCKVRVEEGAPNGIR-----VVATPNSL--KPLSAPRNAKSGGGGGGSVS 175
D+E + +V+E P + ATP+ L P++A K G
Sbjct: 61 LGDKESLLNSLYRFQVDEACPPLVHKPRFTAQATPSGLFPDPMAADELLKQEG------- 113
Query: 176 GGAGVKLPPLASFADVYGDLMSAKELNCGICGDKCGFEHYRSTKD-NFTICMKCFKNGNY 234
A E +C C C + Y K +F +C +CF +G +
Sbjct: 114 ---------------------PAVEYHCNSCSADCSRKRYHCPKQADFDLCTECFNSGKF 152
Query: 235 GEKRSVEDFILNESNENSVKQSTVWTEGETLLLLESVLKHGDDWELVAQSVQTKTKLDCI 294
S DFIL E E S WT+ ETLLLLE++ ++W +A+ V TKTK C+
Sbjct: 153 SSDMSSSDFILMEPAEAPGVGSGKWTDQETLLLLEALEIFKENWNEIAEHVATKTKAQCM 212
Query: 295 SKLIELPFGELMLGSAHRNDNINIANGIVNNAIQVQSSSSDHQ--ETSKTQNQTPELTSE 352
+++P + L D I+ + I + + S D+ + + + + + E
Sbjct: 213 LHFLQMPIEDAFL------DQIDYKDPISKDTTDLAVSKDDNSVLKDAPEEAENKKRVDE 266
Query: 353 KEPNGDAVNESPSKRQRVAALSESSSSLMKQVGLISTSVDPHITAAAADAAITALCDENL 412
E + ++V+ S ++ + A + DEN+
Sbjct: 267 DETMKEVPEPEDGNEEKVSQESSKPGDASEETNEMEAEQKTPKLETAIEERCKDEADENI 326
Query: 413 CPREIFDVEED--YASSANSLISDSERAHEVEGL 444
+ + + ED ++S+ + S ++ + V GL
Sbjct: 327 ALKALTEAFEDVGHSSTPEASFSFADLGNPVMGL 360
Score = 48.1 bits (113), Expect = 1e-05
Identities = 59/229 (25%), Positives = 99/229 (42%), Gaps = 21/229 (9%)
Query: 325 NAIQVQSSSSDHQETSKTQNQTPELTSEKEPNGDAVNESPSKRQRVAALSESSSSLMKQV 384
+ + ++ + E S+ L SEK+P D V K Q A E ++ ++
Sbjct: 536 STVSQSAADASQPEASRDVEMKDTLQSEKDPE-DVVKTVGEKVQ--LAKEEGANDVLSTP 592
Query: 385 GLISTSVDPHITAAA-----ADAAITALCDENLCPREIFDVEEDYASSANSLISDSERAH 439
S S P +A+A A + +L D + + D Y L +
Sbjct: 593 DK-SVSQQPIGSASAPENGTAGSLFLSLSDTLITLPTLHDSFISYRFVIEMLFAGGNP-- 649
Query: 440 EVEGLEMDRSTQAEKDD-RGPKDDIPLPLRLRAGIATALGAAAARAKLMADQEDREIEHL 498
+EG + EKD G KD + RA I+ A+ AAA +AK +A QE+ +I L
Sbjct: 650 NIEG-------KKEKDICEGTKDKYNIEKLKRAAIS-AISAAAVKAKNLAKQEEDQIRQL 701
Query: 499 VATIIEAQ-IGKLQHKVKHFDELELLMKKEHAEIEELKDSILTERIDVL 546
++IE Q + KL+ K+ F+E E L + ++E + + ER ++
Sbjct: 702 SGSLIEKQQLHKLEAKLSIFNEAESLTMRVREQLERSRQRLYHERAQII 750
>At3g07740 unknown protein
Length = 548
Score = 38.9 bits (89), Expect = 0.008
Identities = 27/103 (26%), Positives = 46/103 (44%), Gaps = 16/103 (15%)
Query: 194 DLMSAKELNCGICGDKCGFEHYRSTKDNFTICMKCFKNG-NYGEKRSVEDFILNESNENS 252
DL C +C D F +C++CF G ++ + + ++ S
Sbjct: 59 DLSGLVRFKCAVCMD-------------FDLCVECFSVGVELNRHKNSHPYRVMDNLSFS 105
Query: 253 VKQSTVWTEGETLLLLESVLKHG-DDWELVAQSVQTKTKLDCI 294
+ S W E +LLLE++ +G +W+ VA V +KT +CI
Sbjct: 106 LVTSD-WNADEEILLLEAIATYGFGNWKEVADHVGSKTTTECI 147
>At2g06220 hypothetical protein
Length = 428
Score = 35.4 bits (80), Expect = 0.085
Identities = 32/133 (24%), Positives = 57/133 (42%), Gaps = 12/133 (9%)
Query: 330 QSSSSDHQETSKTQNQTPELTSEKEPNGDAVNESPSKRQRVAALSESSSSLMKQVGLIST 389
+S+ +E S +Q ++SE E +GDA E P++ A S++ K+ I
Sbjct: 14 ESTERPSEEVSTSQT----VSSEIEIDGDAAVEVPTEP---AETDVSANKTPKKTAEIEE 66
Query: 390 SVDPHITAAAADAAITALCDENLCPREIFDVEEDYASSANSLISDSERAHEVEGLEMDRS 449
+++ + A + TA + +EI + E + VE + +
Sbjct: 67 AIEESLDGDEASSKGTADIE-----KEILEANEGDGDELEEEEMVNTTKEGVENINCQDT 121
Query: 450 TQAEKDDRGPKDD 462
+AE DD GP+DD
Sbjct: 122 CEAEDDDSGPEDD 134
>At1g08340 unknown protein
Length = 404
Score = 35.4 bits (80), Expect = 0.085
Identities = 23/72 (31%), Positives = 35/72 (47%), Gaps = 4/72 (5%)
Query: 116 EAEKEEEREDEEEGEGERCKVRVEEGAPNGIRVVATPNSLKPLSAPRNAKSGGGGGGSVS 175
E E+EE+ +++EE EG+ + EE A I+VVA + S ++ G
Sbjct: 334 EEEEEEDEDEDEEEEGDGVYIIKEEEASEIIKVVADEHK----SGSIKSEFEGSSATDSK 389
Query: 176 GGAGVKLPPLAS 187
G GV PP+ S
Sbjct: 390 GDNGVVQPPICS 401
>At5g49390 putative protein
Length = 422
Score = 34.3 bits (77), Expect = 0.19
Identities = 34/142 (23%), Positives = 62/142 (42%), Gaps = 12/142 (8%)
Query: 324 NNAIQVQSSSSDHQETSKTQNQTPELTSEKEPNGDAVNESPSKRQRVAALSESSSSLMKQ 383
+NA S+SS E ++ N P++T+ + + V A S +SS+ M++
Sbjct: 259 SNATAASSASSTQMEDAEPYN--PDVTAASSTCSTQMEVAEPYNPGVTAASSTSSTQMEE 316
Query: 384 V--------GLISTSVDPHITAAAADAAITALCDENLCPREIFDVEEDYASSANSLISDS 435
STS+ P A + +TA ++ P E+ + ++A+S +S S
Sbjct: 317 ADPYNPDVTAASSTSITPMEVAEPYNPDVTAASSTSITPMEVAEPYNPGVTAASS-VSPS 375
Query: 436 ERAHEVEGLEMDRSTQAEKDDR 457
+R+ + E D + EK R
Sbjct: 376 KRSGGLVDYE-DDEDELEKSKR 396
>At5g47390 Myb-related transcription activator-like
Length = 365
Score = 34.3 bits (77), Expect = 0.19
Identities = 40/180 (22%), Positives = 71/180 (39%), Gaps = 33/180 (18%)
Query: 235 GEKRSVEDFILNESNENSVKQSTVWTEGETLLLLESVLKHG-DDWELVAQS-VQTKTKLD 292
G+ + EDF+ S+ K+ T WTE E + L + K G DW ++++ V T+T
Sbjct: 74 GDGYASEDFVAGSSSSRERKKGTPWTEEEHRMFLLGLQKLGKGDWRGISRNYVTTRTPTQ 133
Query: 293 CIS----------------------KLIELPFGELMLG-SAHRNDNINIANGIVNNAIQV 329
S ++ G++ + DNI + ++
Sbjct: 134 VASHAQKYFIRQSNVSRRKRRSSLFDMVPDEVGDIPMDLQEPEEDNIPVET-------EM 186
Query: 330 QSSSSDHQETSKTQNQTPELTSEKE-PNGDAVNESPSKRQRVAALSESSSSLMKQVGLIS 388
Q + S HQ + + P + +E + D+ N + + AA + SSS L + L S
Sbjct: 187 QGADSIHQTLAPSSLHAPSILEIEECESMDSTNSTTGEPTATAAAASSSSRLEETTQLQS 246
>At3g57980 unknown protein
Length = 650
Score = 33.9 bits (76), Expect = 0.25
Identities = 23/63 (36%), Positives = 36/63 (56%), Gaps = 9/63 (14%)
Query: 246 NESNENSVKQSTVWTEGETLLLLESVLKHG-DDWELVAQSV-------QTKTKLDCISKL 297
+E+++NS ++ T W+ E LLL +V +HG D W+ VA V +T T +DC K
Sbjct: 4 SENDKNSPEKQT-WSTMEELLLACAVHRHGTDSWDSVASEVHKQNSTFRTLTAIDCRHKY 62
Query: 298 IEL 300
+L
Sbjct: 63 NDL 65
>At3g18100 putative transcription factor (MYB4R1)
Length = 847
Score = 33.9 bits (76), Expect = 0.25
Identities = 27/112 (24%), Positives = 47/112 (41%), Gaps = 7/112 (6%)
Query: 259 WTEGETLLLLESVLKHGDDWELVAQSVQTKTKLDCISKLIEL-PFGELMLGSAHRNDNIN 317
WTE E L E++ +HG W VA ++ +T C+ + L P +L A R
Sbjct: 601 WTEEEDEKLREAIAEHGYSWSKVATNLSCRTDNQCLRRWKRLYPHQVALLQEARRLQK-- 658
Query: 318 IANGIVNNAIQVQSSSSDHQETSKTQNQTPELTSEKEPNGDAVNESPSKRQR 369
V N + +S + P+++ E EP+ A+ + +Q+
Sbjct: 659 --EASVGNFVDRESERP--ALVTSPILALPDISLEPEPDSVALKKKRKAKQK 706
>At5g45420 unknown protein
Length = 309
Score = 33.5 bits (75), Expect = 0.32
Identities = 21/91 (23%), Positives = 46/91 (50%), Gaps = 14/91 (15%)
Query: 227 KCFKNGNYGE----KRSVEDFILNESNENSVK------QSTVWTEGETLLLLESVLKHGD 276
K +++ +Y + +++ + +++E+ ENS +W+ GE + LL ++
Sbjct: 210 KIYESDDYAQFLKNRKASDPRLVDENEENSGAGGDAEGTKEIWSNGEDIALLNALKAFPK 269
Query: 277 D----WELVAQSVQTKTKLDCISKLIELPFG 303
+ WE +A +V K+K C+ ++ EL G
Sbjct: 270 EAAMRWEKIAAAVPGKSKAACMKRVTELKKG 300
>At3g51770 putative protein
Length = 958
Score = 33.5 bits (75), Expect = 0.32
Identities = 15/44 (34%), Positives = 22/44 (49%)
Query: 136 VRVEEGAPNGIRVVATPNSLKPLSAPRNAKSGGGGGGSVSGGAG 179
+++ EG P++ P P N+ +GGGGGG GG G
Sbjct: 12 LKLAEGCKGTQVYALNPSAPTPPPPPGNSSTGGGGGGGSGGGTG 55
>At3g22790 unknown protein
Length = 1694
Score = 33.5 bits (75), Expect = 0.32
Identities = 54/250 (21%), Positives = 105/250 (41%), Gaps = 27/250 (10%)
Query: 325 NAIQVQSSSSDHQETSKTQNQTPELTSEK--EPNGDAVNESPSKRQRVAALSESSSSLMK 382
N ++ SS ET K + + + EK E +N+S + ++ + L + SL K
Sbjct: 500 NLSELNDSSMIFLETQKCEISSLKEIKEKLEEEVARHINQSSAFQEEIRRLKDEIDSLNK 559
Query: 383 QVGLISTSVD-PHITAAAADAAITALCDENLCPREIFDVEEDYASSANSLISDSERAHE- 440
+ I V+ + + ++ L DEN E+ + + D + + + +
Sbjct: 560 RYQAIMEQVNLAGLDPKSLACSVRKLQDENSKLTELCNHQSDDKDALTEKLRELDNILRK 619
Query: 441 ---VEGLEMDRSTQAEKDDRGPKDDIPLPLRLRAGIATALGAAAARAKLMAD-------- 489
+E L ++ +T+ + KD L R + A RA L++
Sbjct: 620 NVCLEKLLLESNTKLDGSREKTKD---LQERCESLRGEKYEFIAERANLLSQLQIMTENM 676
Query: 490 QEDREIEHLVATIIEAQIGKLQ---HKVKHFDELELLMKKEHAEIEELKDSILT------ 540
Q+ E L+ T + +LQ K K F+E L+K + AE+ + ++S+++
Sbjct: 677 QKLLEKNSLLETSLSGANIELQCVKEKSKCFEEFFQLLKNDKAELIKERESLISQLNAVK 736
Query: 541 ERIDVLQKTF 550
E++ VL+K F
Sbjct: 737 EKLGVLEKKF 746
>At2g44430 unknown protein
Length = 646
Score = 33.5 bits (75), Expect = 0.32
Identities = 19/48 (39%), Positives = 28/48 (57%), Gaps = 2/48 (4%)
Query: 246 NESNENSVKQST-VWTEGETLLLLESVLKHG-DDWELVAQSVQTKTKL 291
+E N N+ K T W E LLL +V +HG DW+ VA V++++ L
Sbjct: 36 DEDNSNTKKSQTQAWGTWEELLLACAVKRHGFGDWDSVATEVRSRSSL 83
>At2g42320 unknown protein
Length = 669
Score = 33.5 bits (75), Expect = 0.32
Identities = 45/219 (20%), Positives = 85/219 (38%), Gaps = 28/219 (12%)
Query: 324 NNAIQVQSSSSDHQETSKTQNQTPELTSEKEPNGDAVNESPSKRQRVAALSESSSSLMKQ 383
NN +Q + S+ ++ K Q + +E+E + D + E SS++
Sbjct: 7 NNVVQSKRSTKSVRKDQKLQKTNSQKKTEQEKHKDLDTK------------EESSNISTV 54
Query: 384 VGLISTSVDPHITAAAADAAITALCDENLCPREIFDVEEDYASSANSLISDSERAHEVEG 443
+ DP + D + L D+N I + S ++S++ +E+ H + G
Sbjct: 55 ASDSTIQSDPSESYETVD--VRYLDDDN---GVIASADAHQESDSSSVVDKTEKEHNLSG 109
Query: 444 ----LEMDRSTQAEKDDRGPKDDIPLPLRLRAGIATALGAAAARAKLMADQEDREIEHLV 499
LE D Q KD KD R GI+ + A+ L A E+ +
Sbjct: 110 SICDLEKDVDEQECKDANIDKD-------AREGISADVWEDASNGALSAGSENEAADVTE 162
Query: 500 ATIIEAQIGKLQHKVKHFDELELLMKKEHAEIEELKDSI 538
+ G + K++ + +++E E+ L+ S+
Sbjct: 163 NNGGNFEDGSSEEKIERLETRIEKLEEELREVAALEISL 201
>At3g52250 putative protein
Length = 1677
Score = 33.1 bits (74), Expect = 0.42
Identities = 14/36 (38%), Positives = 20/36 (54%)
Query: 259 WTEGETLLLLESVLKHGDDWELVAQSVQTKTKLDCI 294
WT E + L + HG D++ +A S+ KT DCI
Sbjct: 889 WTSEEKEIFLNLLAMHGKDFKKIASSLTQKTTADCI 924
>At3g10660 calmodulin-domain protein kinase CDPK isoform 2
Length = 646
Score = 32.7 bits (73), Expect = 0.55
Identities = 15/57 (26%), Positives = 31/57 (54%)
Query: 327 IQVQSSSSDHQETSKTQNQTPELTSEKEPNGDAVNESPSKRQRVAALSESSSSLMKQ 383
+Q +S+ + + SK + PE TSE +P A + P +RV++ + S++++
Sbjct: 121 VQPESAKPETKSESKPETTKPETTSETKPETKAEPQKPKHMRRVSSAGLRTESVLQR 177
>At2g42700 unknown protein
Length = 788
Score = 32.7 bits (73), Expect = 0.55
Identities = 27/96 (28%), Positives = 43/96 (44%), Gaps = 4/96 (4%)
Query: 364 PSKRQRVAALSESSSSLMKQVGLISTSVDPHITAAAADAAITALCDENLCPREIFDVEE- 422
P + + ALS+S SSL+K G+I TAAA D + +A D + +V
Sbjct: 421 PELQAMIKALSQSQSSLLKNKGIIQLGA---ATAAALDESQSAKWDTFSSAEMMLNVSAG 477
Query: 423 DYASSANSLISDSERAHEVEGLEMDRSTQAEKDDRG 458
D + + ISD V L+ ++ + + RG
Sbjct: 478 DTSQGLAAQISDLINKSAVAELQAKKNEKPDSSSRG 513
>At5g20780 putative protein
Length = 818
Score = 32.3 bits (72), Expect = 0.72
Identities = 21/77 (27%), Positives = 35/77 (45%), Gaps = 3/77 (3%)
Query: 477 LGAAAARAKLMADQEDREIEHLVATIIEAQIGKLQHKVKHFDELELLMKKEHAEIEELKD 536
+ A RAK + ++E R + V+ I E L V H +E+E + E + E+ D
Sbjct: 131 MSAVVERAKTIDEEEGRGSKRTVSEIWEINDEDL---VTHVEEVEAALLGEDGKQSEIND 187
Query: 537 SILTERIDVLQKTFRSG 553
IL E + ++ G
Sbjct: 188 DILLEHVQAVELQLLGG 204
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.130 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,135,902
Number of Sequences: 26719
Number of extensions: 600193
Number of successful extensions: 5211
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 59
Number of HSP's that attempted gapping in prelim test: 4882
Number of HSP's gapped (non-prelim): 199
length of query: 565
length of database: 11,318,596
effective HSP length: 104
effective length of query: 461
effective length of database: 8,539,820
effective search space: 3936857020
effective search space used: 3936857020
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0063.10


